Crucial Residues of C-Terminal Oligopeptide C60 to Improve the Yield of Prebiotic Xylooligosaccharides by Truncated Mutation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Chemicals
2.2. Sequence and Protein Structure Analysis
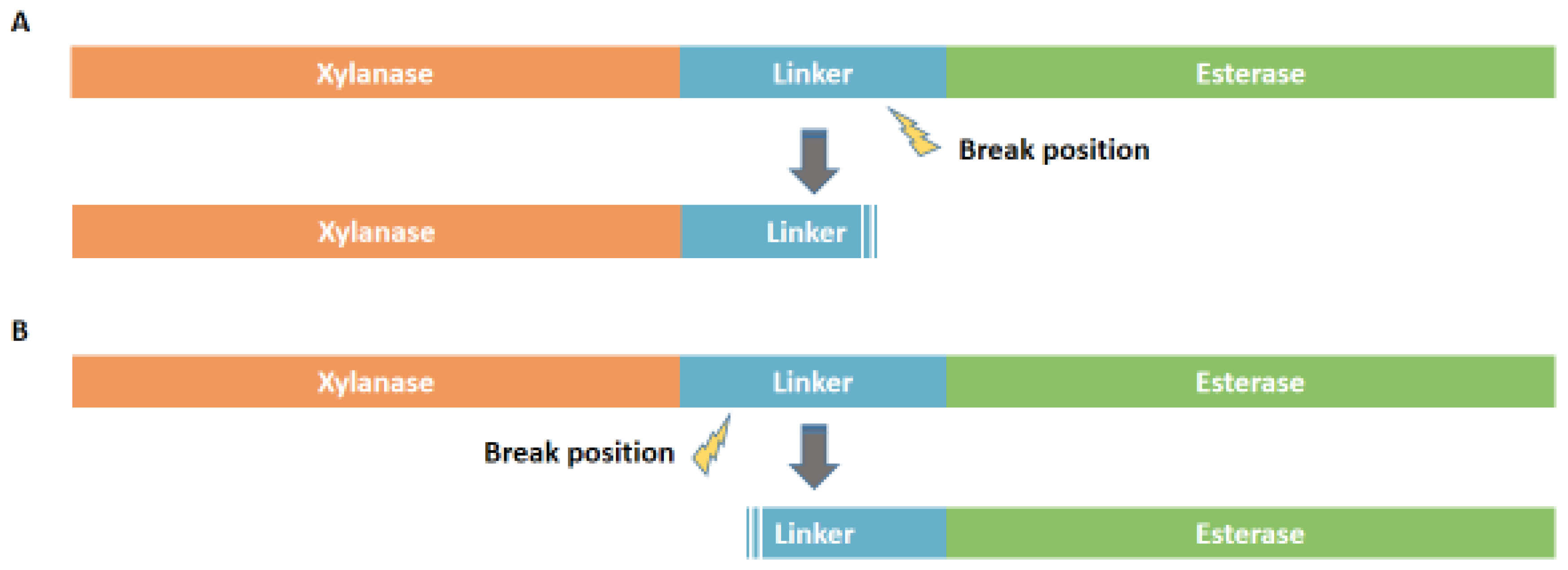
2.3. Construction, Protein Expression, and Purification of Truncated Variants
2.4. Enzymatic Activity Assay and Characterization
2.5. XOS Production Analyzed by High-Performance Anion-Exchange Chromatography
2.6. Statistical Analysis
3. Results and Discussions
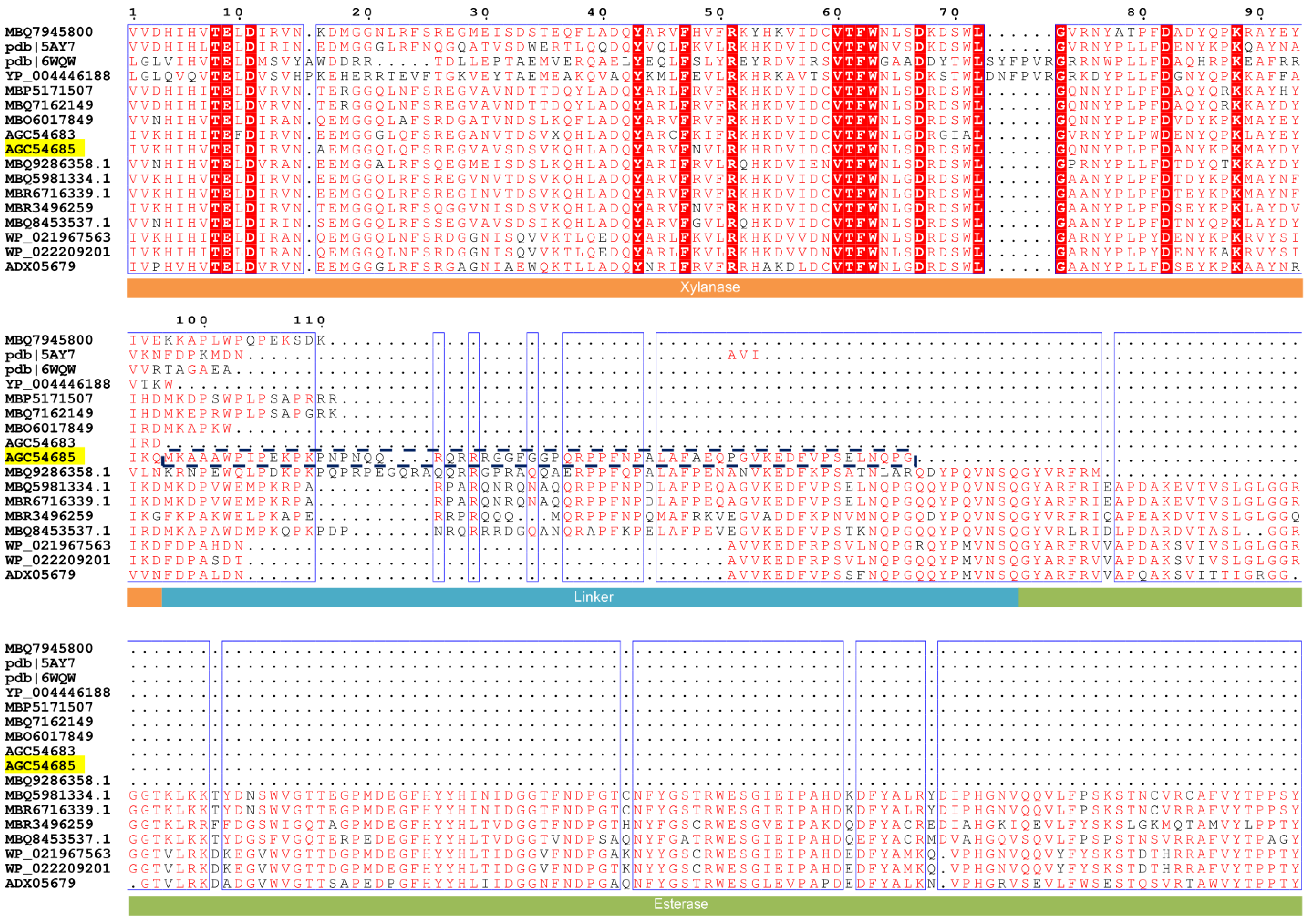
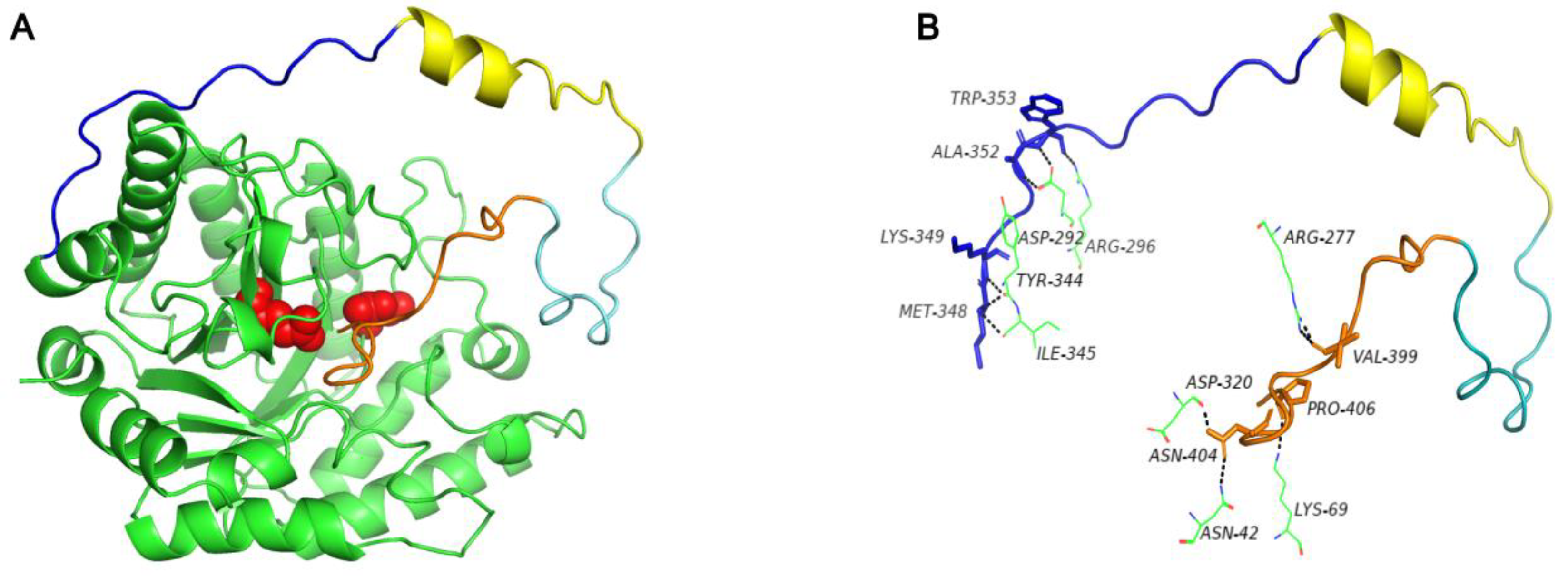
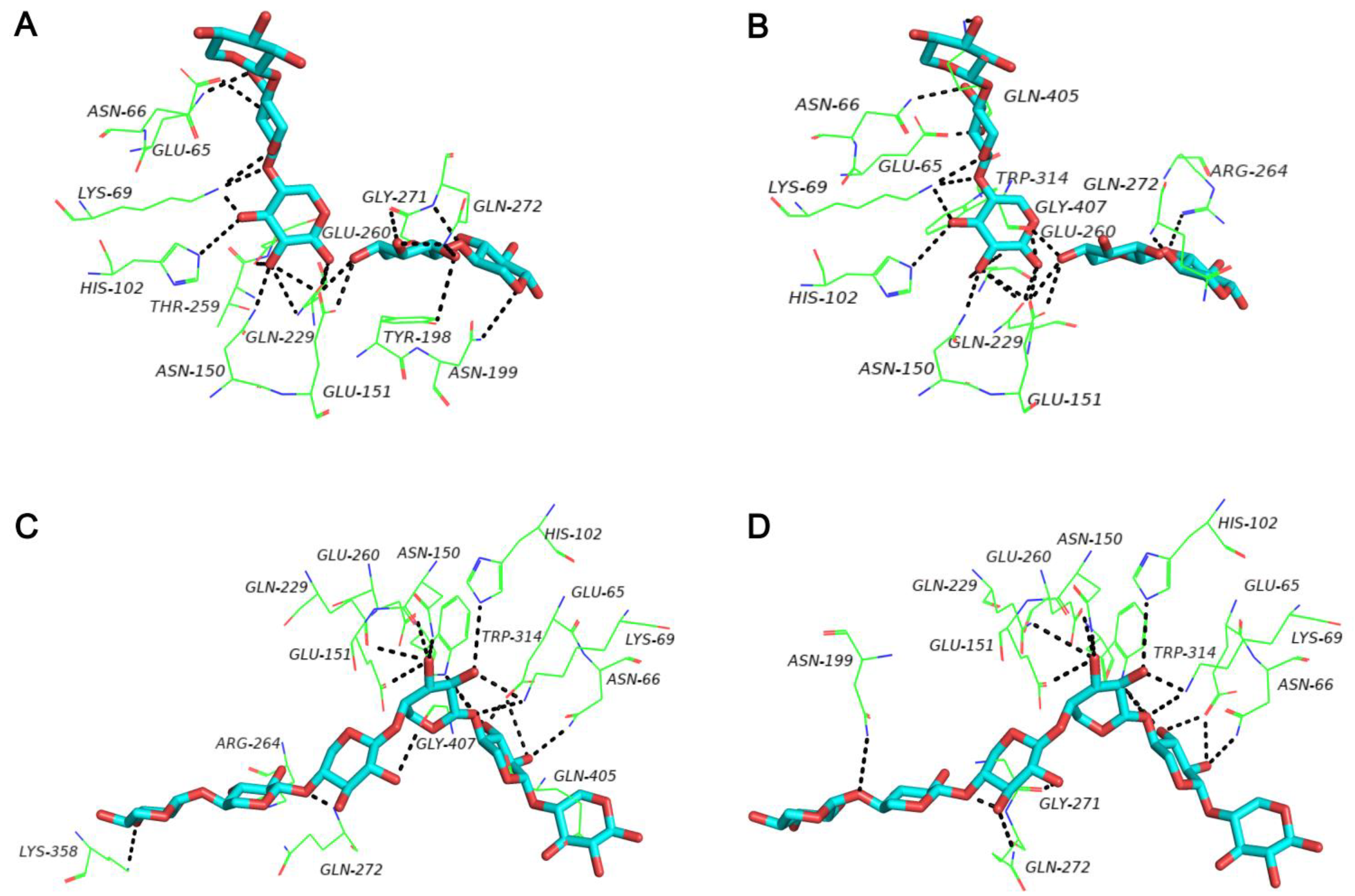
3.1. Sequence Alignment and Protein Structure Analysis of C-Terminal Oligopeptide C60
3.2. Residue Contents Analysis of C-Terminal Oligopeptide C60
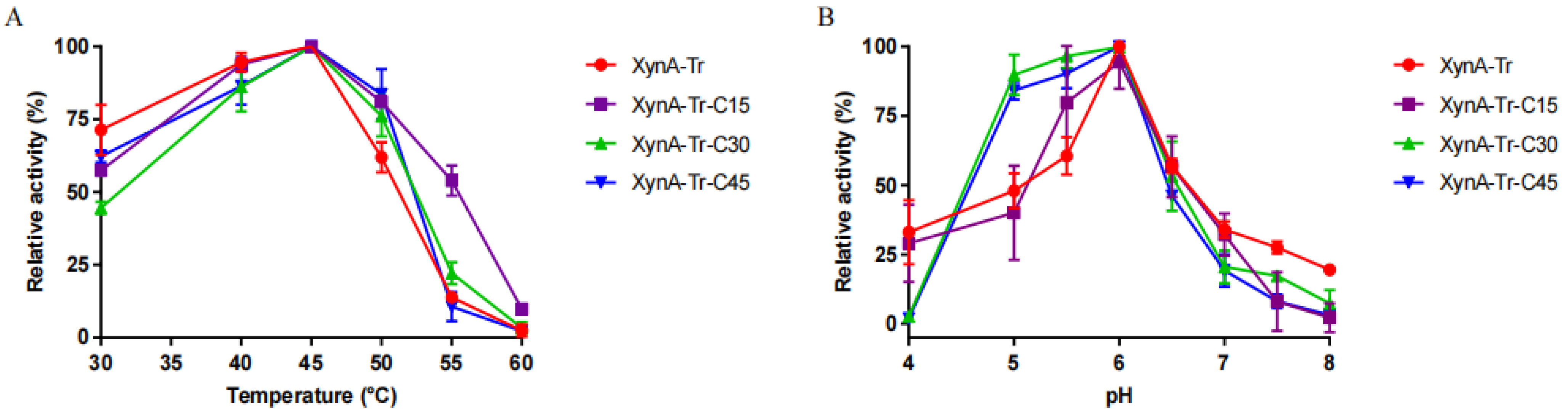
3.3. Enzyme Expression, Purification, and Characterization of Truncated Variants
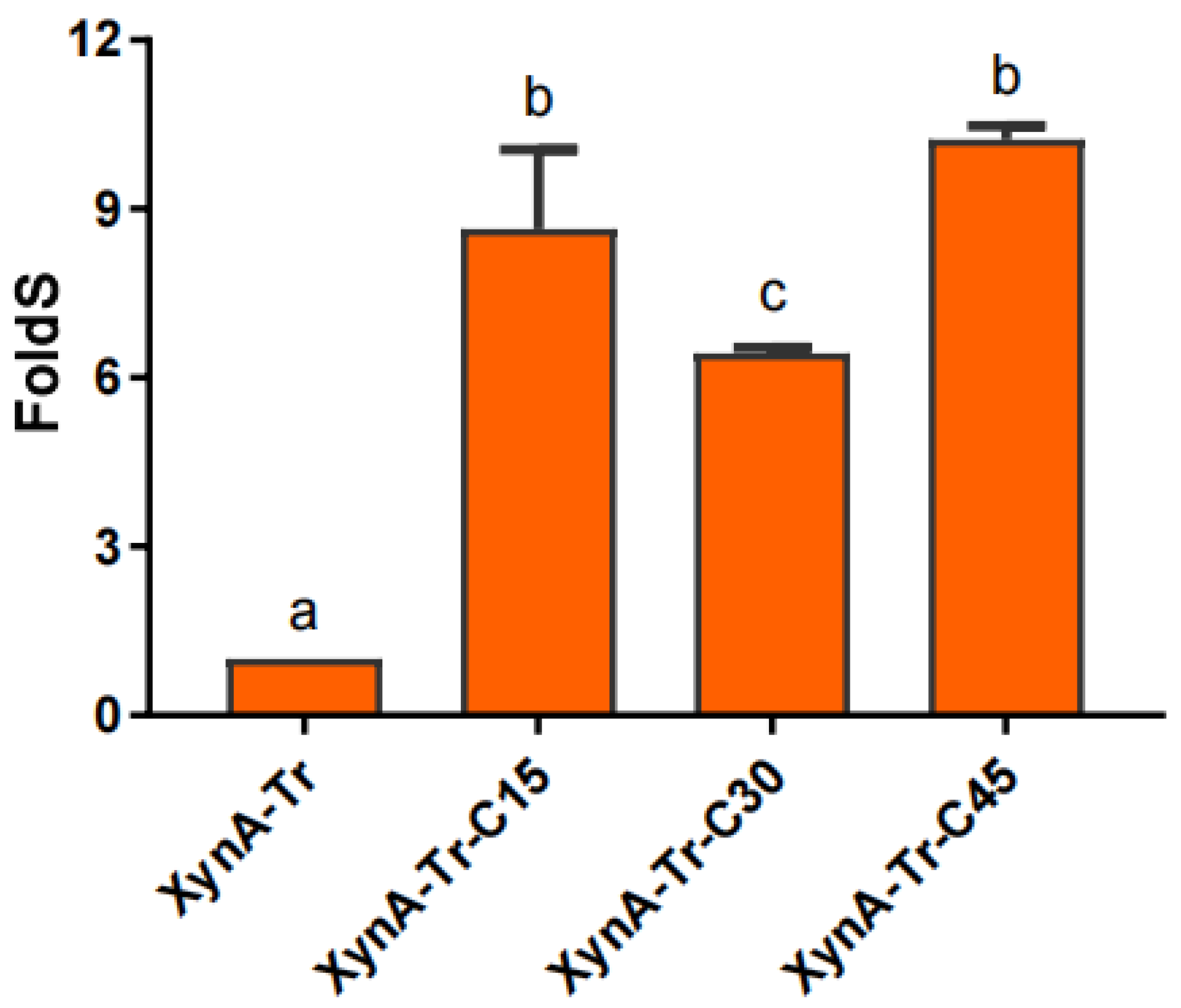
3.4. Analysis of XOS Production of Truncated Variants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Summanen, P.H.; Komoriya, T.; Finegold, S.M. In vitro study of the prebiotic xylooligosaccharide (XOS) on the growth of Bifidobacterium spp. and Lactobacillus spp. Int. J. Food Sci. Nutr. 2015, 66, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Freitas, C.D.; Terrone, C.C.; Masarin, F.; Carmona, E.C.; Brienzo, M. In