Isolation, Identification, and Biological Activity Analysis of Swim Bladder Polypeptides from Acipenser schrencki
Abstract
:1. Introduction
2. Materials and Methods
2.1. Raw Materials and Reagents
2.2. Determination of Basic Content
2.3. Enzymatic Digestion and Ultrafiltration
2.4. Determination of Degree of Hydrolysis (DH)
2.5. Antioxidant Assay
2.5.1. DPPH• Scavenging Activity
2.5.2. O2•- Scavenging Activity
2.5.3. •OH Scavenging Activity
2.5.4. Fe3+ Reducing Power
2.6. Analysis of Target Peptides
2.6.1. AA Composition
2.6.2. UV Scanning Spectrum
2.6.3. Molecular Weight Distribution
2.6.4. Peptide Sequence Identification
2.7. Statistical Analysis
3. Results and Discussions
3.1. Basic Components of Swim Bladders
3.2. Optimization of the Enzymatic Process of Fish Bladder
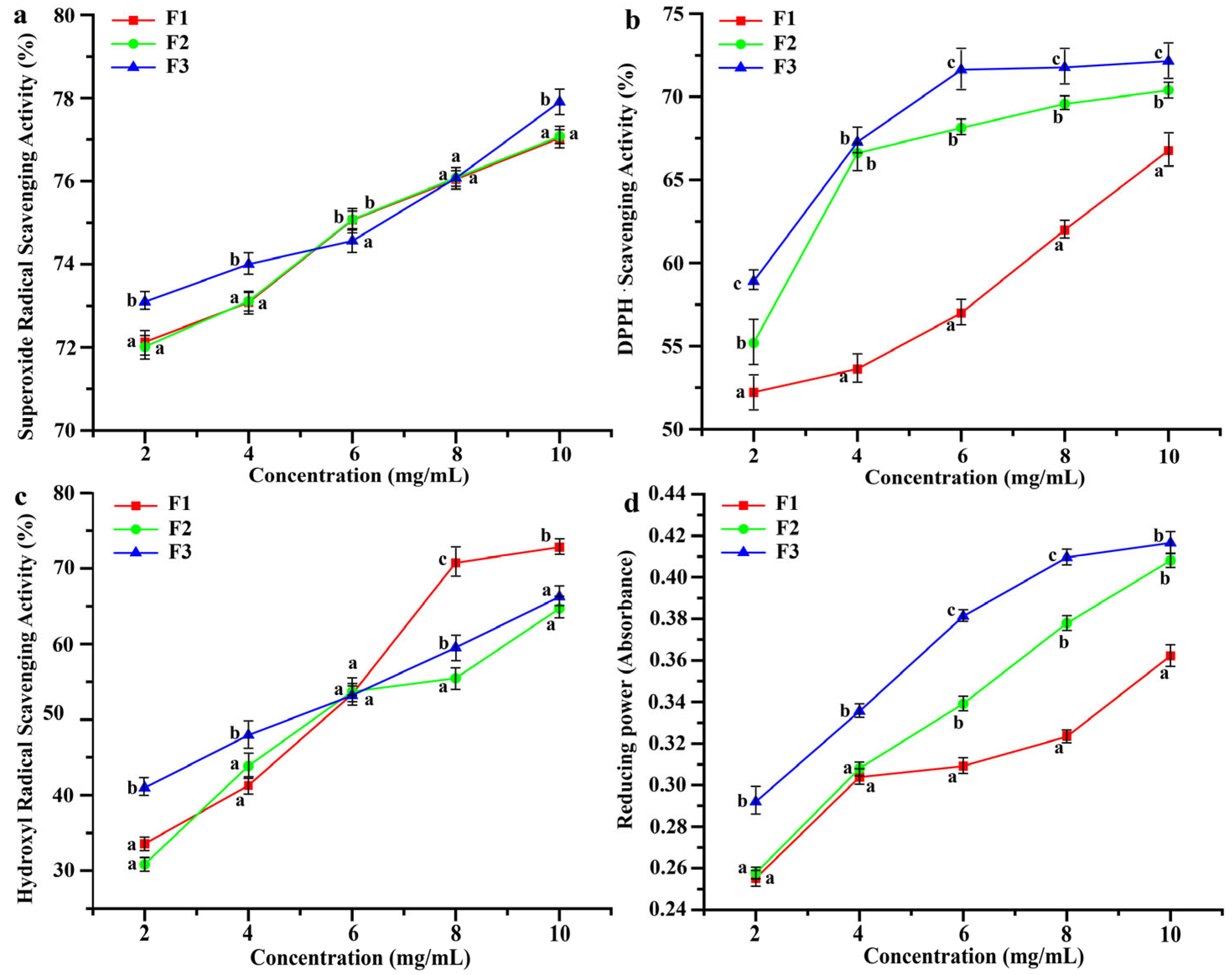
3.3. Antioxidant Properties of Peptides with Different Molecular Weights
3.4. Analysis and Characterization of F3
3.4.1. AA Composition of F3
3.4.2. UV Spectrum of F3
3.4.3. Molecular Weight of F3
3.4.4. Peptide Sequence of F3
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.B.; Zhao, Y.Q.; Wang, Y.M.; Chi, C.F.; Wang, B. Eight Collagen Peptides from Hydrolysate Fraction of Spanish Mackerel Skins: Isolation, Identification, and In Vitro Antioxidant Activity Evaluation. Mar. Drugs 2019, 17, 224. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, R.; Pateiro, M.; Gagaoua, M.; Barba, F.J.; Zhang, W.; Lorenzo, J.M. A Comprehensive Review on Lipid Oxidation in Meat and Meat Products. Antioxidants 2019, 8, 429. [Google Scholar] [CrossRef] [PubMed]
- Acaroz, U.; Ince, S.; Arslan-Acaroz, D.; Gurler, Z.; Demirel, H.H.; Kucukkurt, I.; Eryavuz, A.; Kara, R.; Varol, N.; Zhu, K. Bisphenol-A induced oxidative stress, inflammatory gene expression, and metabolic and histopathological changes in male Wistar albino rats: Protective role of boron. Toxicol. Res. 2019, 8, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef] [PubMed]
- Lv, R.; Dong, Y.; Bao, Z.; Zhang, S.; Lin, S.; Sun, N. Advances in the activity evaluation and cellular regulation pathways of food-derived antioxidant peptides. Trends Food Sci. Technol. 2022, 122, 171–186. [Google Scholar] [CrossRef]
- Gonzalez-Serrano, D.J.; Hadidi, M.; Varcheh, M.; Jelyani, A.Z.; Moreno, A.; Lorenzo, J.M. Bioactive Peptide Fractions from Collagen Hydrolysate of Common Carp Fish Byproduct: Antioxidant and Functional Properties. Antioxidants 2022, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Nurilmala, M.; Hizbullah, H.H.; Karnia, E.; Kusumaningtyas, E.; Ochiai, Y. Characterization and Antioxidant Activity of Collagen, Gelatin, and the Derived Peptides from Yellowfin Tuna (Thunnus albacares) Skin. Mar. Drugs 2020, 18, 98. [Google Scholar] [CrossRef]
