Stress Adaptation Responses of a Listeria monocytogenes 1/2a Strain via Proteome Profiling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Reference Antisera
2.2. Listeria monocytogenes’ Growth for Proteomic Analyses
2.3. Protein Extraction and Immunoblotting
2.4. Mass Spectrometry (MS) and Data Analysis
2.5. Gene Ontology Analysis
2.6. STRING Analysis
3. Results and Discussion
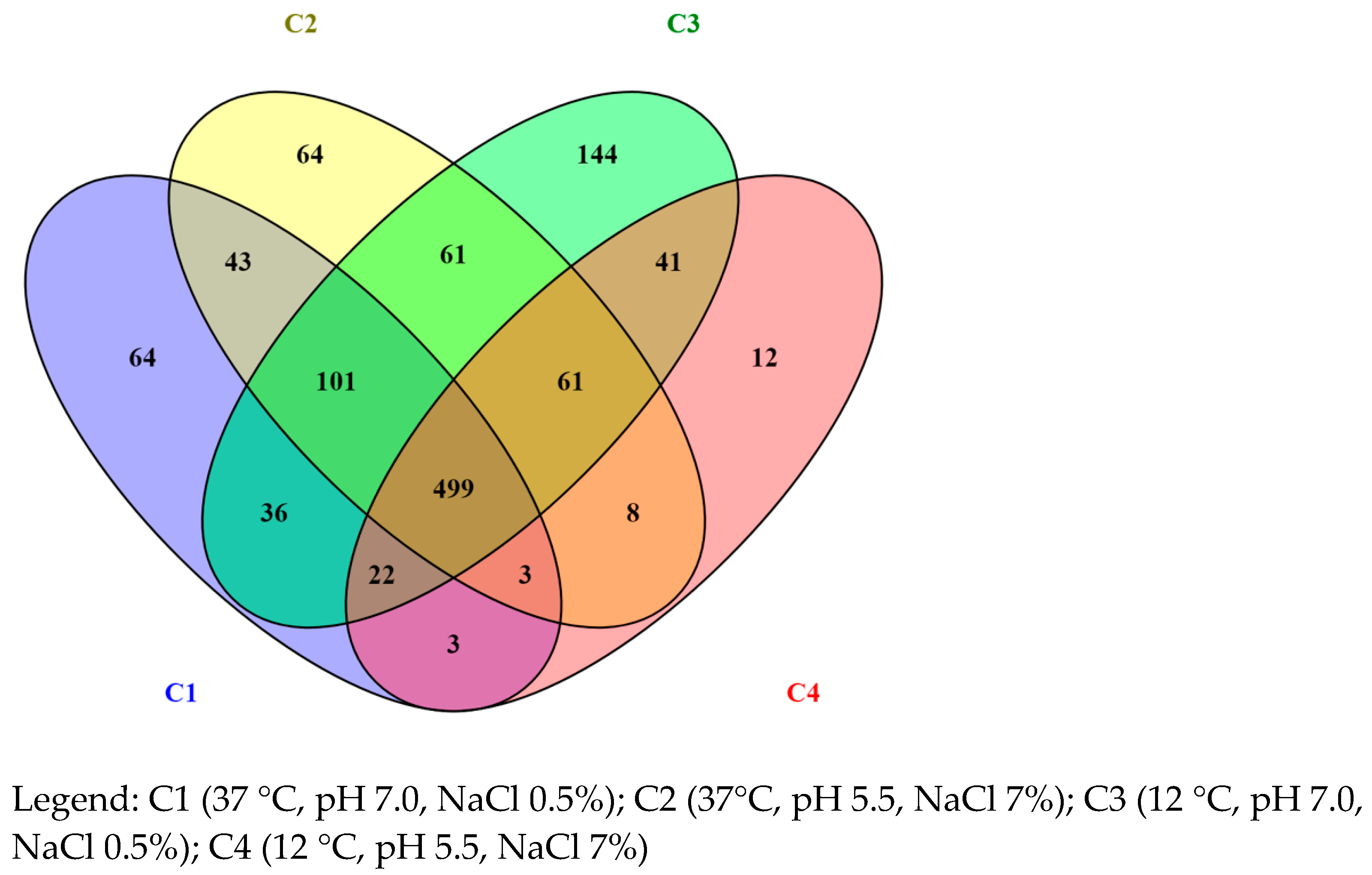
Proteome Profiling Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Valenti, M.; Ranganathan, N.; Moore, L.S.P.; Hughes, S. Listeria monocytogenes infections: Presentation, diagnosis and treatment. Br. J. Hosp. Med. 2021, 82, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Park, Y.J.; Kim, M.; Seo, Y.H.; Kin, Y.A.; Choi, J.Y.; Yong, D.; Jeong, S.H.; Lee, K. Increasing incidence of listeriosis and infection-associated clinical outcomes. Ann. Lab. Med. 2018, 38, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Silk, B.J.; McCoy, M.H.; Iwamoto, M.; Griffin, P.M. Foodborne listeriosis acquired in hospitals. Clin. Infect. Dis. 2014, 59, 532–540. [Google Scholar] [CrossRef]
- Pérez-Baltar, A.; Pérez-Boto, D.; Medina, M.; Montiel, R. Genomic diversity and characterization of Listeria monocytogenes from dry-cured ham processing plants. Food Microbiol. 2021, 99, 103779. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) the European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, 7666.
- Lachmann, R.; Halbedel, S.; Adler, M.; Becker, N.; Allerberger, F.; Holzer, A.; Boone, I.; Falkenhorst, G.; Kleta, S.; Al Dahouk, S.; et al. Nationwide outbreak of invasive listeriosis associated with consumption of meat products in health care facilities, Germany, 2014–2019. Clin. Microbiol. Infect. 2021, 27, 1035.e1–1035.e5. [Google Scholar] [CrossRef]
- National Institute for Communicable Diseases. Situation report on listeriosis outbreak, South Africa, 2017. 2018. Available online: https://www.who.int/csr/don/02-may-2018-listeriosis-south-africa/en/ (accessed on 20 April 2023).