vitro study of the effect of xylooligosaccharides obtained from banana pseudostem xylan by enzymatic hydrolysis on probiotic bacteria. J. Biocatal. Agric. Biotechnol. 2021, 33, 101973. [Google Scholar] [CrossRef]
- Capetti, C.C.d.M.; Vacilotto, M.M.; Dabul, A.N.G.; Sepulchro, A.G.V.; Pellegrini, V.O.A.; Polikarpov, I. Recent advances in the enzymatic production and applications of xylooligosaccharides. World J. Microbiol. Biotechnol. 2021, 37, 169. [Google Scholar] [CrossRef] [PubMed]
- Pinales-Márquez, C.; Rodríguez-Jasso, R.; Araújo, R.; Loredo-Trevio, A.; Ruiz, H.A. Circular bioeconomy and integrated biorefinery in the production of xylooligosaccharides from lignocellulosic biomass: A review. Ind. Crops Prod. 2021, 162, 113274. [Google Scholar] [CrossRef]
- Nur, I.W.A.; Jamaliah, M.J.; Ahmad, F.I.; Siti, F.Z.M.F.; Roshanida, A.R.; Rosli, M.I. High xylooligosaccharides (XOS) production from pretreated kenaf stem by enzyme mixture hydrolysis. Ind. Crops Prod. 2016, 81, 11–19. [Google Scholar]
- Biely, P.; Singh, S.; Puchart, V. Towards enzymatic breakdown of complex plant xylan structures: State of the art. Biotechnol. Adv. 2016, 34, 1260–1274. [Google Scholar] [CrossRef] [PubMed]