- Wang, W.Y.; Zhao, Y.Q.; Zhao, G.X.; Chi, C.F.; Wang, B. Antioxidant Peptides from Collagen Hydrolysate of Redlip Croaker (Pseudosciaena polyactis) Scales: Preparation, Characterization, and Cytoprotective Effects on H2O2-Damaged HepG2 Cells. Mar. Drugs 2020, 18, 156. [Google Scholar] [CrossRef]
- Ding, D.; Du, B.; Zhang, C.; Zaman, F.; Huang, Y. Isolation and identification of an antioxidant collagen peptide from skipjack tuna (Katsuwonus pelamis) bone. RSC Adv. 2019, 9, 27032–27041. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, H.; Yang, F.; Jin, S.; Liu, C.; Zhang, L.; Huang, J.; Wang, S.; Yan, Z.; Cai, X.; et al. Physicochemical, antioxidant properties of giant croaker (Nibea japonica) swim bladders collagen and wound healing evaluation. Int. J. Biol. Macromol. 2019, 138, 483–491. [Google Scholar] [CrossRef]
- Dong, Y.; Dai, Z. Physicochemical, Structural and Antioxidant Properties of Collagens from the Swim Bladder of Four Fish Species. Mar. Drugs 2022, 20, 550. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liang, L.; Regenstein, J.M.; Zhou, P. Extraction and characterisation of pepsin-solubilised collagen from fins, scales, skins, bones and swim bladders of bighead carp (Hypophthalmichthys nobilis). Food Chem. 2012, 133, 1441–1448. [Google Scholar] [CrossRef]
- Cruz-López, H.; Rodríguez-Morales, S.; Enríquez-Paredes, L.M.; Villarreal-Gómez, L.J.; Olivera-Castillo, L.; Cortes-Santiago, Y.; López, L.M. Comparison of collagen characteristic from the skin and swim bladder of Gulf corvina (Cynoscion othonopterus). Tissue Cell 2021, 72, 101593. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Liu, Y.; Adányi, N.; Zhang, X. Effects of waterless live transportation on survivability, physiological responses and flesh quality in Chinese farmed sturgeon (Acipenser schrenckii). Aquaculture 2020, 518, 734834. [Google Scholar] [CrossRef]
- Lopez, A.; Vasconi, M.; Bellagamba, F.; Mentasti, T.; Moretti, V.M. Sturgeon Meat and Caviar Quality from Different Cultured Species. Fishes 2020, 5, 9. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2022; FAO: Rome, Italy, 2022. [Google Scholar]
