Transcriptomics Integrated with Changes in Cell Wall Material of Chestnut (Castanea mollissima Blume) during Storage Provides a New Insight into the “Calcification” Process
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Determination of the Moisture Content
2.3. Determination of Cell Wall Material in Different Calcified Chestnut Seeds
2.3.1. Extraction of the Cell Wall Material
2.3.2. Extraction and Determination of the Lignin and Cellulose Contents
2.3.3. Extraction and Determination of Pectin Content with Different Solubilities
2.4. Transcriptome Analysis of Chestnut with Different Degrees of Calcification
2.4.1. RNA Extraction and Transcriptome Sequencing
2.4.2. Analysis of Differentially Expressed Genes
2.4.3. Gene Set Enrichment Analysis
2.4.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.4.5. Statistical Analyses
3. Results
3.1. Relationship between the Degree of Chestnut Calcification and Moisture Content
3.2. Changes in Cell Wall Material (CWM) Content in Different Stages of Chestnut Calcification
3.3. Changes in the Lignin and Cellulose Content of Different Stages of Calcification
3.4. Changes in the Pectin Content of Chestnut at Different Stages of Calcification
3.5. Transcriptome Sequencing and Assembly
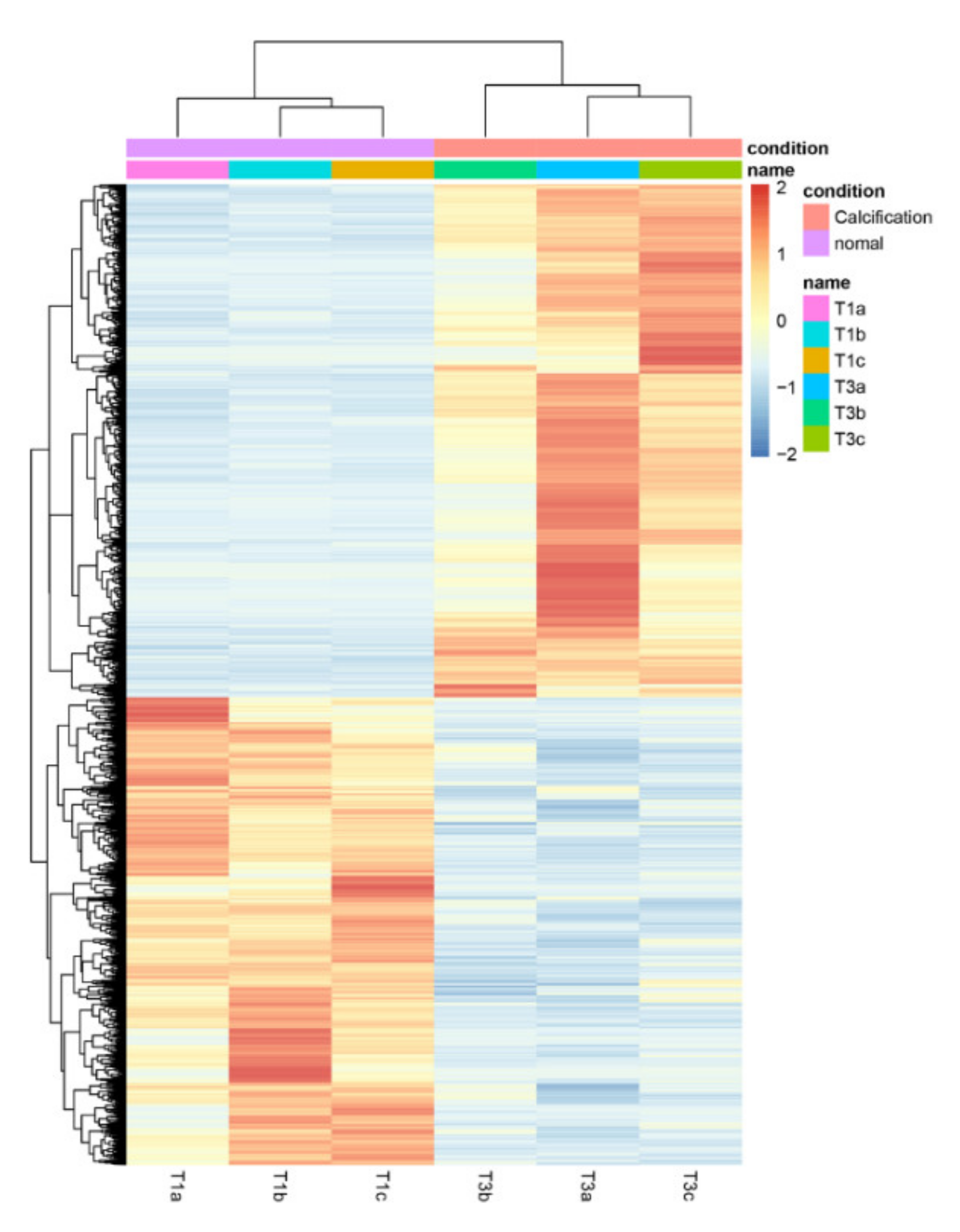
3.6. Comparisons between the 0% and 50% Stages of Calcification
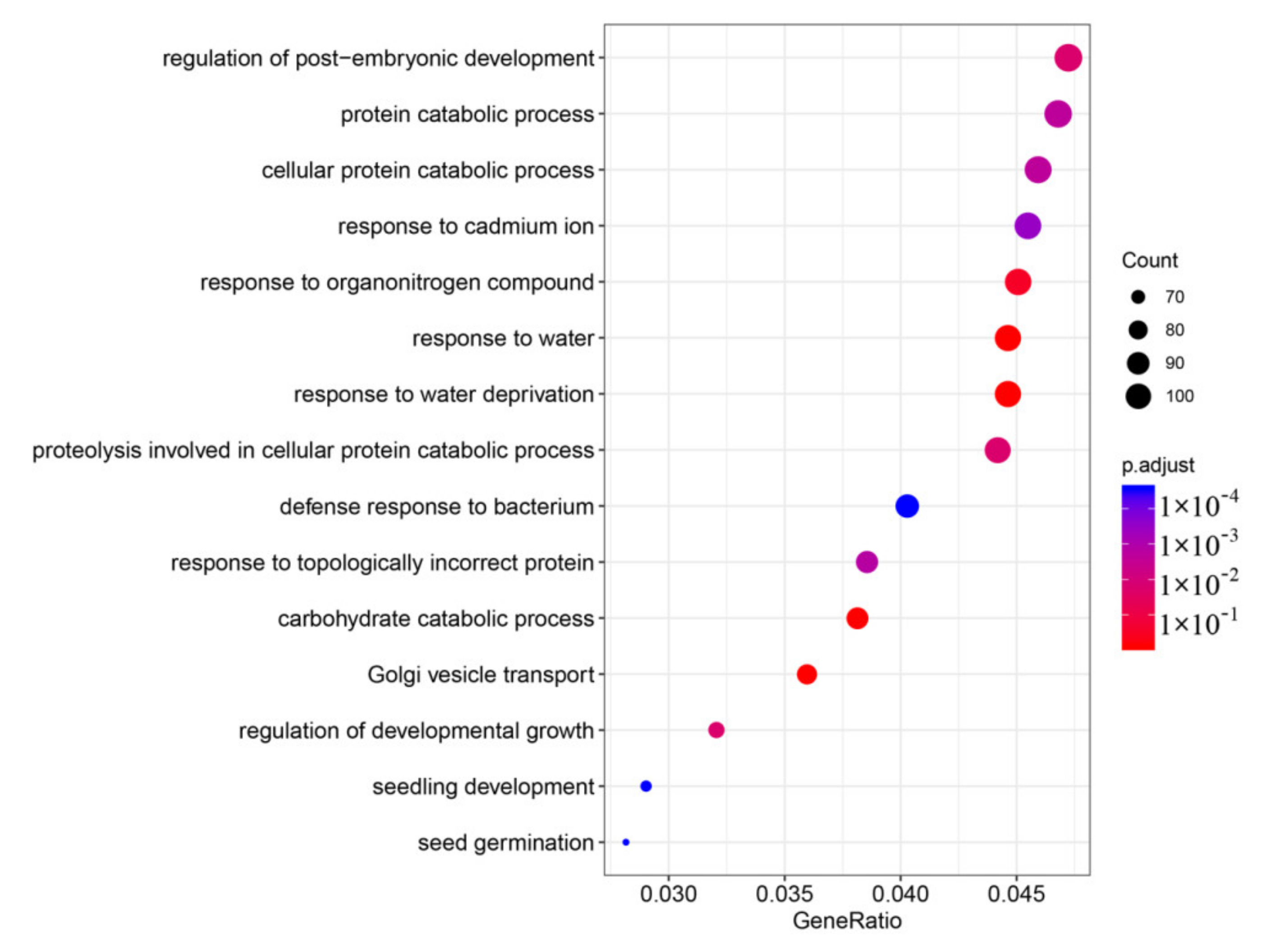
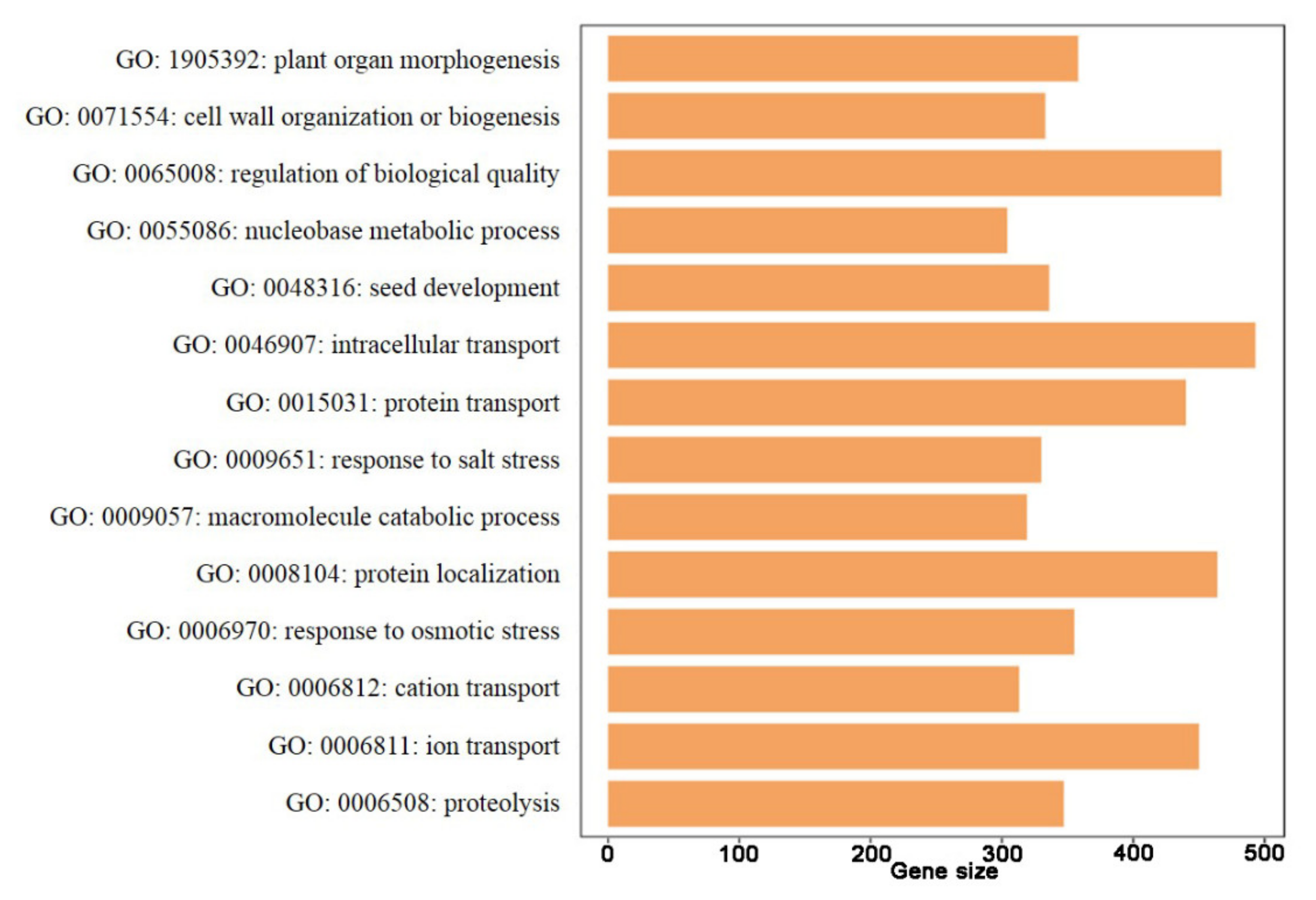
3.7. GO and KEGG Enrichment of the DEGs
3.8. GSEA Enrichment Analysis
3.9. Transcriptomic Insights into Chestnut Calcification and Moisture Content
3.10. Transcriptomic Insights into Cellulose and Lignin Metabolism
3.11. Pectin Metabolism-Related DEGs
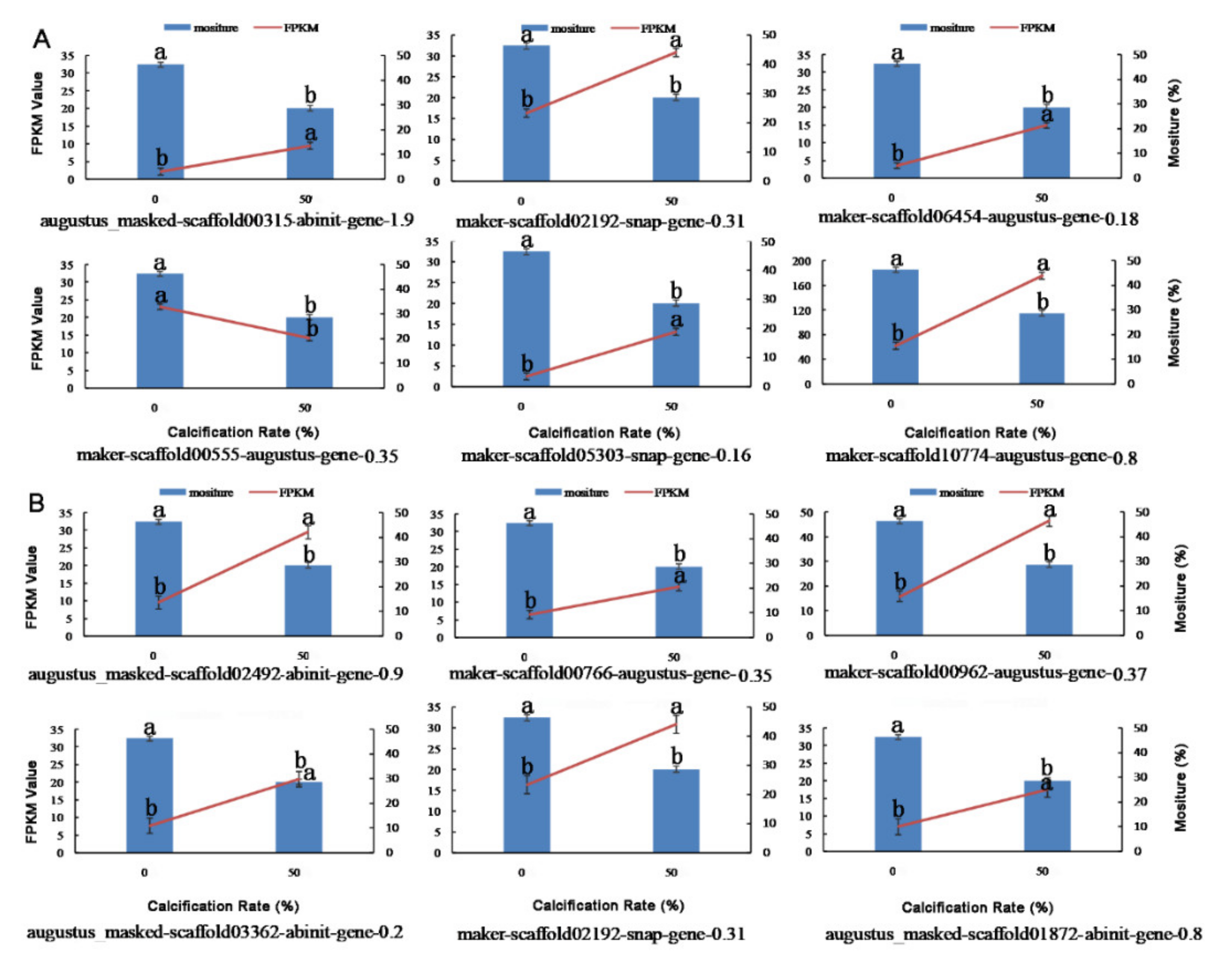
3.12. Validation of Transcriptome Data by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Liu, G.H.; Fang, J.Y. Spatial patterns of chestnut (Castanea millissima) and its species geographical distribution in China. Acta Ecol. Sin. 2001, 21, 164–170. [Google Scholar]
