Combined Anti-Biofilm Enzymes Strengthen the Eradicate Effect of Vibrio parahaemolyticus Biofilm: Mechanism on cpsA-J Expression and Application on Different Carriers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Culture Conditions
2.2. Preparation of Combined Enzymes
2.3. Analysis of the Biofilm Formation of V. parahaemolyticus
2.4. Analysis of the Biofilm Formation and Eradication of V. parahaemolyticus
2.5. Analysis of Exopolysaccharide (EPS)
2.6. Observation of Biofilm by CLSM and SEM
2.7. Analysis of cpsA-J Gene Expression
2.8. Preparation and Characterization of V. parahaemolyticus Biofilm on Different Carriers
2.9. Statistical Analysis
3. Results and Discussion
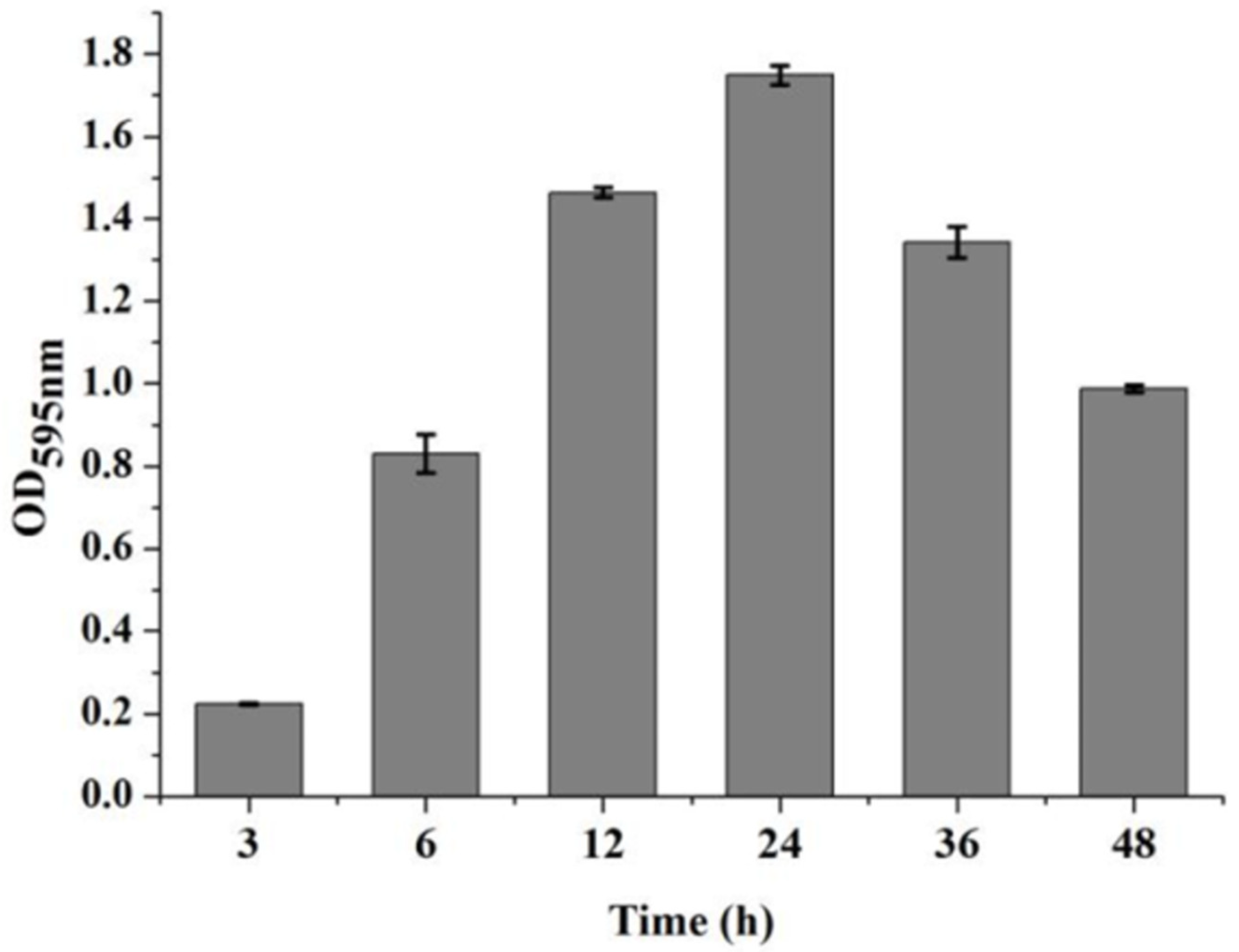
3.1. The Dynamic Development of V. parahaemolyticus Biofilm Formation
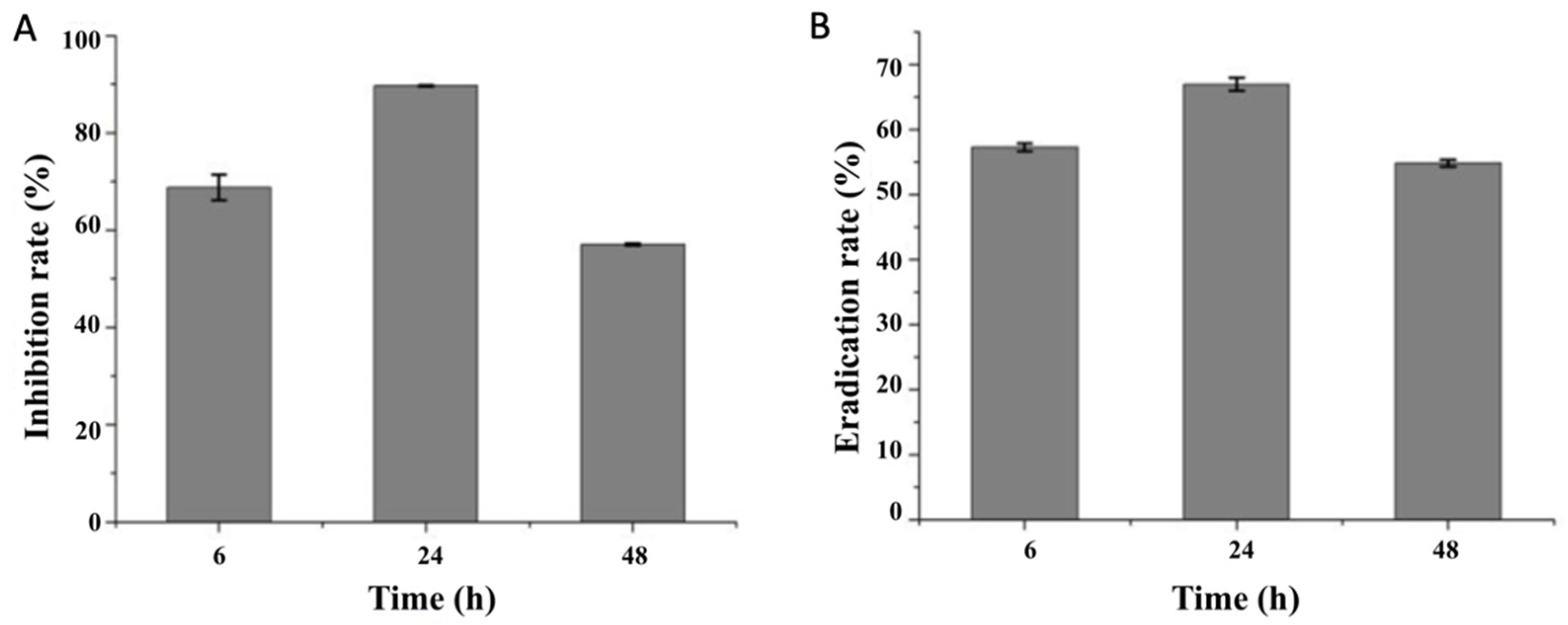
3.2. Effects of Combined Enzymes on the Biofilm Formation and Eradication of V. parahaemolyticus
| Enzymes | Biofilm Inhibition Rate (%) |
|---|---|
| Proteinase K Lipase Cellulase Combined enzymes | 58.6 ± 0.89 a 60.2 ± 1.84 a 80.9 ± 0.67 b 89.7 ± 0.16 c |
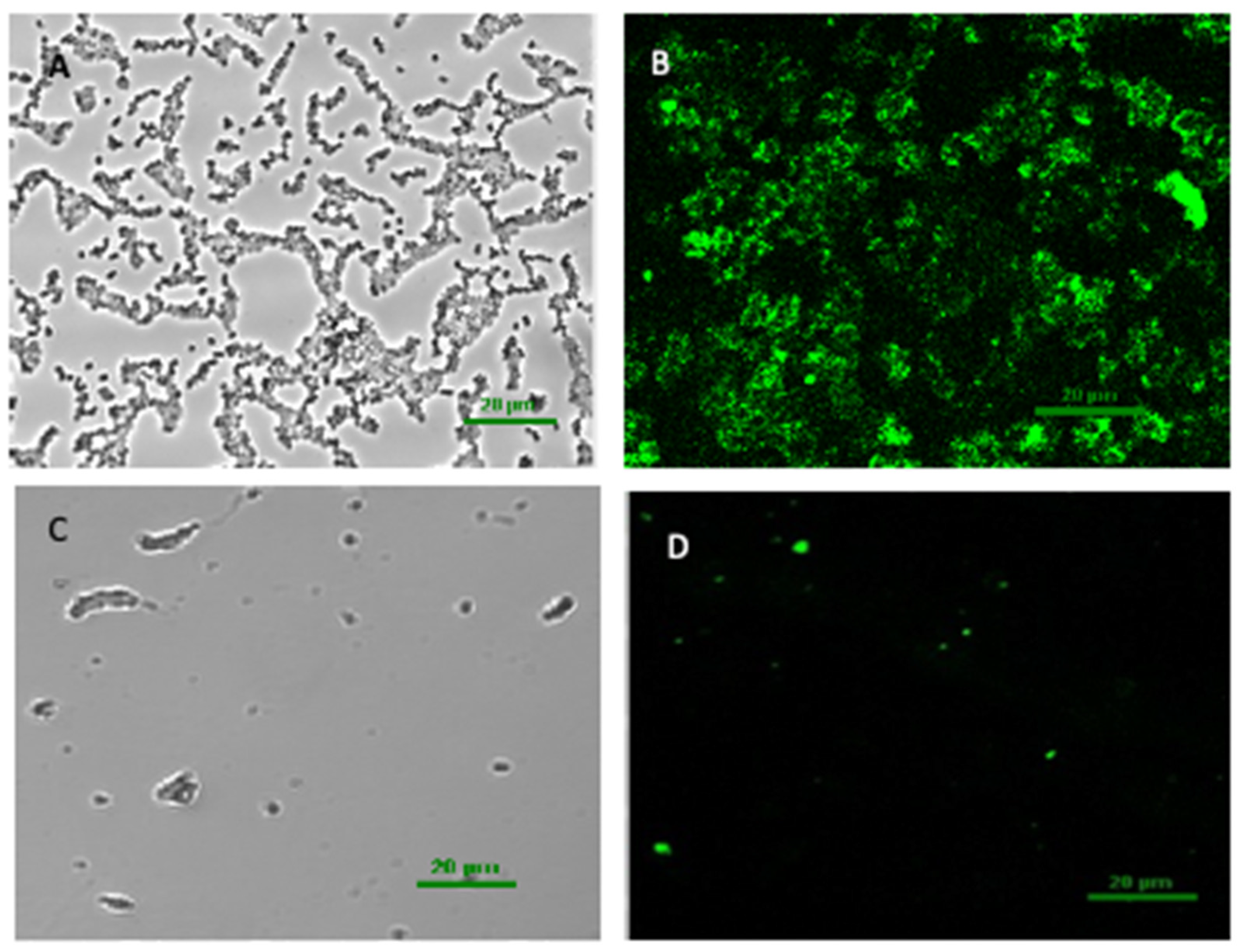