- Chen, R.; Liu, Z.; Wang, J.; Jin, W.; Abdu, H.I.; Pei, J.; Wang, Q.; El-Aty, A.M.A. A review of the nutritional value and biological activities of sturgeon processed byproducts. Front. Nutr. 2022, 9, 1024309. [Google Scholar] [CrossRef] [PubMed]
- In, A.; Latimer, J.W.; Horwitz, W. Official Methods of Analysis, 16th ed.; AOAC International Press: Washington, DC, USA, 2005. [Google Scholar]
- Wasswa, J.; Tang, J.; Gu, X.; Yuan, X. Influence of the extent of enzymatic hydrolysis on the functional properties of protein hydrolysate from grass carp (Ctenopharyngodon idella) skin. Food Chem. 2007, 104, 1698–1704. [Google Scholar] [CrossRef]
- Teng, D.; Fang, Y.; Song, X.; Gao, Y. Optimization of enzymatic hydrolysis parameters for antioxidant capacity of peptide from goat placenta. Food Bioprod. Process. 2011, 89, 202–208. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, F.; Han, F.; Wang, H. Purification and characterization of antioxidative peptides from salmon protamine hydrolysate. J. Food Biochem. 2008, 32, 654–671. [Google Scholar] [CrossRef]
- Xia, Y.; Bamdad, F.; Gänzle, M.; Chen, L. Fractionation and characterization of antioxidant peptides derived from barley glutelin by enzymatic hydrolysis. Food Chem. 2012, 134, 1509–1518. [Google Scholar] [CrossRef]
- González-Félix, M.L.; Perez-Velazquez, M.; Castellanos-Rico, M.; Sachs, A.M.; Gray, L.D.; Gaines, S.D.; Goto, G.M. First report on the swim bladder index, proximate composition, and fatty acid analysis of swim bladder from cultured Totoaba macdonaldi fed compound aquafeeds. Aquac. Rep. 2021, 21, 100901. [Google Scholar] [CrossRef]
- Martins, E.; Fernandes, R.; Alves, A.L.; Sousa, R.O.; Reis, R.L.; Silva, T.H. Skin Byproducts of Reinhardtius hippoglossoides (Greenland halibut) as Ecosustainable Source of Marine Collagen. Appl. Sci. 2022, 12, 11282. [Google Scholar] [CrossRef]
- Zhao, M.; Li, P.; Zhang, Y.; Xi, J.; Ye, Y.; Du, B. Comparison of Nutritional Quality in Fish Maw Products from Different Sources. Food Res. Dev. 2020, 41, 12–17. [Google Scholar]
- Noman, A.; Xu, Y.; Al-Bukhaiti, W.Q.; Abed, S.M.; Ali, A.H.; Ramadhan, A.H.; Xia, W. Influence of enzymatic hydrolysis conditions on the degree of hydrolysis and functional properties of protein hydrolysate obtained from Chinese sturgeon (Acipenser sinensis) by using papain enzyme. Process Biochem. 2018, 67, 19–28. [Google Scholar] [CrossRef]