- Hero, J.S.; Pisa, J.H.; Romero, C.M.; Nordberg Karlsson, E.; Linares-Pastén, J.A.; Martinez, M.A. Endo-xylanases from Cohnella sp. AR92 aimed at xylan and arabinoxylan conversion into value-added products. Appl. Microbiol. Biotechnol. 2021, 105, 6759–6778. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic. Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, J.; Peng, F.; Peng, X.P.; Peng, P.; Xu, F.; Sun, R.C. Structural features and antioxidant activity of xylooligosaccharides enzymatically produced from sugarcane bagasse. Bioresour. Technol. 2013, 127, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xue, X.; Zhao, H.; Yang, P.; Luo, H.; Zhao, J.; Huang, H.; Yao, B. A C-terminal proline-rich sequence simultaneously broadens the optimal temperature and pH ranges and improves the catalytic efficiency of glycosyl hydrolase family 10 ruminal xylanases. Appl. Environ. Microbiol. 2014, 80, 3426–3432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veno, J.; Ahmad Kamarudin, N.H.; Mohamad Ali, M.S.; Masomian, M.; Raja Abd Rahman, R.N.Z. Directed Evolution of Recombinant C-Terminal Truncated Staphylococcus epidermidis Lipase AT2 for the Enhancement of Thermostability. Int. J. Mol. Sci. 2017, 18, 2202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, D.; Satyanarayana, T. Molecular approaches for ameliorating microbial xylanases. Bioresour. Technol. 