- De Vasconcelos, M.C.; Bennett, R.N.; Rosa, E.A.; Ferreira-Cardoso, J.V. Composition of European chestnut (Castanea sativa Mill.) and association with health effects: Fresh and processed products. J. Sci. Food Agric. 2010, 90, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Li, H.; Wang, Y.; Li, S.; Xue, C. Structural characterization and biological activity of galactoglucan from Castanea mollissima Blume. J. Carbohydr. Chem. 2019, 38, 398–411. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Z.; Liu, G.; Wu, Y.; Ouyang, J. Inhibitory Effect of Chestnut (Castanea mollissima Blume) Inner Skin Extract on the Activity of α-Amylase, α-Glucosidase, Dipeptidyl Peptidase IV and In Vitro Digestibility of Starches. Food Chem. 2020, 324, 126847. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Gu, C.; Zhu, D.; Liu, P.; Lai, Y.; Zeng, Q. Water stress affects on cell membrane lipid oxidation and calcification of chestnut (Castanea mollissima Bl.). Postharvest Biol. Technol. 2017, 126, 34–39. [Google Scholar] [CrossRef]
- Gu, C.; Lai, Y.; Zhu, D.; Liu, P.; Zeng, Q. Changes of the metabolism of starch and sucrose content in chestnuts at different degrees of calcification. Sci. Agric. Sin. 2013, 46, 1676–1681. [Google Scholar]
- Van Buren, J. The chemistry of texture in fruits and vegetables. J. Texture Stud. 1979, 10, 1–23. [Google Scholar] [CrossRef]
- Ji, Y.; Hu, W.; Liao, J.; Xiu, Z.; Jiang, A.; Guan, Y.; Yang, X.; Feng, K. Ethanol vapor delays softening of postharvest blueberry by retarding cell wall degradation during cold storage and shelf life. Postharvest Biol. Technol. 2021, 177, 111538. [Google Scholar] [CrossRef]
- Somerville, C. Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 2006, 22, 53–78. [Google Scholar] [CrossRef]
- Welker, C.M.; Balasubramanian, V.K.; Petti, C.; Rai, K.M.; DeBolt, S.; Mendu, V. Engineering plant biomass lignin content and composition for biofuels and bioproducts. Energies 2015, 8, 7654–7676. [Google Scholar] [CrossRef] [Green Version]