- Zhao, W.H.; Luo, Q.B.; Pan, X.; Chi, C.F.; Sun, K.L.; Wang, B. Preparation, identification, and activity evaluation of ten antioxidant peptides from protein hydrolysate of swim bladders of miiuy croaker (Miichthys miiuy). J. Funct. Foods 2018, 47, 503–511. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.M.; Chi, C.F.; Luo, H.Y.; Deng, S.G.; Ma, J.Y. Isolation and characterization of collagen and antioxidant collagen peptides from scales of croceine croaker (Pseudosciaena crocea). Mar. Drugs 2013, 11, 4641–4661. [Google Scholar] [CrossRef]
- Jin, J.E.; Ahn, C.B.; Je, J.Y. Purification and characterization of antioxidant peptides from enzymatically hydrolyzed ark shell (Scapharca subcrenata). Process Biochem. 2018, 72, 170–176. [Google Scholar] [CrossRef]
- Chen, J.; Cui, C.; Zhao, H.; Wang, H.; Zhao, M.; Wang, W.; Dong, K. The effect of high solid concentrations on enzymatic hydrolysis of soya bean protein isolate and antioxidant activity of the resulting hydrolysates. Int. J. Food Sci. Technol. 2018, 53, 954–961. [Google Scholar] [CrossRef]
- Li, N.; Lv, S.; Ma, Y.; Liu, N.; Wang, S.; Zhou, D. In vitro antioxidant and anti-aging properties of swim bladder peptides from Atlantic cod (Gadus morhua). Int. J. Food Prop. 2020, 23, 1416–1429. [Google Scholar] [CrossRef]
- Nagai, T.; Suzuki, N.; Nagashima, T. Collagen from common minke whale (Balaenoptera acutorostrata) unesu. Food Chem. 2008, 111, 296–301. [Google Scholar] [CrossRef]
- Wu, R.; Wu, C.; Liu, D.; Yang, X.; Huang, J.; Zhang, J.; Liao, B.; He, H.; Li, H. Overview of Antioxidant Peptides Derived from Marine Resources: The Sources, Characteristic, Purification, and Evaluation Methods. Appl. Biochem. Biotechnol. 2015, 176, 1815–1833. [Google Scholar] [CrossRef] [PubMed]
- Shazly, A.B.; He, Z.; El-Aziz, M.A.; Zeng, M.; Zhang, S.; Qin, F.; Chen, J. Fractionation and identification of novel antioxidant peptides from buffalo and bovine casein hydrolysates. Food Chem. 2017, 232, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.B.; He, T.P.; Li, H.B.; Tang, H.W.; Xia, E.Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.H.; Chi, C.F.; Zhao, Y.Q.; Wang, B. Preparation, Physicochemical and Antioxidant Properties of Acid- and Pepsin-Soluble Collagens from the Swim Bladders of Miiuy Croaker (Miichthys miiuy). Mar. Drugs 2018, 16, 161. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, D.; Liu, M.; Li, Y.; Lv, R.; Li, X.; Wang, Q.; Ren, D.; Wu, L.; Zhou, H. Identification of Antioxidant Peptides Derived from Tilapia (Oreochromis niloticus) Skin and Their Mechanism of Action by Molecular Docking. Foods 2022, 11, 2576. [Google Scholar] [CrossRef] [PubMed]
