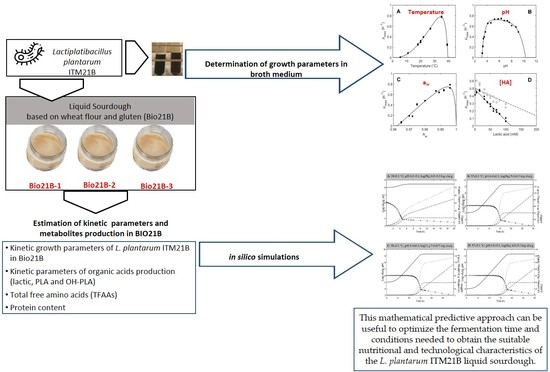
Modeling of Growth and Organic Acid Kinetics and Evolution of the Protein Profile and Amino Acid Content during Lactiplantibacillus plantarum ITM21B Fermentation in Liquid Sourdough
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Growth Kinetic of L. plantarum in Broth
2.3. Growth Kinetic of L. plantarum in Liquid Sourdough (Bio21B) Samples
2.4. Microbiological and Physicochemical Analyses of Bio21B during L. plantarum Fermentation
2.5. Determination of Organic Acids in Bio21B
2.6. Total Free Amino Acids, Protein Content, and Profile of Bio21B
2.7. Mathematical Modeling
2.7.1. Determination of the Maximum Specific Growth Rates
2.7.2. Temperature, pH, aw, and Lactic Acid Models
2.7.3. Effects of L. plantarum Growth on the Nutritional Profile