3.3. The CLSM Images of the Biofilm Structure of V. parahaemolyticus
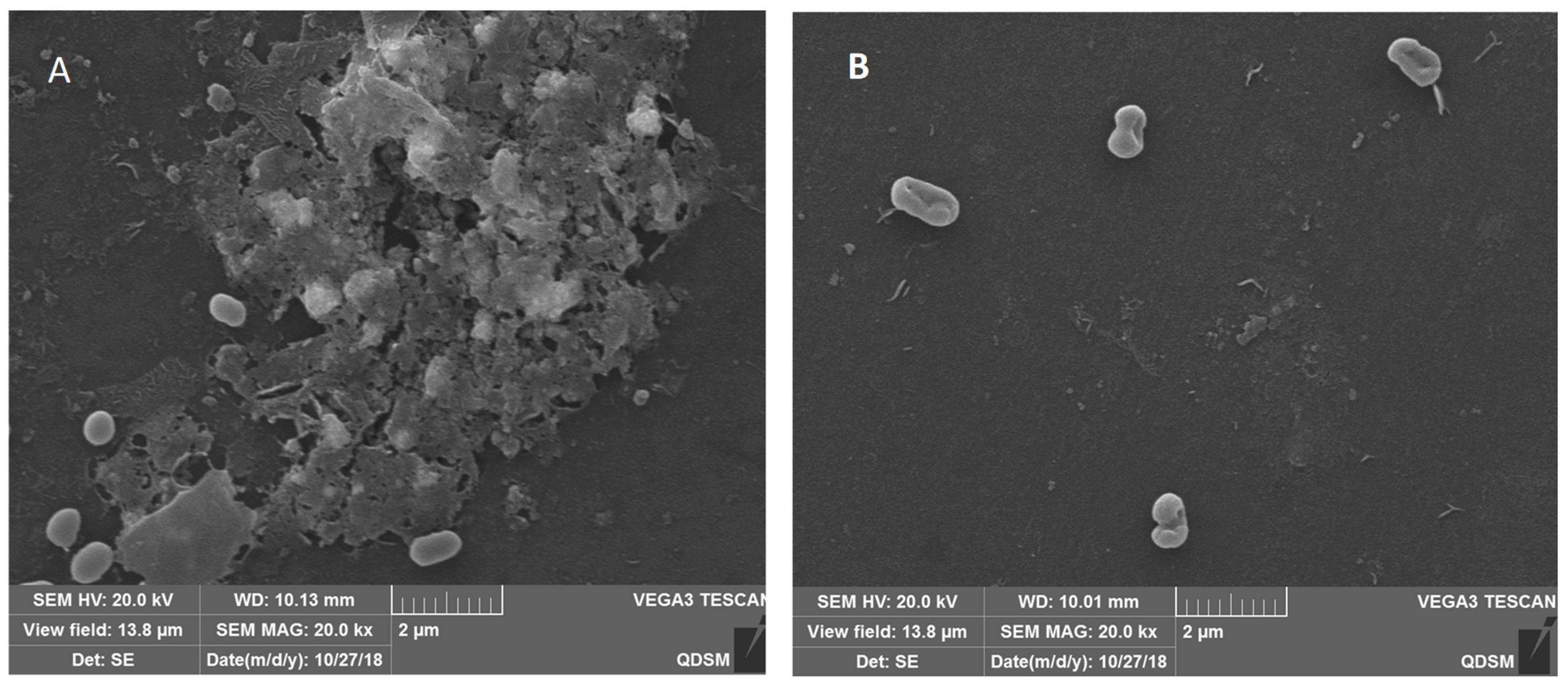
3.4. The SEM Images of the Biofilm Structure of V. parahaemolyticus
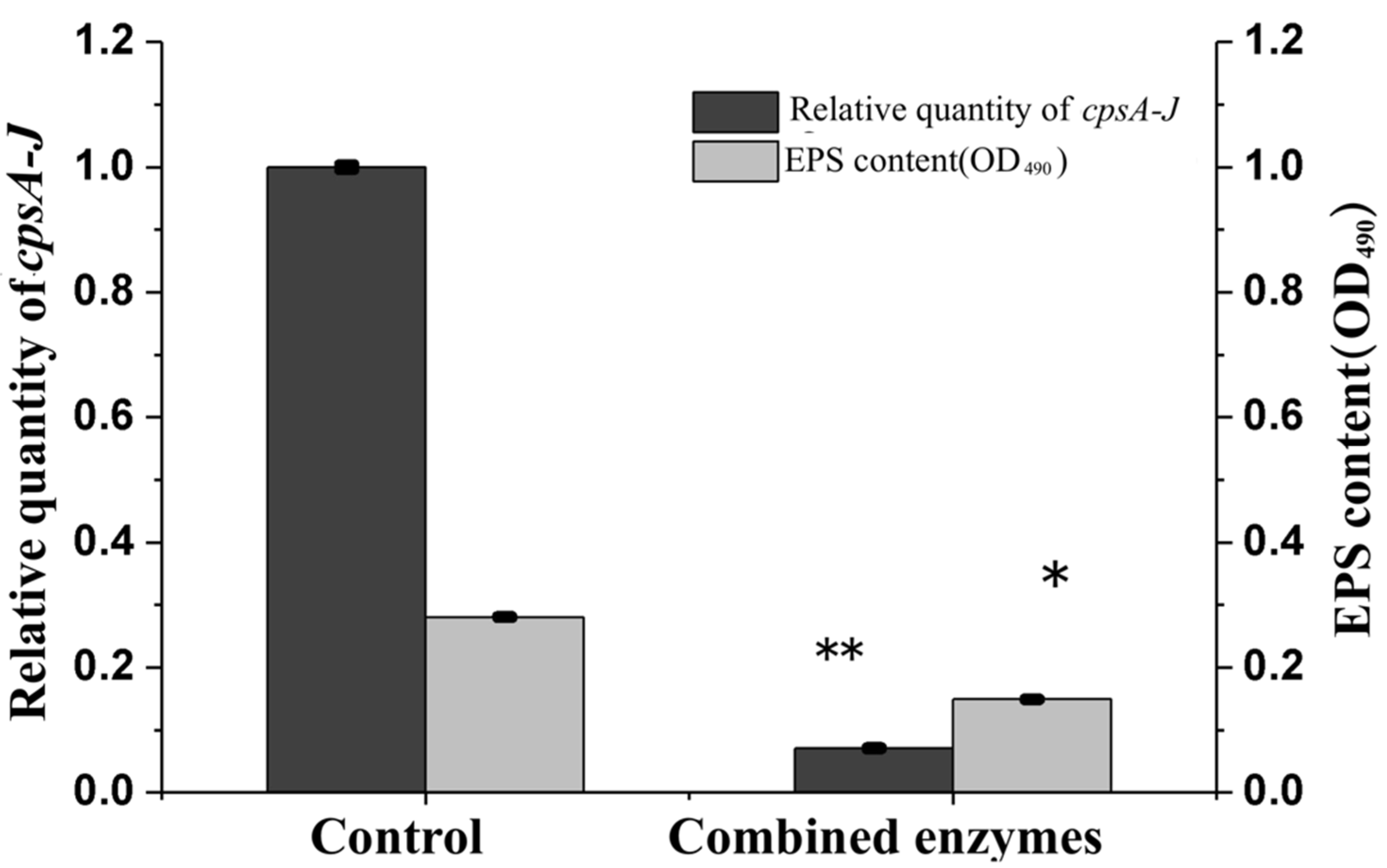
3.5. Effects of Combined Enzymes on EPS Concentration and cpsA-J mRNA Relative Expression of V. parahaemolyticus
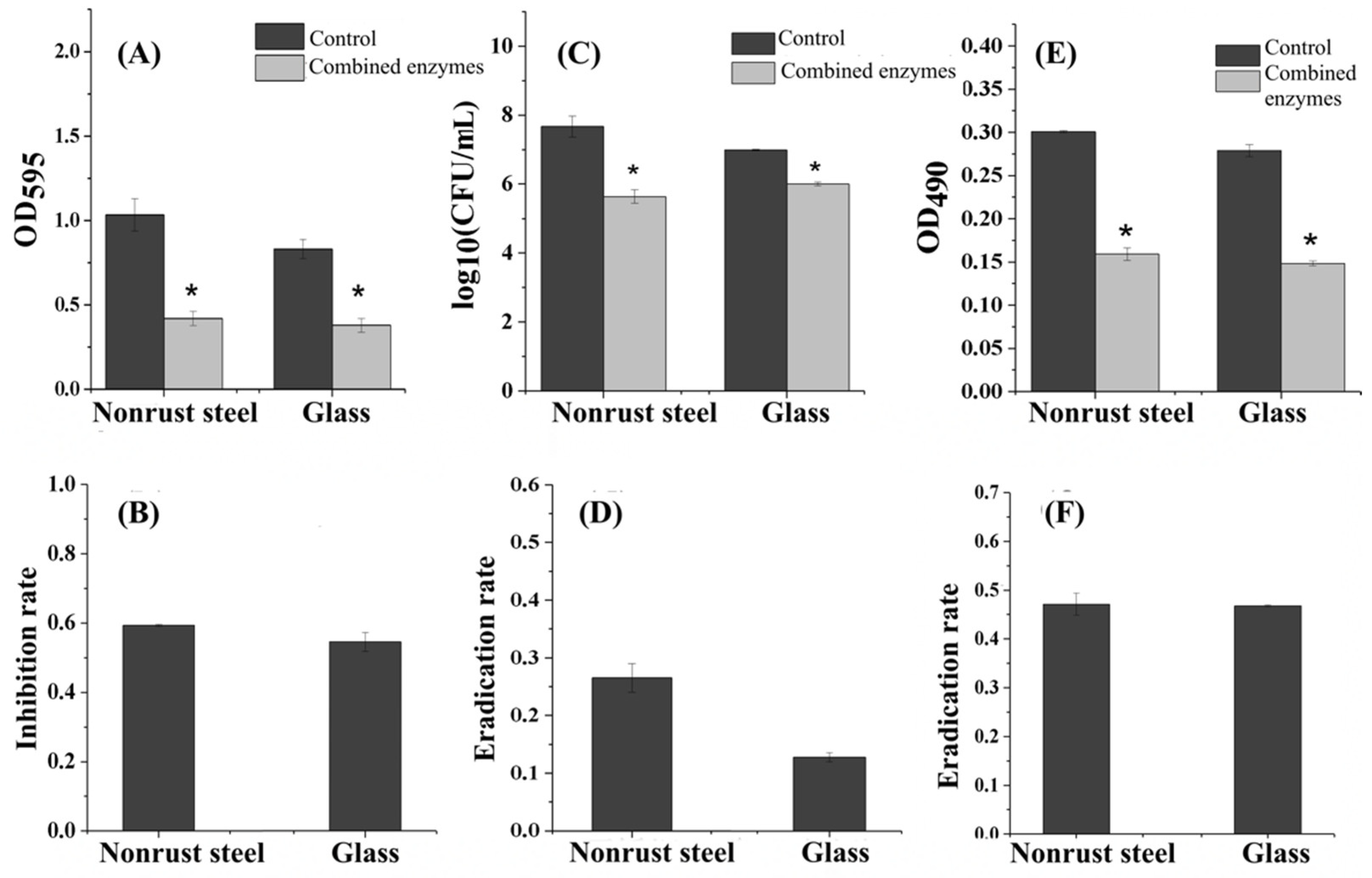
3.6. Inhibition Effect of the Combined Enzymes on Different Carriers
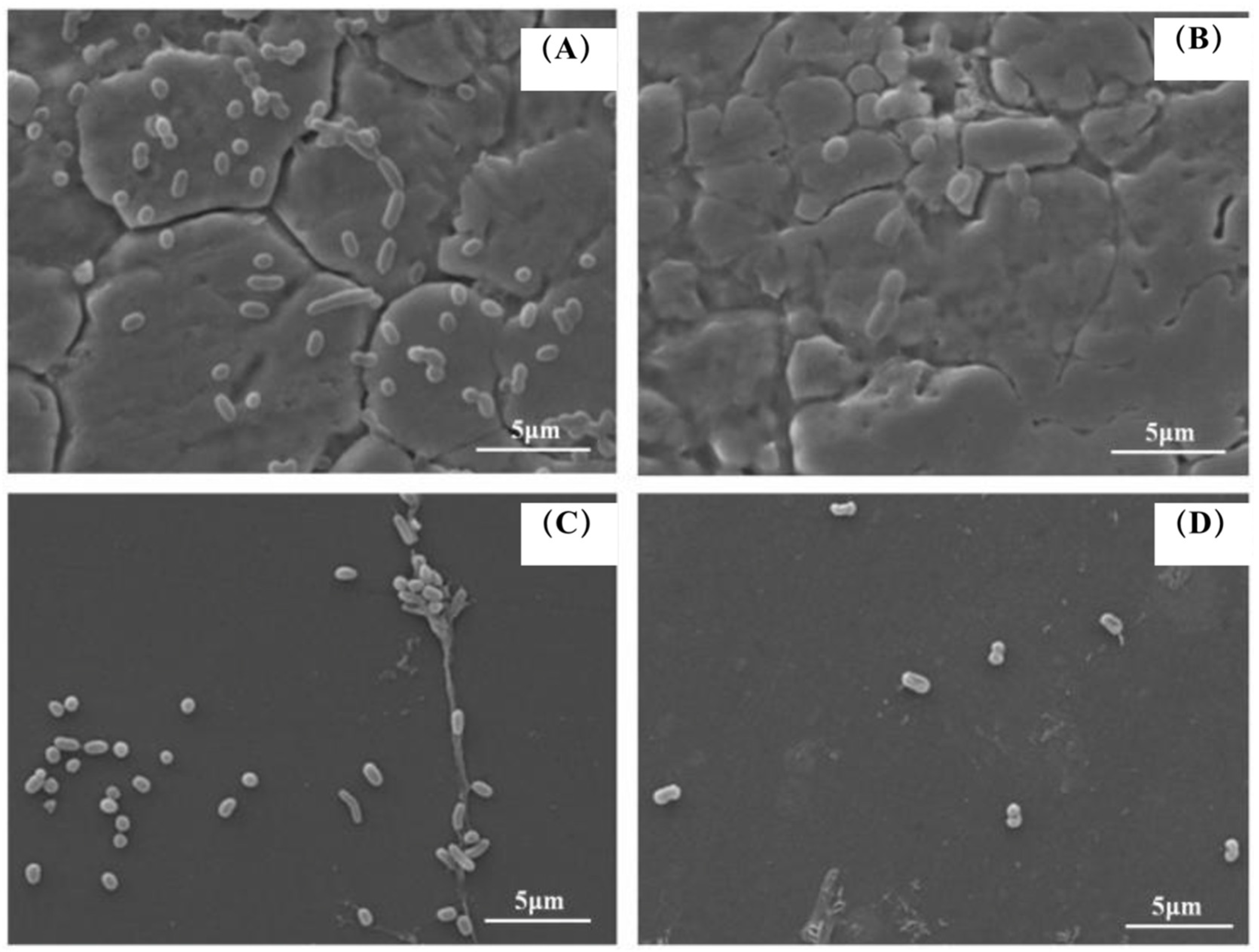
3.7. SEM Images of Combined Enzymes-Inoculated Carriers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berlanga, M.; Guerrero, R. Living together in biofilms: The microbial cell factory and its biotechnological implications. Microb. Cell Fact. 2016, 15, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Q.; Song, X.; Zhang, Z.; Fu, J.; Wang, X.; Malakar, P.K.; Liu, H.; Pan, Y.; Zhao, Y. Removal of Foodborne Pathogen Biofilms by Acidic Electrolyzed Water. Front. Microbiol. 2017, 8, 988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alabdullatif, M.; Atreya, C.D.; Ramirez-Arcos, S. Antimicrobial peptides: An effective approach to prevent bacterial biofilm formation in platelet concentrates. Transfusion 2018, 58, 2013–2021. [Google Scholar] [CrossRef] [PubMed]