2012, 117, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, N.; Leow, A.T.C.; Abd Rahman, R.N.Z.R.; Mohd Shariff, F. Single Residue Substitution at N-Terminal Affects Temperature Stability and Activity of L2 Lipase. Molecules 2020, 25, 3433. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L.; Blum, R.; Glennon, W.E.; Burton, A.L. Measurement of carboxymethylcellulase activity. Anal. Biochem. 1960, 1, 127–132. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, H.; Yang, P.; Zhao, J.; Huang, H.; Xue, X.; Zhang, X.; Diao, Q.; Yao, B. Comparative quantitative analysis of gene expression profiles of glycoside hydrolase family 10 xylanases in the sheep rumen during a feeding cycle. Appl. Environ. Microbiol. 2013, 79, 1212–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.H.; Jiang, N.; Li, C.X.; Luo, X.M.; Zhao, S.; Feng, J.X. Purification and characterization of an endo-xylanase from Trichoderma sp., with xylobiose as the main product from xylan hydrolysis. World J. Microbiol. Biotechnol. 2019, 35, 171. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jiang, Y.; Tong, X.; Zhao, L.; Pei, J. Co-production of Xylooligosaccharides and Xylose from Poplar Sawdust by Recombinant Endo-1,4-β-Xylanase and β-Xylosidase Mixture Hydrolysis. Front. Bioeng. Biotechnol. 2021, 8, 637397. [Google Scholar] [CrossRef] [PubMed]