- Sila, A.; Bougatef, A. Antioxidant peptides from marine by-products: Isolation, identification and application in food systems. A review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- Najafian, L.; Babji, A.S. Isolation, purification and identification of three novel antioxidative peptides from patin (Pangasius sutchi) myofibrillar protein hydrolysates. LWT Food Sci. Technol. 2015, 60, 452–461. [Google Scholar] [CrossRef]
- Girgih, A.T.; He, R.; Malomo, S.; Offengenden, M.; Wu, J.; Aluko, R.E. Structural and functional characterization of hemp seed (Cannabis sativa L.) protein-derived antioxidant and antihypertensive peptides. J. Funct. Foods 2014, 6, 384–394. [Google Scholar] [CrossRef]
- Ma, R.; Chen, Q.; Dai, Y.; Huang, Y.; Hou, Q.; Huang, Y.; Zhong, K.; Huang, Y.; Gao, H.; Bu, Q. Identification of novel antioxidant peptides from sea squirt (Halocynthia roretzi) and its neuroprotective effect in 6-OHDA-induced neurotoxicity. Food Funct. 2022, 13, 6008–6021. [Google Scholar] [CrossRef]
- Li, Y.; Yu, J. Research progress in structure-activity relationship of bioactive peptides. J. Med. Food 2015, 18, 147–156. [Google Scholar] [CrossRef]
- Alemán, A.; Giménez, B.; Pérez-Santin, E.; Gómez-Guillén, M.C.; Montero, P. Contribution of Leu and Hyp residues to antioxidant and ACE-inhibitory activities of peptide sequences isolated from squid gelatin hydrolysate. Food Chem. 2011, 125, 334–341. [Google Scholar] [CrossRef]


| Solid-Liquid Ratio (g:mL) | Time (h) | Temperature (°C) | Enzyme/Substrate(U/g) |
|---|---|---|---|
| 1:5 | 2 | 40 | 3000 |
| 1:10 | 3 | 45 | 4000 |
| 1:15 | 4 | 50 | 5000 |
| 1:20 | 5 | 55 | 6000 |
| 1:25 | 6 | 60 | 7000 |
| Components | Protein | Moisture | Ash | Fat |
|---|---|---|---|---|
| (%) | 78.61 ± 4.30 | 15.17 ± 0.03 | 0.99 ± 0.02 | 2.97 ± 0.07 |
| AA (%) | F3 |
|---|---|
| HydroPro | 5.28 ± 0.17 |
| Asp | 7.18 ± 0.08 |
| Thr | 3.56 ± 0.13 |
| Ser | 5.52 ± 0.24 |
| Glu | 12.21 ± 0.06 |
| Pro | 6.17 ± 0.15 |
| Gly | 21.57 ± 0.16 |
| Ala | 10.42 ± 0.07 |
| Val | 2.79 ± 0.11 |
| Met | 1.57 ± 0.03 |
| Ile | 2.29 ± 0.04 |
| Leu | 3.96 ± 0.12 |
| Tyr | 1.64 ± 0.07 |
| Phe | 2.62 ± 0.03 |
| Lys | 3.82 ± 0.02 |
| His | 1.29 ± 0.01 |
| Arg | 8.12 ± 0.05 |
| HAA | 51.39 ± 0.29 |
| EAA | 30.02 ± 0.15 |
| Peptide Sequence | Protein ID. | Score | Peptide Ranker a | Potential Bioactive Peptides b | Biological Functions b |
|---|---|---|---|---|---|
| MFGF | A0A444U3D5 | 65.81 | 0.9943 | FGF | Antioxidative |