- Li, W.; Bai, L.; Fu, P.; Han, H.; Liu, J.; Guo, Y. The epidemiology of Listeria monocytogenes in China. Foodborne Path. Dis. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- McCollum, J.T.; Cronquist, A.B.; Silk, B.J.; Jackson, K.A.; O’Connor, K.A.; Cosgrove, S.; Gossack, J.P.; Parachini, S.S.; Jain, N.S.; Ettestad, P.; et al. Multistate outbreak of listeriosis associated with cantaloupe. N. Engl. J. Med. 2013, 369, 944–953. [Google Scholar] [CrossRef]
- Huang, Y.-T.; Ko, W.-C.; Chan, Y.-J.; Lu, J.-J.; Tsai, H.-Y.; Liao, C.-H.; Sheng, W.-H.; Teng, L.-J.; Hsueh, P.-R. Molecular characterization of clinical isolates in Taiwan, 2000–2013. PLoS ONE 2015, 10, e0141241. [Google Scholar]
- Chafsey, I.; Ostrowski, R.; Guilbaud, M.; Teixeira, P.; Herry, J.M.; Caccia, N.; Chambon, C.; Hébraud, M.; Azeredo, J.; Bellon-Fontaine, M.N.; et al. Deep impact of the inactivation of the SecA2-only protein export pathway on the proteosurfaceome of Listeria monocytogenes. J. Proteomics 2022, 250, 104388. [Google Scholar] [CrossRef] [PubMed]
- Lakicevic, B.Z.; Den Besten, H.M.W.; De Biase, D. Landscape of stress response and virulence genes among Listeria monocytogenes strains. Front. Microbiol. 2022, 12, 738470. [Google Scholar] [CrossRef] [PubMed]
- Berrang, M.E.; Meinersmann, R.J.; Frank, J.F.; Ladely, S.R. Colonization of a newly constructed commercial chicken further processing plant with Listeria monocytogenes. J. Food Protect. 2010, 73, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.; Alvarez-Ordóñez, A.; Jordan, K. Monitoring occurrence and persistence of Listeria monocytogenes in foods and food processing environments in the Republic of Ireland. Front. Microbiol. 2014, 5, 436. [Google Scholar] [CrossRef] [PubMed]
- Bolocan, A.S.; Nicolau, A.I.; Álvarez-Ordóñez, A.; Borda, D.; Oniciuc, E.A.; Stessl, B.; Gurgu, L.; Wagner, M. Dynamics of Listeria monocytogenes colonisation in a newly opened meat processing facility. Meat Sci. 2016, 113, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Tiensuu, T.; Guerreiro, D.N.; Oliveira, A.H.; O’Byrne, C.; Johansson, J. Flick of a switch: Regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer. Microbiology 2019, 165, 819–833. [Google Scholar] [CrossRef]