2.7.4. Determination of Kinetic Growth Parameters of L. plantarum ITM21B in Bio21B for the In Silico Simulations
2.8. Statistical Analysis
3. Results and Discussion
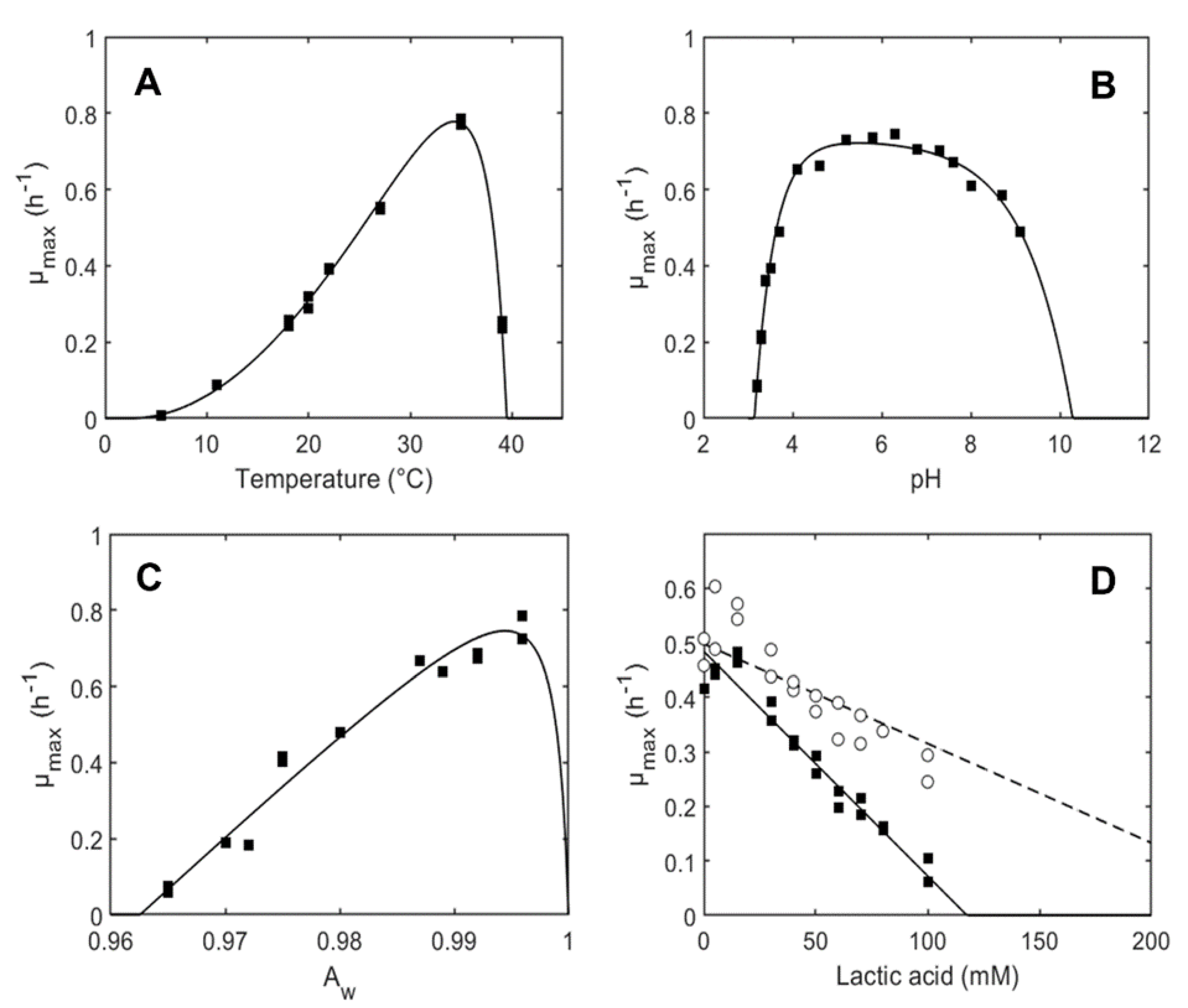
3.1. Growth Model Parameters of L. plantarum ITM21B in Liquid Medium
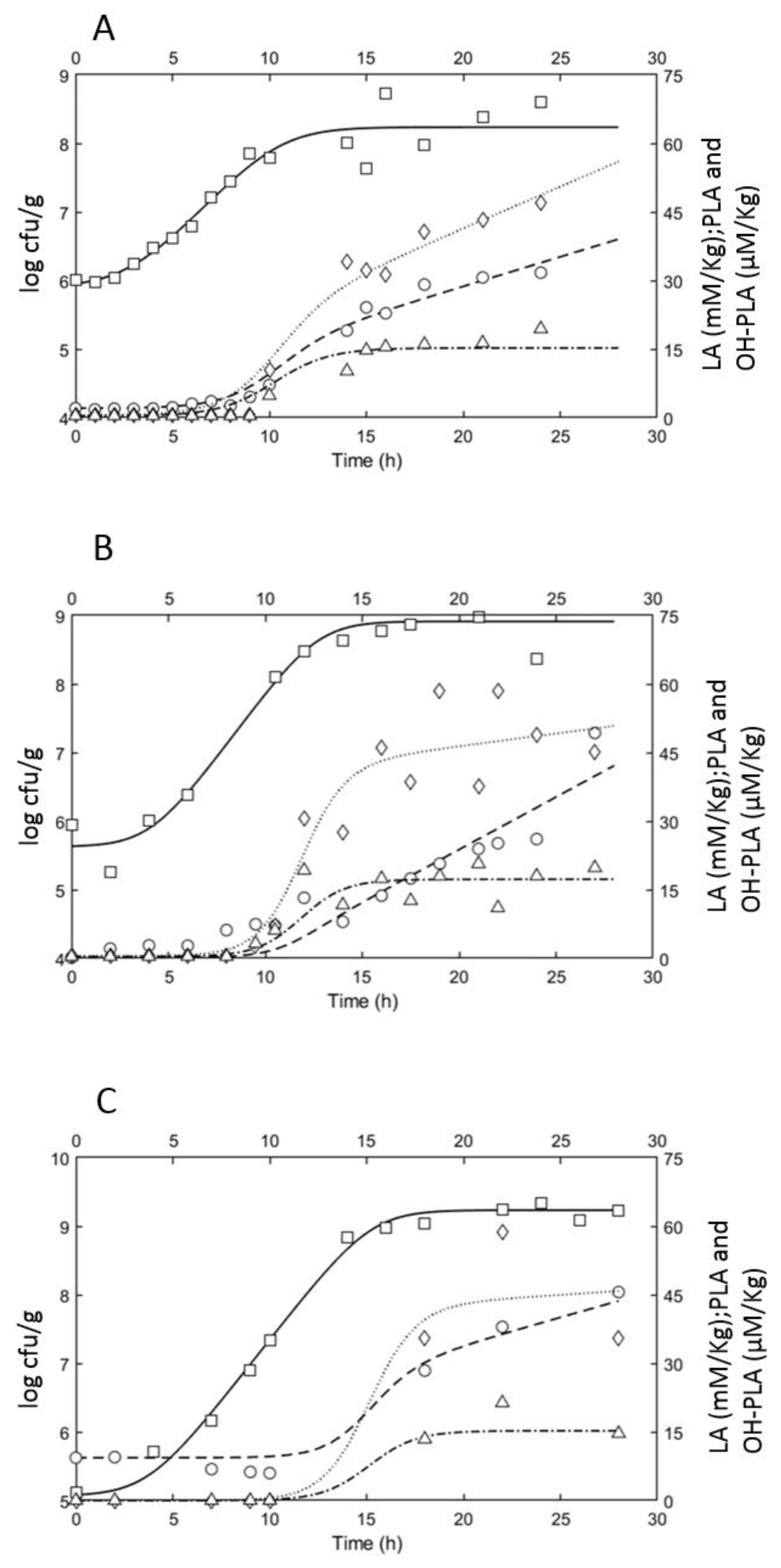
3.2. Growth Performances of L. plantarum ITM21B in Bio21B during Fermentation and Evolution of Metabolites Produced
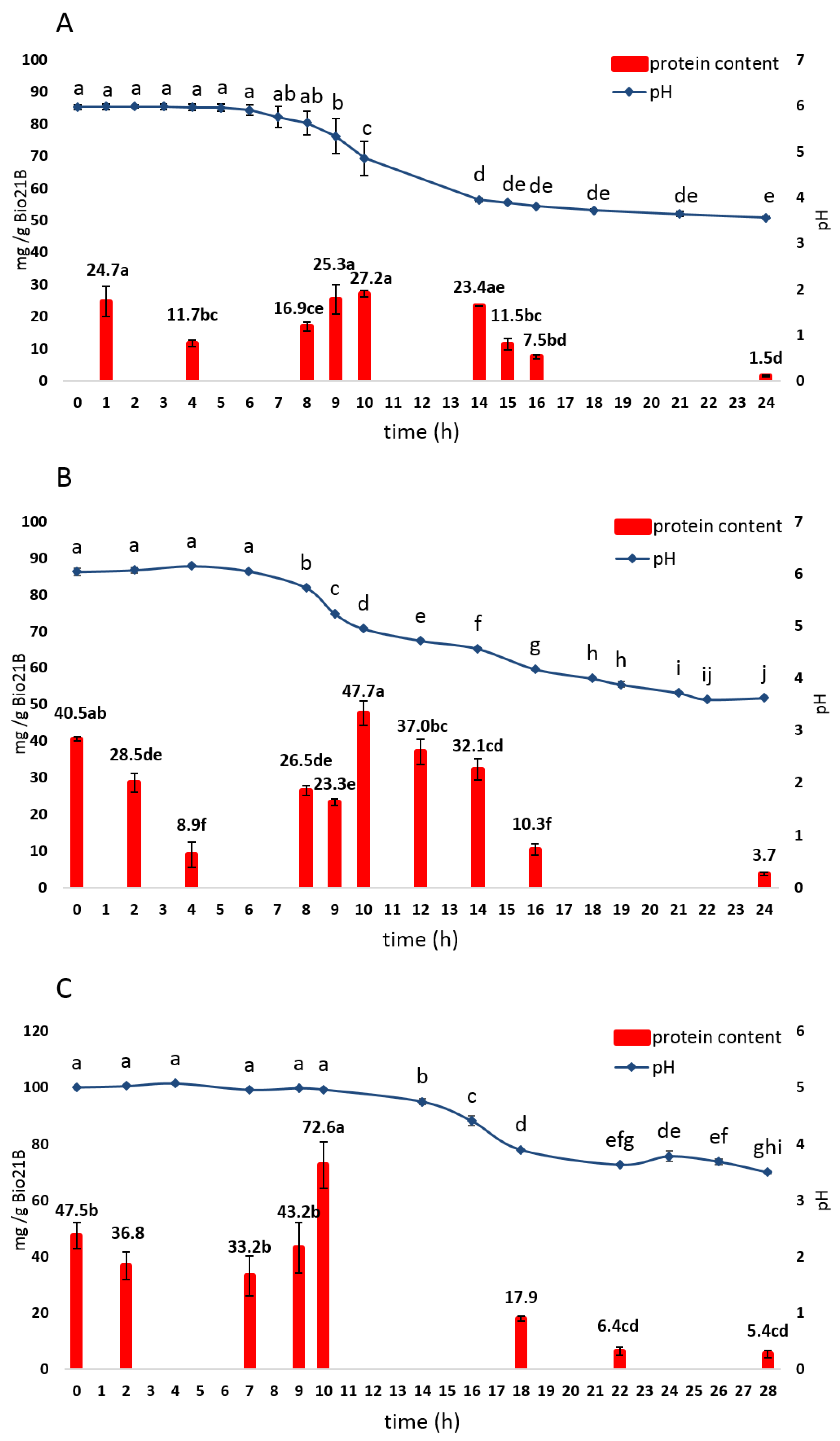
3.2.1. Microbiological and Physicochemical Parameters Evolution
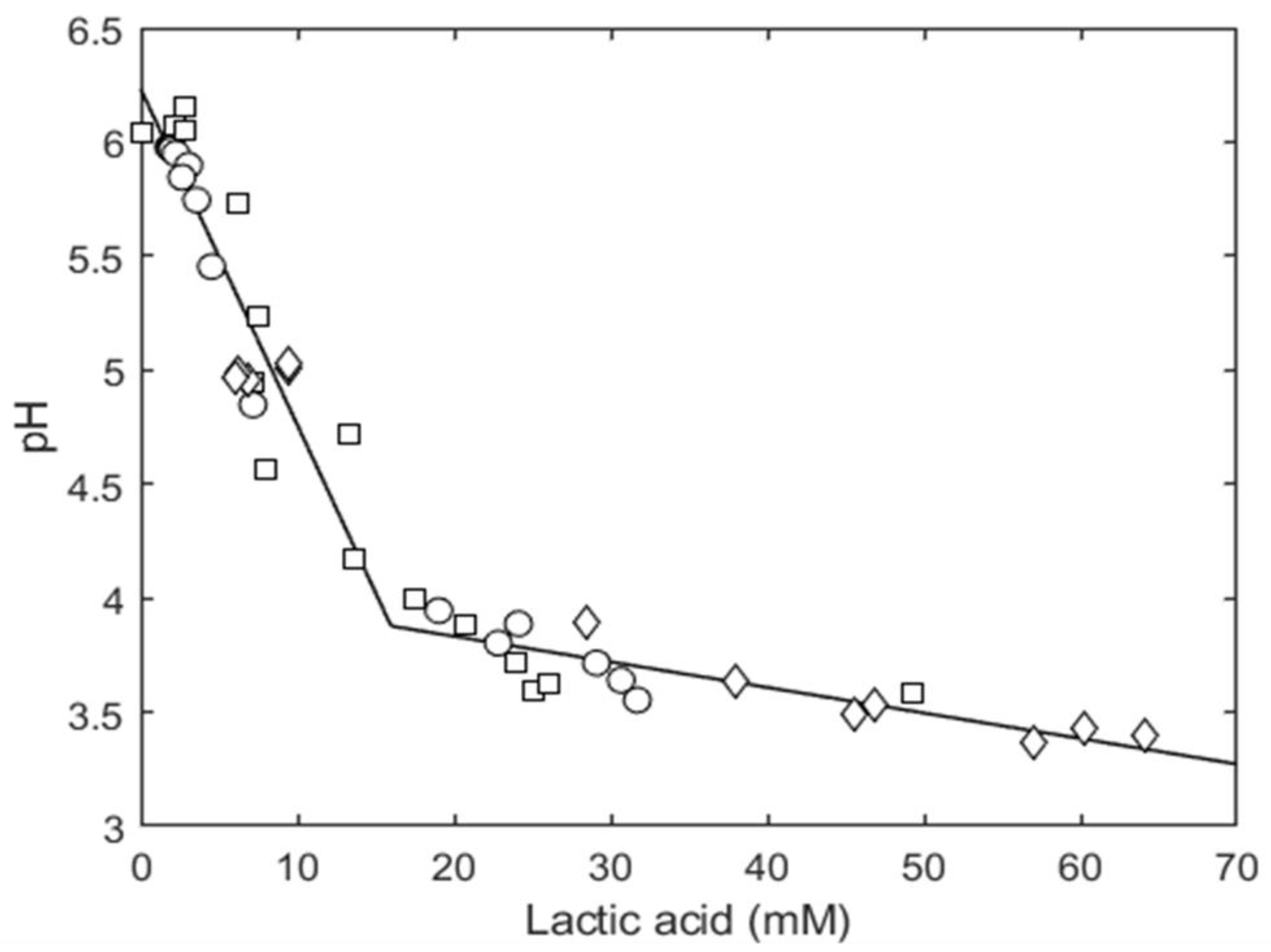
3.2.2. Organic Acid Content Evolution
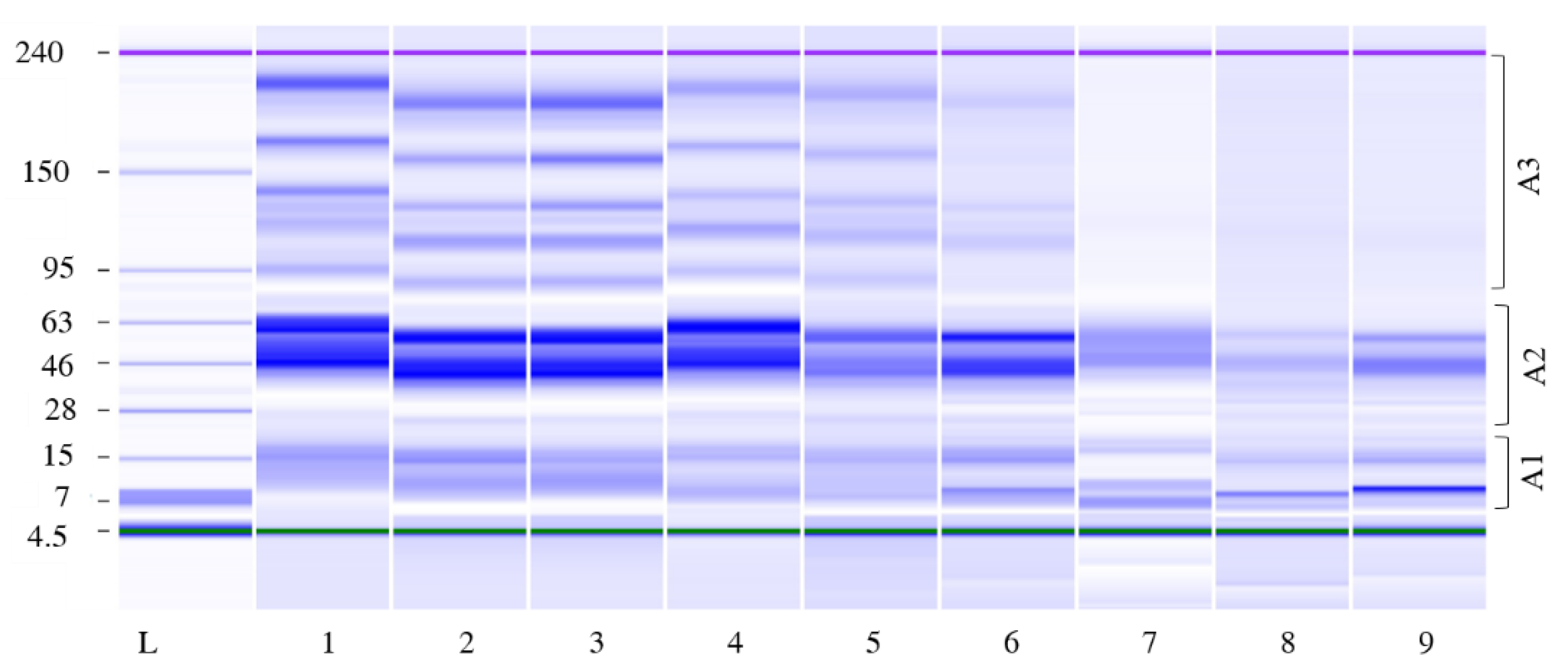
3.2.3. Protein Content Evolution
3.2.4. Amino Acid Content Evolution
3.3. Modeling Results
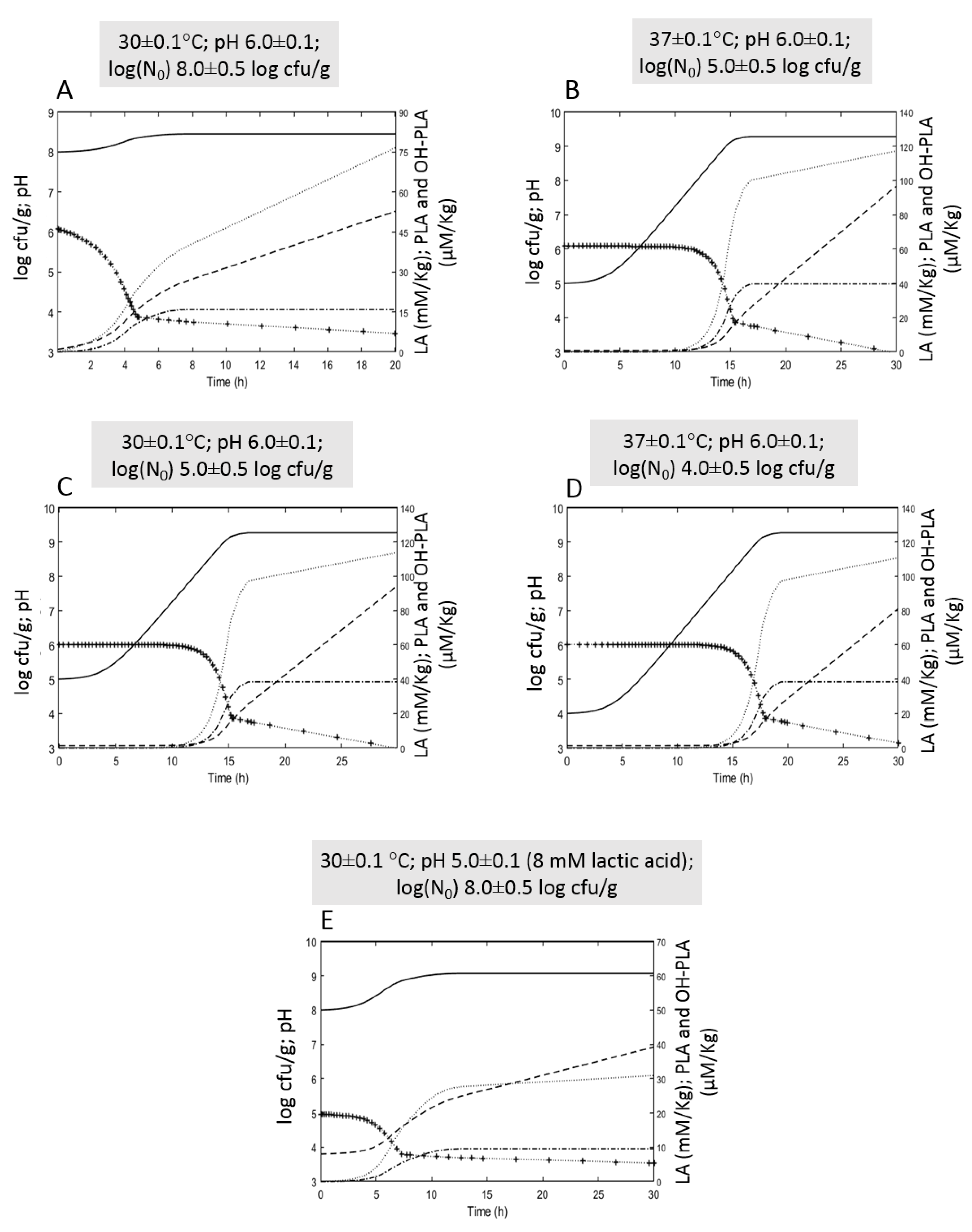