- Voragen, A.G.; Coenen, G.J.; Verhoef, R.P.; Schols, H.A. Pectin, a versatile polysaccharide present in plant cell walls. Struct. Chem. 2009, 20, 263–275. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Gu, C.; He, S.; Zhu, D.; Huang, Y.; Zhou, Q. Widely targeted metabolomics analysis reveals new biomarkers and mechanistic insights on chestnut (Castanea mollissima Bl.) calcification process. Food Res. Int. 2021, 141, 110128. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Shi, F.; Chen, Y.; Wang, M.; Zhao, Y.; Geng, G. Transcriptome analysis of Chinese chestnut (Castanea mollissima Blume) in response to Dryocosmus kuriphilus Yasumatsu infestation. Int. J. Mol. Sci. 2019, 20, 855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gu, C.; Zhang, Q.; Zhu, D.; Zheng, Z.; Qiu, H. Effects of tea polyphenol on calcification and reactive oxygen metabolism of chestnuts. Sci. Agric. Sin. 2011, 44, 1000–1005. [Google Scholar]
- Babiker, A.Z.; Dulloo, M.E.; El Balla, M.A.M.; El Tahir, L. Effects of low cost drying methods on seed quality of Sorghum bicolor (L.) Monech. Afr. J. Plant Sci. 2010, 4, 339–345. [Google Scholar]
- Gerber, L.; Zhang, B.; Roach, M.; Rende, U.; Gorzsás, A.; Kumar, M.; Burgert, I.; Niittylä, T.; Sundberg, B. Deficient sucrose synthase activity in developing wood does not specifically affect cellulose biosynthesis, but causes an overall decrease in cell wall polymers. New Phytol. 2014, 203, 1220–1230. [Google Scholar] [CrossRef]
- Femenia, A.; Garcia-Conesa, M.; Simal, S.; Rosselló, C. Characterisation of the cell walls of loquat (Eriobotrya japonica L.) fruit tissues. Carbohydr. Polym. 1998, 35, 169–177. [Google Scholar] [CrossRef]
- Huang, W.; Zhu, N.; Zhu, C.; Wu, D.; Chen, K. Morphology and cell wall composition changes in lignified cells from loquat fruit during postharvest storage. Postharvest Biol. Technol. 2019, 157, 110975. [Google Scholar] [CrossRef]