- Gautério, G.; Silva, L.G.D.; Hübner, T.; Ribeiro, T.; Kalil, S.J. Xylooligosaccharides production by crude and partially purified xylanase from Aureobasidium pullulans: Biochemical and thermodynamic properties of the enzymes and their application in xylan hydrolysis. Process Biochem. 2021, 104, 161–170. [Google Scholar] [CrossRef]
- Marasinghe, S.D.; Jo, E.; Hettiarachchi, S.A.; Lee, Y.; Eom, T.Y.; Gang, Y.; Kang, Y.H.; Oh, C. Characterization of glycoside hydrolase family 11 xylanase from Streptomyces sp. strain J103; its synergetic effect with acetyl xylan esterase and enhancement of enzymatic hydrolysis of lignocellulosic biomass. Microb. Cell Fact. 2021, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhang, C.; Xing, X.H. Design and construction of chimeric linker library with controllable flexibilities for precision protein engineering. Methods Enzymol. 2021, 647, 23–49. [Google Scholar] [PubMed]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 297, 249–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scocchi, M.; Tossi, A.; Gennaro, R. Proline-rich antimicrobial peptides: Converging to a non-lytic mechanism of action. Cell Mol. Life Sci. 2011, 68, 2317–2330. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Guler, H.I.; Ozer, A.; Sapmaz, M.T.; Belduz, A.O.; Hasan, F.; Shah, A.A. C-Terminal proline-rich sequence broadens the optimal temperature and pH ranges of recombinant xylanase from Geobacillus thermodenitrificans C5. Enzym. Microb. Technol. 