3.4. Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arora, K.; Ameur, H.; Polo, A.; Di Cagno, R.; Rizzello, C.G.; Gobbetti, M. Thirty years of knowledge on sourdough fermentation: A systematic review. Trends Food Sci. Technol. 2021, 108, 71–83. [Google Scholar] [CrossRef]
- Gänzle, M. The periodic table of fermented foods: Limitations and opportunities. Appl. Microbiol. Biotechnol. 2022, 106, 2815–2826. [Google Scholar] [CrossRef]
- Kumar, S.; Rattu, R.; Mitharwal, S.; Chandra, A.; Kumar, S.; Kaushik, A.; Mishra, V.; Nema, P.K. Trends in non-dairy-based probiotic food products: Advances and challenges. J. Food Process. Pres. 2022, 46, e16578. [Google Scholar] [CrossRef]
- Bartkiene, E.; Özogul, F.; Rocha, J.M. Bread Sourdough Lactic Acid Bacteria—Technological, Antimicrobial, Toxin-Degrading, Immune System-, and Faecal Microbiota-Modelling Biological Agents for the Preparation of Food, Nutraceuticals and Feed. Foods 2022, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Fugaban, J.I.I.; Holzapfel, W.H.; Todorov, S. The Overview of Natural by-Products of Beneficial Lactic Acid Bacteria as Promising Antimicrobial Agents. Appl. Food Biotechnol. 2022, 9, 127–143. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Valerio, F.; Evidente, A.; Lazzaroni, S.; Corsetti, A.; Gobbetti, M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl. Environ. Microb. 2000, 66, 4084–4090. [Google Scholar] [CrossRef] [PubMed]
- Valerio, F.; De Bellis, P.; Lonigro, S.L.; Visconti, A.; Lavermicocca, P. Use of Lactobacillus plantarum fermentation products in bread-making to prevent Bacillus subtilis ropy spoilage. Int. J. Food Microbiol. 2008, 122, 328–332. [Google Scholar] [CrossRef]
- Di Biase, M.; Bavaro, A.R.; Lonigro, S.L.; Pontonio, E.; Conte, A.; Padalino, L.; Minisci, A.; Lavermicocca, P.; Valerio, F. Lactobacillus plantarum ITM21B fermentation product and chickpea flour enhance the nutritional profile of salt reduced bakery products. Int. J. Food Sci. Nutr. 2019, 70, 701–713. [Google Scholar] [CrossRef]
- Valerio, F.; Conte, A.; Di Biase, M.; Lattanzio, V.M.; Lonigro, S.L.; Padalino, L.; Pontonio, E.; Lavermicocca, P. Formulation of yeast-leavened bread with reduced salt content by using a Lactobacillus plantarum fermentation product. Food Chem. 2017, 221, 582–589. [Google Scholar] [CrossRef]
- Regulation (EC) no 2073/2005 on Microbiological Criteria for Foodstuffs. 2005. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:02005R2073-20200308 (accessed on 18 October 2022).