- Kintner, P.K.; Van Buren, J.P. Carbohydrate interference and its correction in pectin analysis using the m-hydroxydiphenyl method. J. Food Sci. 1982, 47, 756–759. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, L.; Hu, S.; Luo, X.; Cao, Y. Overexpression of the bamboo sucrose synthase gene (BeSUS5) improves cellulose production, cell wall thickness and fiber quality in transgenic poplar. Tree Genet. Genom. 2020, 16, 75. [Google Scholar] [CrossRef]
- Lam, P.Y.; Wang, L.; Lui, A.C.; Liu, H.; Takeda-Kimura, Y.; Chen, M.-X.; Zhu, F.Y.; Zhang, J.; Umezawa, T.; Tobimatsu, Y. Deficiency in flavonoid biosynthesis genes CHS, CHI, and CHIL alters rice flavonoid and lignin profiles. Plant Physiol. 2021, 188, 1993–2011. [Google Scholar] [CrossRef] [PubMed]
- Babbar, N.; Dejonghe, W.; Gatti, M.; Sforza, S.; Elst, K. Pectic oligosaccharides from agricultural by-products: Production, characterization and health benefits. Crit. Rev. Biotechnol. 2016, 36, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Amos, R.A.; Pattathil, S.; Yang, J.Y.; Atmodjo, M.A.; Urbanowicz, B.R.; Moremen, K.W.; Mohnen, D. A two-phase model for the non-processive biosynthesis of homogalacturonan polysaccharides by the GAUT1: GAUT7 complex. J. Biol. Chem. 2018, 293, 19047–19063. [Google Scholar] [CrossRef] [Green Version]
- Christiaens, S.; Van Buggenhout, S.; Houben, K.; Jamsazzadeh Kermani, Z.; Moelants, K.R.; Ngouémazong, E.D.; Van Loey, A.; Hendrickx, M.E. Process structure function relations of pectin in food. Crit. Rev. Food Sci. Nutr. 2016, 56, 1021–1042. [Google Scholar] [CrossRef]
- Schuch, W.; Kanczler, J.; Robertson, D.; Hobson, G.; Tucker, G.; Grierson, D.; Bright, S.; Bird, C. Fruit quality characteristics of transgenic tomato fruit with altered polygalacturonase activity. HortScience 1991, 26, 1517–1520. [Google Scholar] [CrossRef]
- Yang, B.; Jiang, G.; Gu, C.; Yang, H.; Jiang, Y. Structural changes in polysaccharides isolated from chestnut (Castanea mollissima Bl.) fruit at different degrees of hardening. Food Chem. 2010, 119, 1211–1215. [Google Scholar] [CrossRef]