- Kataky, D.; Knowles, E. Biofilm formation on abiotic surfaces and their redox activity. Curr. Opinn. Electrochem. 2018, 12, 121–128. [Google Scholar] [CrossRef]
- Meireles, A.; Borges, A.; Giaouris, E.; Simões, M. The current knowledge on the application of anti-biofilm enzymes in the food industry. Food Res. Int. 2016, 86, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Malone, M.; Goeres, D.M.; Gosbell, I.; Vickery, K.; Jensen, S.; Stoodley, P. Approaches to biofilm-associated infections: The need for standardized and relevant biofilm methods for clinical applications. Expert Rev. Anti Infect. Ther. 2017, 15, 147–156. [Google Scholar] [CrossRef]
- Burton, E.; Gawande, P.V.; Yakandawala, N.; LoVetri, K.; Zhanel, G.G.; Romeo, T.; Friesen, A.D.; Madhyastha, S. Antibiofilm Activity of GlmU Enzyme Inhibitors against Catheter-Associated Uropathogens. Antimicrob. Agents Chemother. 2006, 50, 1835–1840. [Google Scholar] [CrossRef] [Green Version]
- Solihin, J.; Waturangi, D.E.; Purwadaria, T. Induction of amylase and protease as antibiofilm agents by starch, casein, and yeast extract in Arthrobacter sp. CW01. BMC Microbiol. 2021, 21, 232. [Google Scholar] [CrossRef]
- Loiselle, M.; Anderson, K.W. The Use of Cellulase in Inhibiting Biofilm Formation from Organisms Commonly Found on Medical Implants. Biofouling 2003, 19, 77–85. [Google Scholar] [CrossRef]
- Kalpana, B.J.; Aarthy, S.; Pandian, S.K. Antibiofilm Activity of α-Amylase from Bacillus subtilis S8-18 Against Biofilm Forming Human Bacterial Pathogens. Appl. Biochem. Biotech. 2012, 167, 1778–1794. [Google Scholar] [CrossRef]
- Araújo, P.; Machado, I.; Meireles, A.; Leiknes, T.; Mergulhão, F.; Melo, L.; Simões, M. Combination of selected enzymes with cetyltrimethylammonium bromide in biofilm inactivation, removal and regrowth. Food Res. Int. 2017, 95, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, S.S.; Carvalho, J.; Aires, C.; Nitschke, M. Disruption of Staphylococcus aureus biofilms using rhamnolipid biosurfactants. J. Dairy Sci. 2017, 100, 7864–7873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Chen, G.W.; Yang, Y.P.; Wu, S.Y.; Liu, Q. Reactive Oxygen Species (ROS) Regulates Listeria monocytogenes Biofilm Formation. Food Sci. 2013, 34, 189–193. (In Chinese) [Google Scholar]