2016, 91, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Shirzad, F.; Sajedi, R.H.; Shahangian, S.S.; Rasti, B.; Mosadegh, B.; Taghdir, M.; Hosseinkhani, S. Deletion of extra C-terminal segment and its effect on the function and structure of artemin. Int. J. Biol. Macromol. 2011, 49, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sun, X.; Yan, P.; Wang, L.; Chen, H. Non-structured amino-acid impact on GH11 differs from GH10 xylanase. PLoS ONE 2012, 7, e45762. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Cloning, Expression, and Characterization of a New Xylanase from Alkalophilic Paenibacillus sp. 12–11. J. Microbiol. Biotechnol. 2011, 21, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linares-Pasten, J.A.; Anna, A.; Nordberg, K.E. Structural considerations on the use of endo-xylanases for the production of prebiotic xylooligosaccharides from biomass. Curr. Protein Pept. Sci. 2018, 19, 48–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.W.; Kim, M.J.; Seo, S.; Yoo, S.; Hong, J.H. Relative sweetness and sweetness quality of Xylobiose. Food Sci. Biotechnol. 2017, 26, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Yu, B.Z.; Rogers, J.; Byeon, I.J.; Sekar, K.; Chen, X.; Sundaralingam, M.; Tsai, M.D.; Jain, M.K. Phospholipase A2 engineering. Deletion of the C-terminus segment changes substrate specificity and uncouples calcium and substrate binding at the zwitterionic interface. Biochemistry 1996, 35, 12164–12174. [Google Scholar] [CrossRef]
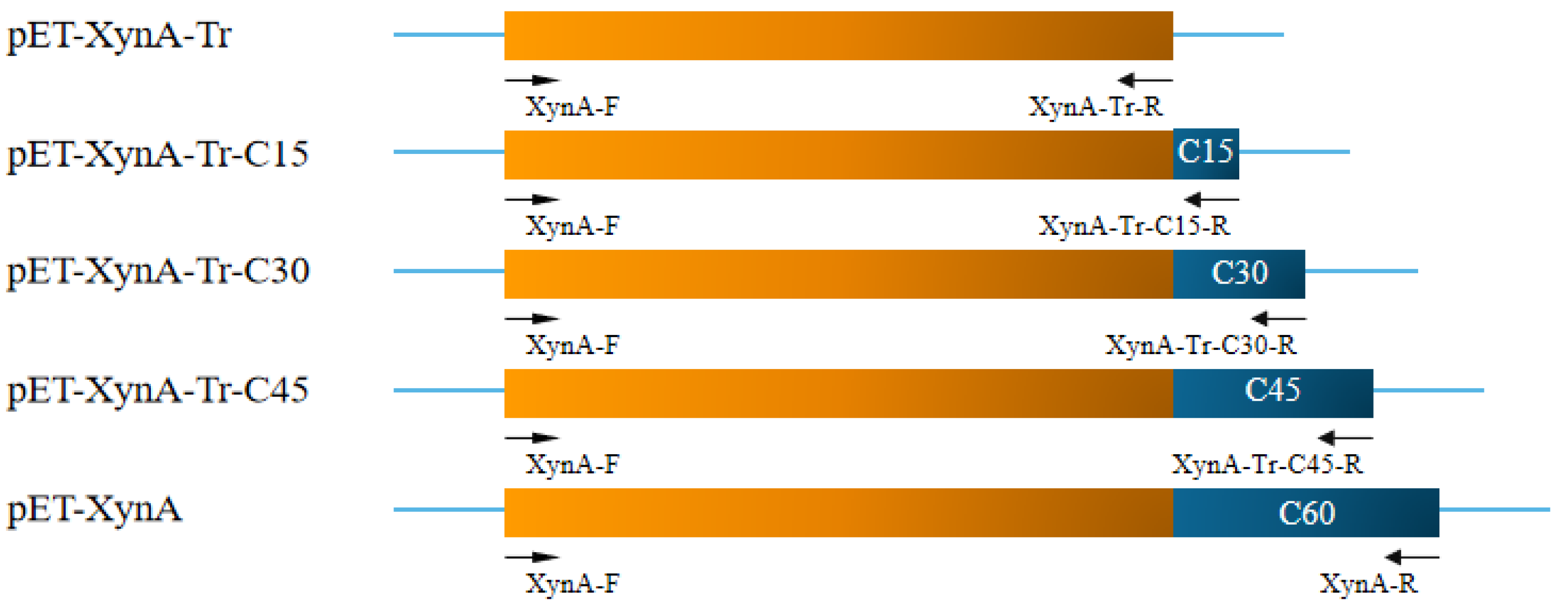

| Gene | Proline (P) | Alanine (A) | Glycine (G) | Glutamine (Q) | Arginine (R) | Glutamic Acid (E) | Phenylalanine (F) | Lysine (K) | Asparagine (N) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |
| XynA-Tr | 15 | 4.34 | 30 | 8.67 | 16 | 4.62 | 17 | 4.91 | 17 | 4.91 | 20 | 5.78 | 15 | 4.34 | 24 | 6.94 | 22 | 6.36 |
| C60 | 12 | 20 | 6 | 10 | 6 | 10 | 6 | 10 | 5 | 8.33 | 4 | 6.67 | 4 | 6.67 | 4 | 6.67 | 4 | 6.67 |
| MBQ5981334 | 11 | 17.46 | 5 | 7.94 | 2 | 3.17 | 10 | 15.87 | 5 | 7.94 | 4 | 6.35 | 3 | 4.76 | 3 | 4.76 | 5 | 7.94 |
| MBR6716339 | 11 | 17.46 | 5 | 7.94 | 2 | 3.17 | 10 | 15.87 | 5 | 7.94 | 4 | 6.35 | 3 | 4.76 | 3 | 4.76 | 5 | 7.94 |
| MBR3496259 | 10 | 16.67 | 4 | 6.67 | 2 | 3.33 | 9 | 15 | 5 | 8.33 | 3 | 5 | 4 | 6.67 | 5 | 8.33 | 4 | 6.67 |
| MBQ8453537 | 11 | 16.67 | 5 | 7.58 | 3 | 4.55 | 9 | 13.64 | 5 | 7.58 | 4 | 6.06 | 3 | 4.55 | 6 | 9.09 | 4 | 6.06 |
| MBQ9286358 | 13 | 17.81 | 8 | 10.96 | 2 | 2.74 | 11 | 15.07 | 8 | 10.96 | 5 | 6.85 | 3 | 4.11 | 5 | 6.85 | 5 | 6.85 |
| Hydrolysis Products | % Hydrolysis Products a | ||||
|---|---|---|---|---|---|