- Konno, H.; Nakashima, S.; Katoh, K. Metal-tolerant moss scopelophila cataractae accumulates copper in the cell wall pectin of the protonema. J. Plant Physiol. 2010, 167, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Konno, H.; Nakato, T.; Nakashima, S.; Katoh, K. Lygodium japonicum fern accumulates copper in the cell wall pectin. J. Exp. Bot. 2005, 56, 1923–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, N.W.; Huang, Y.P. The RNA degradation pathway is involved in PPARα-modulated anti-oral tumorigenesis. Biomedicine 2019, 9, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, J.P.; Hoy, J.W.; Appezzato-da-Glória, B.; Viveros, A.F.; Vieira, M.L.; Baisakh, N. Sugarcane cell wall-associated defense responses to infection by Sporisorium scitamineum. Front. Plant Sci. 2018, 9, 698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ketsa, S.; Atantee, S. Phenolics, lignin, peroxidase activity and increased firmness of damaged pericarp of mangosteen fruit after impact. Postharvest Biol. Technol. 1998, 14, 117–124. [Google Scholar] [CrossRef]
- Piro, G.; Leucci, M.R.; Waldron, K.; Dalessandro, G. Exposure to water stress causes changes in the biosynthesis of cell wall polysaccharides in roots of wheat cultivars varying in drought tolerance. Plant Sci. 2003, 165, 559–569. [Google Scholar] [CrossRef]
- Sweet, W.J.; Morrison, J.C.; Labavitch, J.M.; Matthews, M.A. Altered synthesis and composition of cell wall of grape (Vitis vinifera L.) leaves during expansion and growth-inhibiting water deficits. Plant Cell Physiol. 1990, 31, 407–414. [Google Scholar]
- Salmon, S.; Hudson, S.M. Crystal morphology, biosynthesis, and physical assembly of cellulose, chitin, and chitosan. J. Macromol. Sci. Part C Polym. Rev. 1997, 37, 199–276. [Google Scholar] [CrossRef]
- Wang, Y.; Chantreau, M.; Sibout, R.; Hawkins, S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013, 4, 220. [Google Scholar] [CrossRef] [Green Version]