- Delignette-Muller, M.L. Principles of Predictive Modeling. In Safety of Meat and Processed Meat. Food Microbiology and Food Safety; Toldrá, F., Ed.; Springer: New York, NY, USA, 2009; pp. 535–558. [Google Scholar] [CrossRef]
- Tenenhaus-Aziza, F.; Ellouze, M. Software for predictive microbiology and risk assessment: A description and comparison of tools presented at the ICPMF8 Software Fair. Food microbiol. 2015, 45, 290–299. [Google Scholar] [CrossRef]
- Dorota, Z.; Danuta, K.K.; Antoni, G.; Motyl, I. Predictive modelling of Lactobacillus casei KN291 survival in fermented soy beverage. J. Microbiol. 2014, 52, 169–178. [Google Scholar] [CrossRef]
- Nikmaram, P.; Mousavi, S.M.; Kiani, H.; Emamdjomeh, Z.; Razavi, S.H.; Mousavi, Z. Modeling the effect of Inulin, pH and storage time on the viability of selected Lactobacillus in a probiotic fruity yogurt drink using the Monte Carlo simulation. J. Food Qual. 2016, 39, 362–369. [Google Scholar] [CrossRef]
- Gänzle, M.G.; Ehmann, M.; Hammes, W.P. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in re-sponse to process parameters of sourdough fermentation. Appl. Environ. Microb. 1998, 64, 2616–2623. [Google Scholar] [CrossRef]
- Altilia, S.; Foschino, R.; Grassi, S.; Antoniani, D.; Dal Bello, F.; Vigentini, I. Investigating the growth kinetics in sour-dough microbial associations. Food Microbiol. 2021, 99, 103837. [Google Scholar] [CrossRef] [PubMed]
- Di Biase, M.; Le Marc, Y.; Bavaro, A.R.; De Bellis, P.; Lonigro, S.L.; Lavermicocca, P.; Postollec, F.; Valerio, F. A predictive growth model for pro-technological and probiotic Lacticaseibacillus paracasei strains fermenting white cabbage. Front. Microbiol. 2022, 13, 907393. [Google Scholar] [CrossRef] [PubMed]
- Luedeking, R.; Piret, E.L. A kinetic study of the lactic acid fermentation. Batch process at controlled pH. J. Biochemical Microbiol. Technol. Eng. 1959, 1, 393–412. [Google Scholar] [CrossRef]
- Poschet, F.; Vereecken, K.M.; Geeraerd, A.H.; Nicolaï, B.M.; Van Impe, J.F. Analysis of a novel class of predictive microbial growth models and application to coculture growth. Int. J. Food Microbiol. 2005, 100, 107–124. [Google Scholar] [CrossRef]
- Vereecken, K.M.; Van Impe, J.F. Analysis and practical implementation of a model for combined growth and metabolite production of lactic acid bacteria. Int. J. Food Microbiol. 2002, 73, 239–250. [Google Scholar] [CrossRef]
- Coroller, L.; Guerrot, V.; Huchet, V.; Le Marc, Y.; Mafart, P.; Sohier, D.; Thuault, D. Modelling the influence of single acid and mixture on bacterial growth. Int. J. Food Microbiol. 2005, 100, 167–178. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.; Harris, H.M.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Micr. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Corsetti, A.; Lavermicocca, P.; Morea, M.; Baruzzi, F.; Tosti, N.; Gobbetti, M. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of southern Italy. Int. J. Food Microbiol. 2001, 64, 95–104. [Google Scholar] [CrossRef]
- Valerio, F.; Di Biase, M.; Lattanzio, V.M.; Lavermicocca, P. Improvement of the antifungal activity of lactic acid bacteria by addition to the growth medium of phenylpyruvic acid, a precursor of phenyllactic acid. Int. J. Food Microbiol. 2016, 222, 1–7. [Google Scholar] [CrossRef]
- Valerio, F.; Bavaro, A.R.; Di Biase, M.; Lonigro, S.L.; Logrieco, A.F.; Lavermicocca, P. Effect of amaranth and quinoa flours on exopolysaccharide production and protein profile of liquid sourdough fermented by Weissella cibaria and Lactobacillus plantarum. Front. Microbiol. 2020, 11, 967. [Google Scholar] [CrossRef] [PubMed]
- Cuppers, H.; Smelt, J. Time to turbidity measurement as a tool for modeling spoilage by Lactobacillus. J. Ind. Microbiol. 1993, 12, 168–171. [Google Scholar] [CrossRef]
- Le Marc, Y.; Baert, L.; Da Silva, N.B.; Postollec, F.; Huchet, V.; Baranyi, J.; Ellouze, M. The effect of pH on the growth rate of Bacillus cereus sensu lato: Quantifying strain variability and modelling the combined effects of temperature and pH. Int. J. Food Microbiol. 2021, 360, 109420. [Google Scholar] [CrossRef] [PubMed]
- Membré, J.M.; Leporq, B.; Vialette, M.; Mettler, E.; Perrier, L.; Zwietering, M.H. Experimental protocols and strain variability of cardinal values (pH and aw) of bacteria using Bioscreen C: Microbial and statistical aspects. In Microbial Adaptation to Changing Environments; Alexon, L., Tronrud, E.S., Merok, K.J., Eds.; Matforsk Norwegian Food Research Institute: Lillehammer, Norway, 2002; pp. 143–146. [Google Scholar]
- Leporq, B.; Membré, J.-M.; Dervin, C.; Buche, P.; Guyonnet, J.P. The “Sym’Previus” software, a tool to support decisions to the foodstuff safety. Int. J. Food Microbiol. 2005, 100, 231–237. [Google Scholar] [CrossRef] [PubMed]