- Xie, T.; Liao, Z.; Lei, H.; Fang, X.; Wang, J.; Zhong, Q. Antibacterial activity of food-grade chitosan against Vibrio parahaemolyticus biofilms. Microb. Pathog. 2017, 110, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, U.T.; Burrows, L.L. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int. J. Food Microbiol. 2014, 187, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Pechaud, Y.; Marcato-Romain, C.; Girbal-Neuhauser, E.; Queinnec, I.; Bessiere, Y.; Paul, E. Combining hydrodynamic and enzymatic treatments to improve multi-species thick biofilm removal. Chem. Eng. Sci. 2012, 80, 109–118. [Google Scholar] [CrossRef]
- Özmen, I. Optimization for coproduction of protease and cellulase from Bacillus subtilis M-11 by the Box–Behnken design and their detergent compatibility. Braz. J. Chem. Eng. 2020, 37, 49–59. [Google Scholar] [CrossRef]
- Naganthran, A.; Masomian, M.; Rahman, R.N.Z.R.A.; Ali, M.S.M.; Nooh, H.M. Improving the Efficiency of New Automatic Dishwashing Detergent Formulation by Addition of Thermostable Lipase, Protease and Amylase. Molecues 2017, 22, 1577. [Google Scholar] [CrossRef] [Green Version]