| GPPGPRGPPGL | A0A444U5J5 | 99.37 | 0.9560 | GPP | Antioxidative |
| GPGGPSGERGPPGPM | A0A444TZY1 | 128.81 | 0.9038 | GPP | Antioxidative |
| FDRPSPPPWAA | A0A444V784 | 107.15 | 0.8992 | PWA, PW | Antioxidative |
| SGPPGFPGSPGPKGE | A0A662YXI2 | 81.33 | 0.8845 | GPP | Antioxidative |
| GLPGPIGPPGPR | A0A444TZY1 | 70.85 | 0.8811 | GPP | Antioxidative |
| FGGRPIPGSPF | A0A444TZH1 | 135.83 | 0.8642 | GGRP | Antioxidative |
| GPRGPPGEPGL | A0A662YUK1 | 101.25 | 0.8567 | GPP | Antioxidative |
| AVPGPPGEPGRL | A0A444UE44 | 68.22 | 0.8412 | GPP | Antioxidative |
| GPPGKDGQPGHPGPIGPA | A0A0S3P5T6 | 179.59 | 0.8358 | KD, GPP | Antioxidative |
| LPLL | A0A444UQF3 | 62.26 | 0.7150 | LPL | Antioxidative |
| PGIPGPEGPR | A0A662YTX1 | 138.26 | 0.7121 | GPE | Antioxidative |
| GIGPEGPHLGIV | A0A444V058 | 143.11 | 0.6989 | HL | Antioxidative |
| AGDDAPRAVFPSIVGRPR | A0A444V306 | 172.93 | 0.6843 | AGDDAPR | Antioxidative |
| SLYPPSEKPIMK | A0A444UV48 | 81.625 | 0.6822 | LY, KP | Antioxidative |
| LLPL | A0A444UQC2 | 62.263 | 0.6800 | LPL | Antioxidative |
| DVVDFPRFPHR | A0A444UZ64 | 100.48 | 0.6657 | PHR | Antioxidative |
| GFAGDDAPRAVFPSIVGRPR | A0A444V306 | 130.27 | 0.6456 | AGDDAPR | Antioxidative |
| LVFL | A0A444UQC2 | 69.29 | 0.6430 | VFL | Antioxidative |
| VFLR | A0A662Z298 | 70.86 | 0.6161 | VFL | Antioxidative |
| GIGPEGPHLGIVQ | A0A444V058 | 102.01 | 0.6093 | HL, PHL, GPE | Antioxidative |
| FRF | A0A444UQC2 | 62.74 | 0.9954 | RF, FR | ACE inhibitor |
| FR | Dipeptidyl peptidase IV inhibitor | ||||
| RF, FR | Dipeptidyl peptidase III inhibitor | ||||
| FPFL | A0A662YSN4 | 60.00 | 0.9930 | FP | ACE inhibitor |
| FP, FL, PF | Dipeptidyl peptidase IV inhibitor | ||||
| FL, PF | Dipeptidyl peptidase III inhibitor | ||||
| FGLF | A0A444UCV7 | 83.87 | 0.9903 | LF, GL, FG | ACE inhibitor |
| GL | Dipeptidyl peptidase IV inhibitor | ||||
| GLF | Immunomodulating | ||||
| GLF | Regulation | ||||
| FPAF | A0A444UQF3 | 83.87 | 0.9898 | FP, AF | ACE inhibitor |
| PA, FP, AF | Dipeptidyl peptidase IV inhibitor | ||||
| GFFGL | A0A662YXS9 | 64.52 | 0.9815 | GF, GL, FG, FF | ACE inhibitor |
| GL, GF, FF | Dipeptidyl peptidase IV inhibitor | ||||
| GF | Dipeptidyl peptidase III inhibitor | ||||
| FPVF | A0A662YZB8 | 69.29 | 0.9749 | VF, FP | ACE inhibitor |
| FP, PV, VF | Dipeptidyl peptidase IV inhibitor | ||||
| VGFF | A0A444UCW2 | 62.26 | 0.9674 | GF, VG, FF | ACE inhibitor |
| GF, VG, FF | Dipeptidyl peptidase IV inhibitor | ||||
| GF | Dipeptidyl peptidase III inhibitor | ||||