- De Bellis, P.; Valerio, F.; Sisto, A.; Lonigro, S.L.; Lavermicocca, P. Probiotic table olives: Microbial populations adhering on olive surface in fermentation sets inoculated with the probiotic strain Lactobacillus paracasei IMPC2.1 in an industrial plant. Int. J. Food Microbiol. 2010, 140, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Pinon, A.; Zwietering, M.H.; Perrier, L.; Membré, J.M.; Leporq, B.; Mettler, E.; Thuault, D.; Coroller, L.; Stahl, V.; Vialette, M. Development and validation of experimental protocols for use of cardinal models for prediction of microorganism growth in food products. Appl. Environ. Microbiol. 2004, 70, 1081–1087. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis; Association of Official Analytical Chemists: Washington, DC, USA, 1990. [Google Scholar]
- Osborne, T.B. The Proteins of the Wheat Kernel (Monograph); Carnegie Inst: Washington, DC, USA, 1907. [Google Scholar]
- Weiss, W.; Vogelmeier, C.; Gorg, A. Electrophoretic characterization of wheat grain allergens from different cultivars involved in bakers’ asthma. Electrophoresis 1993, 14, 805–816. [Google Scholar] [CrossRef]
- Doi, E.; Shibata, D.; Matoba, T. Modified colorimetric ninhydrin methods for peptidase assay. Anal Biochem. 1981, 118, 173–184. [Google Scholar] [CrossRef]
- Džunková, M.; Janovská, D.; Cepková, P.H.; Prohasková, A.; Kolár, M. Glutelin protein fraction as a tool for clear identification of Amaranth accessions. J. Cereal Sci. 2011, 53, 198–205. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Rosso, L.; Bajard, S.; Flandrois, J.P.; Lahellec, C.; Fournaud, J.; Veit, P. Differential growth of Listeria monocytogenes at 4 and 8 °C: Consequences for the shelf life of chilled products. J. Food Protect. 1996, 59, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Rosso, L.; Lobry, J.R.; Flandrois, J.P. An unexpected correlation between cardinal temperatures of microbial growth highlighted by a new model. J. Theor. Biol. 1993, 162, 447–463. [Google Scholar] [CrossRef]
- Presser, K.A.; Ratkowsky, D.A.; Ross, T. Modelling the growth rate of Escherichia coli as a function of pH and lactic acid concentration. Appl. Environ. Microbiol. 1997, 63, 2355–2360. [Google Scholar] [CrossRef] [PubMed]
- Presser, K.A. Physiology and modelling of Escherichia coli growth inhibition due to pH, organic acids temperature and water activity. Ph.D. Thesis, University of Tasmania, Hobart, Australia, 2001. [Google Scholar]
- Couvert, O.; Pinon, A.; Bergis, H.; Bourdichon, F.; Carlin, F.; Cornu, M.; Denis, C.; Gnanou Besse, N.; Guillier, L.; Jamet, E.; et al. Validation of a stochastic modelling approach for Listeria monocytogenes growth in refrigerated foods. Int. J. Food Microbiol. 2010, 144, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Le Marc, Y.; Huchet, V.; Bourgeois, C.M.; Guyonnet, J.P.; Mafart, P.; Thuault, D. Modelling the growth kinetics of Listeria as a function of temperature, pH and organic acid concentration. Int. J. Food Microbiol. 2002, 73, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.P.; Ocio, M.J.; Kaloti, A.; Mackey, B.M. The effect of the growth environment on the lag phase of Listeria monocytogenes. Int. J. Food Microbiol. 1998, 44, 83–92. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 227–294. [Google Scholar] [CrossRef]
- Aryani, D.C.; den Besten, H.M.W.; Zwietering, M.H. Quantifying variability in growth and thermal inactivation kinetics of Lactobacillus plantarum. Appl. Environ. Microbiol. 2016, 82, 4896–4908. [Google Scholar] [CrossRef]
- Valerio, F.; Di Biase, M.; Caputo, L.; Creanza, T.M.; Ancona, N.; Visconti, A.; Lavermicocca, P. Effect of Lactobacillus brevis-based bioingredient and bran on microbiological, physico-chemical and textural quality of yeast leavened bread during storage. Innov Food Sci Emerg Technol. 2014, 25, 2–8. [Google Scholar] [CrossRef]
- Bavaro, A.R.; Di Biase, M.; Conte, A.; Lonigro, S.L.; Caputo, L.; Cedola, A.; Del Nobile, M.A.; Logrieco, A.F.; Lavermicocca, P.; Valerio, F. Weissella cibaria short-fermented liquid sourdoughs based on quinoa or amaranth flours as fat replacer in focaccia bread formulation. Int. J. Food Sci. Technol. 2021, 56, 3197–3208. [Google Scholar] [CrossRef]
- Holzapfel, W.H. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 2002, 75, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, N.; Gánzle, M.G.; Vogel, R.F. Influence of peptide supply and cosubstrates on phenylalanine metabolism of Lactobacillus sanfranciscensis DSM20451T and Lactobacillus plantarum TMW1. 468. J. Agric. Food Chem. 2006, 54, 3832–3839. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, N.; Pavlovic, M.; Ehrmann, M.A.; Gänzle, M.G.; Vogel, R.F. Functional characterization of the proteolytic system of Lactobacillus sanfranciscensis DSM 20451T during growth in sourdough. Appl. Environ. Microb. 2005, 71, 6260–6266. [Google Scholar] [CrossRef] [PubMed]