- Moura, J.C.M.S.; Bonine, C.A.V.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Pandey, A.; Rajamani, U.; Verma, J.; Subba, P.; Chakraborty, N.; Datta, A.; Chakraborty, S.; Chakraborty, N. Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: A proteomic approach. J. Proteome Res. 2010, 9, 3443–3464. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Szakacs-Dobozi, M.; Hemmat, M.; Hrazdina, G. Endopolygalacturonase in apples (Malus domestica) and its expression during fruit ripening. Plant Physiol. 1993, 102, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, W.; Liu, C.-M.; Li, T.; Liang, R.-H.; Luo, S.J. Pectin modifications: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1684–1698. [Google Scholar] [CrossRef] [PubMed]
- Imsabai, W.; Ketsa, S.; van Doorn, W.G. Effect of temperature on softening and the activities of polygalacturonase and pectinesterase in durian fruit. Postharvest Biol. Technol. 2002, 26, 347–351. [Google Scholar] [CrossRef]
- Dong, W.; Li, L.; Cao, R.; Xu, S.; Cheng, L.; Yu, M.; Lv, Z.; Lu, G. Changes in cell wall components and polysaccharide-degrading enzymes in relation to differences in texture during sweetpotato storage root growth. J. Plant Physiol. 2020, 254, 153–282. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Liu, M.; Liu, Y.; Strappe, P.; Sun, H.; Zhou, Z. Comparative non-targeted metabolomic analysis reveals insights into the mechanism of rice yellowing. Food Chem. 2020, 308, 125–621. [Google Scholar] [CrossRef]
- Zhu, G.; Xiao, H.; Guo, Q.; Zhang, Z.; Zhao, J.; Yang, D. Effects of cadmium stress on growth and amino acid metabolism in two Compositae plants. Ecotoxicol. Environ. Saf. 2018, 158, 300–308. [Google Scholar] [CrossRef]
- Kuznetsov, V.V.; Shevyakova, N.I. Polyamines and plant adaptation to saline environments. Desert Plants 2010, 8, 261–298. [Google Scholar]
- Gupta, K.; Dey, A.; Gupta, B. Plant polyamines in abiotic stress responses. Acta Physiol. Plant. 2013, 35, 2015–2036. [Google Scholar] [CrossRef]
- Minocha, R.; Majumdar, R.; Minocha, S.C. Polyamines and abiotic stress in plants: A complex relationship. Front. Plant Sci. 2014, 5, 175. [Google Scholar] [CrossRef] [Green Version]