| GYGF | A0A444UD25 | 62.26 | 0.9652 | GY, YG, GF | ACE inhibitor |
| GF, GY, YG | Dipeptidyl peptidase IV inhibitor | ||||
| GF, YG | Dipeptidyl peptidase III inhibitor | ||||
| YG | Immunomodulating | ||||
| MFLL | A0A444UFC8 | 62.26 | 0.9636 | MF | ACE inhibitor |
| LL, FL, MF | Dipeptidyl peptidase IV inhibitor | ||||
| FL | Dipeptidyl peptidase III inhibitor | ||||
| LL | Stimulating | ||||
| FLGM | A0A444UQD2 | 83.87 | 0.9620 | GM, LG | ACE inhibitor |
| FL | Dipeptidyl peptidase IV inhibitor | ||||
| FL | Dipeptidyl peptidase III inhibitor | ||||
| GFVF | A0A444UFC8 | 62.26 | 0.9611 | VF, GF | ACE inhibitor |
| GF, VF | Dipeptidyl peptidase IV inhibitor | ||||
| GF | Dipeptidyl peptidase III inhibitor | ||||
| KGMF | A0A662YVD5 | 72.08 | 0.9506 | MF, GM, KG | ACE inhibitor |
| KG, MF | dipeptidyl peptidase IV inhibitor | ||||
| LPGLF | A0A444UIZ4 | 83.66 | 0.9486 | LF, PGL, LPG, GL, PG, LP | ACE inhibitor |
| LP, GL, PG | Dipeptidyl peptidase IV inhibitor | ||||
| GLF | Immunomodulating | ||||
| GLF, PG | Regulating | ||||
| LL | Stimulating | ||||
| PG | Antiamnestic | ||||
| PG | Antithrombotic | ||||
| GLLF | A0A444UQC8 | 62.26 | 0.9433 | LF, GL, LLF | ACE inhibitor |
| LL, GL | Dipeptidyl peptidase IV inhibitor | ||||
| GFGGL | A0A444U3D5 | 97.74 | 0.9319 | GF, GL, FG, GG, FGG | ACE inhibitor |
| GL, GF, GG | Dipeptidyl peptidase IV inhibitor | ||||
| GF | Dipeptidyl peptidase III inhibitor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zu, X.-Y.; Liu, W.-B.; Xiong, G.-Q.; Liao, T.; Li, H.-L. Isolation, Identification, and Biological Activity Analysis of Swim Bladder Polypeptides from Acipenser schrencki. Foods 2023, 12, 1934. https://doi.org/10.3390/foods12101934
Zu X-Y, Liu W-B, Xiong G-Q, Liao T, Li H-L. Isolation, Identification, and Biological Activity Analysis of Swim Bladder Polypeptides from Acipenser schrencki. Foods. 2023; 12(10):1934. https://doi.org/10.3390/foods12101934
Chicago/Turabian StyleZu, Xiao-Yan, Wen-Bo Liu, Guang-Quan Xiong, Tao Liao, and Hai-Lan Li. 2023. "Isolation, Identification, and Biological Activity Analysis of Swim Bladder Polypeptides from Acipenser schrencki" Foods 12, no. 10: 1934. https://doi.org/10.3390/foods12101934
APA StyleZu, X.-Y., Liu, W.-B., Xiong, G.-Q., Liao, T., & Li, H.-L. (2023). Isolation, Identification, and Biological Activity Analysis of Swim Bladder Polypeptides from Acipenser schrencki. Foods, 12(10), 1934. https://doi.org/10.3390/foods12101934