- Van der Meulen, R.; Scheirlinck, I.; Van Schoor, A.; Huys, G.; Vancanneyt, M.; Vandamme, P.; De Vuyst, L. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl. Environ. Microb. 2007, 73, 4741–4750. [Google Scholar] [CrossRef] [PubMed]
- Gänzle, M.G.; Loponen, J.; Gobbetti, M. Proteolysis in sourdough fermentations: Mechanisms and potential for im-proved bread quality. Trends Food Sci. Technol. 2008, 19, 513–521. [Google Scholar] [CrossRef]
- Fraberger, V.; Ladurner, M.; Nemec, A.; Grunwald-Gruber, C.; Call, L.M.; Hochegger, R.; Domig, K.J.; D’Amico, S. In-sights into the potential of sourdough-related lactic acid bacteria to degrade proteins in wheat. Microorganisms 2020, 8, 1689. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Piraino, P.; Ricciardi, A.; McSweeney, P.L.; Parente, E. Proteolysis in model sourdough fermentations. J. Agr. Food Chem. 2006, 54, 2567–2574. [Google Scholar] [CrossRef] [PubMed]
- Valerio, F.; Lavermicocca, P.; Pascale, M.; Visconti, A. Production of phenyllactic acid by lactic acid bacteria: An approach to the selection of strains contributing to food quality and preservation. FEMS Microbiol. Lett. 2004, 233, 289–295. [Google Scholar] [CrossRef]

| Starting Fermentation Conditions | ||||
|---|---|---|---|---|
| Liquid Sourdough | log(N0) ± SD | T (°C) ± SD | pH ± SD | LA (mM/Kg) ± SD |
| Bio21B-1 | 6.00 ± 0.32 a | 30.0 ± 0.1 b | 5.97 ± 0.06 a | 2.06 ± 0.60 b |
| Bio21B-2 | 5.95 ± 0.00 a | 37.0 ± 0.1 a | 6.04 ± 0.07 a | <LOD b |
| Bio21B-3 | 5.11 ± 0.00 b | 30.0 ± 0.1 b | 5.00 ± 0.00 b | 9.35 ± 4.36 a |
| ITM21B Mean Value (Confidence Interval) | |
|---|---|
| µopt,MRS (h−1) for T | 0.78 (0.74–0.81) |
| Tmin (°C) | 2.40 (1.43–3.36) |
| Topt (°C) | 34.35 (33.77–34.93) |
| Tmax (°C) | 39.47 (39.32–39.62) |
| Number of data for T | 14 |
| R2 | 0.99 |
| pHmin | 3.14 (3.13–3.16) |
| pHmax | 10.29 (9.54–11.03) |
| Q | 0.41 (0.17–0.64) |
| Number of data for pH | 19 |
| R2 | 0.99 |
| aw,min | 0.963 (0.961–0.964) |
| aw,opt | 0.994 (0.992–0.997) |
| Number of data for aw | 13 |
| R2 | 0.97 |
| MICU (mM) | 14.8 (14.0–15.7) |
| Number of data for [HA] | 38 (19 at pH 4.7 and 19 at pH 5.1) |
| Kinetic Growth Parameters of ITM21B in Liquid Sourdough | ||||
|---|---|---|---|---|
| lag (h) | log (Nmax) | µmax (h−1) | µmax pred (h−1) | |
| Bio21B-1 | 2.2 | 8.25 | 0.70 | 0.88 |
| Bio21B-2 | 3.5 | 8.92 | 0.88 | 0.88 |
| Bio21B-3 | 3.4 | 9.24 | 0.83 | 0.85 |
| Estimated Kinetic Parameters | |||
|---|---|---|---|
| Liquid Sourdough | Acid | YP (mM/kg/cell.h or μM/kg/cell.h) | mp (mM/kg/cell.h or μM/kg/cell.h) |
| Bio21B-1 | LA | 8.24 × 10−8 (6.37 × 10−8–10.11 × 10−8) | 7.49 × 10−9 (5.30 × 10−9–9.68 × 10−9) |
| PLA | 1.35 × 10−7 (9.92 × 10−7–1.70 × 10−7) | 1.06 × 10−8 (0.64 × 10−8–1.48 × 10−8) | |
| OH-PLA | 8.70 × 10−8 (7.66 × 10−8–9.73 × 10−8) | 0 | |
| Bio21B-2 | LA | 6.08 × 10−9 (−4.43 × 10−9–16.60 × 10−9) | 2.80 × 10−9 (1.65 × 10−9–3.96 × 10−9) |
| PLA | 5.06 × 10−8 (3.65 × 10−8–6.47 × 10−8) | 6.57 × 10−10 (−8.82 × 10−10–21.96 × 10−10) | |
| OH-PLA | 2.12 × 10−8 (1.78 × 10−8–2.45 × 10−8) | 0 | |
| Bio21B-3 | LA | 1.10 × 10−8 (0.81 × 10−8–1.39 × 10−8) | 7.05 × 10−10 (5.11 × 10−10–8.99 × 10−10) |
| PLA | 2.45 × 10−8 (1.63 × 10−8–3.26 × 10−8) | 1.95 × 10−10 (−0.20 × 10−10–0.59 × 10−10) | |
| OH-PLA | 1.00 × 10−8 (0.91 × 10−8–1.10 × 10−8) | 0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Biase, M.; Le Marc, Y.; Bavaro, A.R.; Lonigro, S.L.; Verni, M.; Postollec, F.; Valerio, F. Modeling of Growth and Organic Acid Kinetics and Evolution of the Protein Profile and Amino Acid Content during Lactiplantibacillus plantarum ITM21B Fermentation in Liquid Sourdough. Foods 2022, 11, 3942. https://doi.org/10.3390/foods11233942
Di Biase M, Le Marc Y, Bavaro AR, Lonigro SL, Verni M, Postollec F, Valerio F. Modeling of Growth and Organic Acid Kinetics and Evolution of the Protein Profile and Amino Acid Content during Lactiplantibacillus plantarum ITM21B Fermentation in Liquid Sourdough. Foods. 2022; 11(23):3942. https://doi.org/10.3390/foods11233942
Chicago/Turabian StyleDi Biase, Mariaelena, Yvan Le Marc, Anna Rita Bavaro, Stella Lisa Lonigro, Michela Verni, Florence Postollec, and Francesca Valerio. 2022. "Modeling of Growth and Organic Acid Kinetics and Evolution of the Protein Profile and Amino Acid Content during Lactiplantibacillus plantarum ITM21B Fermentation in Liquid Sourdough" Foods 11, no. 23: 3942. https://doi.org/10.3390/foods11233942
APA StyleDi Biase, M., Le Marc, Y., Bavaro, A. R., Lonigro, S. L., Verni, M., Postollec, F., & Valerio, F. (2022). Modeling of Growth and Organic Acid Kinetics and Evolution of the Protein Profile and Amino Acid Content during Lactiplantibacillus plantarum ITM21B Fermentation in Liquid Sourdough. Foods, 11(23), 3942. https://doi.org/10.3390/foods11233942