- Eisazadeh, B.; Mirzajani, F.; Sefidbakht, Y. How is the Effect of Silver Nanoparticles and Lipase/Cellulase Enzymes on Each Other? Iran. J. Sci. Technol. Trans. A Sci. 2020, 44, 27–35. [Google Scholar] [CrossRef]
- Akiba, S.; Kimura, Y.; Yamamoto, K.; Kumagai, H. Purification and characterization of a protease-resistant cellulase from Aspergillus niger. J. Ferment. Bioeng. 1995, 79, 125–130. [Google Scholar] [CrossRef]
- Barnes, A.M.T.; Ballering, K.S.; Leibman, R.S. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. Microbiology 2012, 3, 193–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kean, R.; McKloud, E.; Townsend, E.; Sherry, L.; Delaney, C.; Jones, B.L.; Williams, C.; Ramage, G. The comparative efficacy of antiseptics against Candida auris biofilms. Int. J. Antimicrob. Agents 2018, 52, 673–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayram, P. Enzyme treated CNF biofilms: Characterization. Int. J. Biol. Macromol. 2018, 117, 713–720. [Google Scholar] [CrossRef]
- Song, X.; Ma, Y.; Fu, J.; Zhao, A.; Guo, Z.; Malakar, P.K.; Pan, Y.; Zhao, Y. Effect of temperature on pathogenic and non-pathogenic Vibrio parahaemolyticus biofilm formation. Food Control. 2017, 73, 485–491. [Google Scholar] [CrossRef]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapia-Rodriguez, M.R.; Hernandez-Mendoza, A.; Gonzalez-Aguilar, G.A.; Martinez-Tellez, M.A.; Martins, C.M.; Ayala-Zavala, J.F. Carvacrol as potential quorum sensing inhibitor of Pseudomonas aeruginosa and biofilm production on nonrust steel surfaces. Food Control. 2017, 75, 255–261. [Google Scholar] [CrossRef]
- Cai, L.-L.; Hu, H.-J.; Lu, Q.; Wang, H.-H.; Xu, X.-L.; Zhou, G.-H.; Kang, Z.-L.; Ma, H.-J. Morphophysiological responses of detached and adhered biofilms of Pseudomonas fluorescens to acidic electrolyzed water. Food Microbiol. 2019, 82, 89–98. [Google Scholar] [CrossRef]
- Lekbach, Y.; Li, Z.; Xu, D.; El Abed, S.; Dong, Y.; Liu, D.; Gu, T.; Koraichi, S.I.; Yang, K.; Wang, F. Salvia officinalis extract mitigates the microbiologically influenced corrosion of 304L stainless steel by Pseudomonas aeruginosa biofilm. Bioelectrochemistry 2019, 128, 193–203. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Dong, R.; Ma, L.; Qian, Y.; Liu, Z. Combined Anti-Biofilm Enzymes Strengthen the Eradicate Effect of Vibrio parahaemolyticus Biofilm: Mechanism on cpsA-J Expression and Application on Different Carriers. Foods 2022, 11, 1305. https://doi.org/10.3390/foods11091305
Li Y, Dong R, Ma L, Qian Y, Liu Z. Combined Anti-Biofilm Enzymes Strengthen the Eradicate Effect of Vibrio parahaemolyticus Biofilm: Mechanism on cpsA-J Expression and Application on Different Carriers. Foods. 2022; 11(9):1305. https://doi.org/10.3390/foods11091305
Chicago/Turabian StyleLi, Yuan, Ruyue Dong, Lei Ma, Yilin Qian, and Zunying Liu. 2022. "Combined Anti-Biofilm Enzymes Strengthen the Eradicate Effect of Vibrio parahaemolyticus Biofilm: Mechanism on cpsA-J Expression and Application on Different Carriers" Foods 11, no. 9: 1305. https://doi.org/10.3390/foods11091305
APA StyleLi, Y., Dong, R., Ma, L., Qian, Y., & Liu, Z. (2022). Combined Anti-Biofilm Enzymes Strengthen the Eradicate Effect of Vibrio parahaemolyticus Biofilm: Mechanism on cpsA-J Expression and Application on Different Carriers. Foods, 11(9), 1305. https://doi.org/10.3390/foods11091305