- Biswas, J.; Sinha, D.; Mukherjee, S.; Roy, S.; Siddiqi, M.; Roy, M. Curcumin protects DNA damage in a chronically arsenic-exposed population of West Bengal. Hum. Exp. Toxicol. 2010, 29, 513–524. [Google Scholar] [CrossRef] [PubMed]






| Samples | Clean Reads | Clean Data | GC content (%) | ≥Q30 |
|---|---|---|---|---|
| T1a | 32,619,737 b | 8,154,934,250 b | 44.91 | 93.50 |
| T1b | 30,425,950 d | 7,606,487,500 d | 44.95 | 93.57 |
| T1c | 23,793,560 e | 5,948,390,000 e | 45.06 | 93.13 |
| T3a | 31,853,069 c | 7,963,267,250 c | 45.22 | 93.73 |
| T3b | 34,018,941 a | 8,504,735,250 a | 45.29 | 93.36 |
| T3c | 32,735,082 b | 8,183,770,500 b | 45.06 | 93.36 |
| Samples | Total Reads | Mapped Reads | Unique Mapped | Multiple Mapped |
|---|---|---|---|---|
| T1a | 65,239,474 b | 54,516,406 c | 53,664,614 c | 851,792 b |
| T1b | 60,851,900 d | 50,891,629 d | 49,885,627 d | 1,006,002 a |
| T1c | 47,587,120 e | 39,889,854 e | 39,123,856 e | 765,998 c |
| T3a | 63,706,138 c | 54,636,733 c | 53,975,213 c | 661,520 d |
| T3b | 68,037,882 a | 58,347,962 a | 57,572,054 a | 775,908 c |
| T3c | 65,470,164 b | 56,715,906 b | 56,031,375 b | 684,531 d |
| Gene | FPKM (0%) | FPKM (50%) | FDR | Log2FC | Regulated |
|---|---|---|---|---|---|
| Cellulose biosynthetic process | |||||
| Scaffold01281-gene-0.39 | 7.13 | 1.59 | 0.0034 | −1.84 | Down |
| Scaffold01202-gene-0.30 | 112.48 | 42.79 | 0.00012 | −2.00 | Down |
| Scaffold03287-gene-0.16 | 2.05 | 0.43 | 0.00100 | −4.46 | Down |
| Scaffold00980-gene-0.41 | 0.08 | 0.03 | 0.00681 | −1.51 | Down |
| Scaffold02563-gene-0.4 | 33.76 | 10.17 | 0.00549 | −1.95 | Down |
| Scaffold11300-gene-0.9 | 4.83 | 16.05 | 0.00676 | 1.73 | Up |
| Sucrose synthase activity | |||||
| Scaffold0125-gene-0.31 | 78.45 | 30.52 | 0.00358 | −1.44 | Down |
| Lignin catabolic process | |||||
| Scaffold34956-gene-0.3 | 8.24 | 1.04 | 0.00128 | −3.09 | Down |