| XynA-Tr | XynA-Tr-C15 | XynA-Tr-C30 | XynA-Tr-C45 | XynA | |
| Xylose | 11.98 | 8.25 | 13.24 | 8.75 | 14.43 |
| Xylobiose | 32.43 | 46.33 | 43.41 | 49.60 | 45.82 |
| Xylotriose | 38.00 | 32.36 | 30.41 | 30.80 | 32.45 |
| Xylotetraose | 5.37 | 1.39 | 1.40 | 0.37 | 2.03 |
| Xylobiose + Xylotriose + Xylotetraose | 75.80 | 80.08 | 75.22 | 80.78 | 80.31 |
| Other xylooligosaccharides | 12.22 | 11.67 | 11.54 | 10.48 | 5.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, K.; Jin, S.; Wang, Y.; Yu, Z.; Sun, J.; Liu, T.; Zhang, Z.; Zhang, T.; Li, Z.; Zhao, J. Crucial Residues of C-Terminal Oligopeptide C60 to Improve the Yield of Prebiotic Xylooligosaccharides by Truncated Mutation. Foods 2022, 11, 862. https://doi.org/10.3390/foods11060862
Pan K, Jin S, Wang Y, Yu Z, Sun J, Liu T, Zhang Z, Zhang T, Li Z, Zhao J. Crucial Residues of C-Terminal Oligopeptide C60 to Improve the Yield of Prebiotic Xylooligosaccharides by Truncated Mutation. Foods. 2022; 11(6):862. https://doi.org/10.3390/foods11060862
Chicago/Turabian StylePan, Kungang, Shanzheng Jin, Yue Wang, Zhao Yu, Junhao Sun, Tianhui Liu, Zhengjie Zhang, Tongcun Zhang, Zhongyuan Li, and Junqi Zhao. 2022. "Crucial Residues of C-Terminal Oligopeptide C60 to Improve the Yield of Prebiotic Xylooligosaccharides by Truncated Mutation" Foods 11, no. 6: 862. https://doi.org/10.3390/foods11060862