| Scaffold08019-gene-0.16 | 0.71 | 0.05 | 0.00021 | −4.35 | Down |
| Gene | FPKM (0%) | FPKM (50%) | FDR | Log2FC | Regulated |
|---|---|---|---|---|---|
| Pectin biosynthetic process | |||||
| Scaffold02462-gene-0.13 | 1.15 | 0.13 | 1.34 × 10−7 | −3.60 | Down |
| Scaffold00897-gene-0.33 | 20.93 | 7.75 | 0.00201 | −1.53 | Down |
| Scaffold02056-gene-0.28 | 2.7 | 0.28 | 0.00554 | −2.74 | Down |
| Scaffold04120-gene-0.22 | 2.19 | 25.97 | 4.74 × 10−6 | 3.66 | Up |
| Pectinesterase activity | |||||
| Scaffold11321-gene-0.6 | 33.66 | 0.22 | 2.92 × 10−16 | −7.86 | Down |
| Scaffold01256-gene-0.30 | 33.61 | 0.02 | 4.48 × 10−9 | −4.36 | Down |
| Scaffold00325-gene-0.6 | 0.25 | 0.03 | 0.00563 | −4.85 | Down |
| Polygalacturonase activity | |||||
| Scaffold02071-gene-0.36 | 6.75 | 1.15 | 0.00016 | −2.43 | Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhu, C.; Zhao, Y.; Zhang, S.; Wang, W. Transcriptomics Integrated with Changes in Cell Wall Material of Chestnut (Castanea mollissima Blume) during Storage Provides a New Insight into the “Calcification” Process. Foods 2022, 11, 1136. https://doi.org/10.3390/foods11081136
Chen Y, Zhu C, Zhao Y, Zhang S, Wang W. Transcriptomics Integrated with Changes in Cell Wall Material of Chestnut (Castanea mollissima Blume) during Storage Provides a New Insight into the “Calcification” Process. Foods. 2022; 11(8):1136. https://doi.org/10.3390/foods11081136
Chicago/Turabian StyleChen, Yu, Cancan Zhu, Yuqiang Zhao, Shijie Zhang, and Wu Wang. 2022. "Transcriptomics Integrated with Changes in Cell Wall Material of Chestnut (Castanea mollissima Blume) during Storage Provides a New Insight into the “Calcification” Process" Foods 11, no. 8: 1136. https://doi.org/10.3390/foods11081136
APA StyleChen, Y., Zhu, C., Zhao, Y., Zhang, S., & Wang, W. (2022). Transcriptomics Integrated with Changes in Cell Wall Material of Chestnut (Castanea mollissima Blume) during Storage Provides a New Insight into the “Calcification” Process. Foods, 11(8), 1136. https://doi.org/10.3390/foods11081136