- Sibanda, T.; Buys, E.M. Listeria monocytogenes pathogenesis: The role of stress adaptation. Microorganisms 2022, 10, 1522. [Google Scholar] [CrossRef]
- Li, Z.; Pérez-Osorio, A.; Wang, Y.; Eckmann, K.; Glover, W.A.; Allard, M.W.; Brown, E.W.; Chen, Y. Whole genome sequencing analyses of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington State. BMC Microbiol. 2017, 17, 134. [Google Scholar] [CrossRef]
- Cimbalo, A.; Frangiamone, M.; Font, G.; Manyes, L. The importance of transcriptomics and proteomics for studying molecular mechanisms of mycotoxin exposure: A review. Food Chem. Toxicol. 2022, 169, 113396. [Google Scholar] [CrossRef]
- Agregán, R.; Pateiro, M.; Kumar, M.; Franco, D.; Capanoglu, E.; Dhama, K.; Lorenzo, J.M. The potential of proteomics in the study of processed meat products. J. Proteomics 2023, 270, 104744. [Google Scholar] [CrossRef]
- Dessaux, C.; Guerreiro, D.N.; Pucciarelli, M.G.; O’Byrne, C.P.; García-del Portillo, F. Impact of osmotic stress on the phosphorylation and subcellular location of Listeria monocytogenes stressosome proteins. Sci. Rep. 2020, 10, 20837. [Google Scholar] [CrossRef]
- Schaumburg, J.; Diekmann, O.; Hagendorff, P.; Bergmann, S.; Rohde, M.; Hammerschmidt, S.; Jänsch, L.; Wehland, J.; Kärst, U. The cell wall subproteome of Listeria monocytogenes. Proteomics 2004, 4, 2991–3006. [Google Scholar] [CrossRef]
- García-del Portillo, F.; Calvo, E.; D’Orazio, V.; Pucciarelli, M.G. Association of ActA to peptidoglycan revealed by cell wall proteomics of intracellular Listeria monocytogenes. J. Biol. Chem. 2011, 286, 34675–34689. [Google Scholar] [CrossRef]
- Lee, Y.J.; Wang, C. Proteomic analysis reveals the temperature-dependent presence of extracytoplasmic peptidases in the biofilm exoproteome of Listeria monocytogenes EGD-e. J. Microbiol. 2020, 58, 761–771. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, F.; Visciano, P.; Krasteva, I.; Torresi, M.; Tittarelli, M.; Pomilio, F.; Iannetti, L.; Di Febo, T.; Paparella, A.; Schirone, M.; et al. Immunoproteome profiling of Listeria monocytogenes under mild acid and salt stress conditions. Proteomics 2022, 22, e2200082. [Google Scholar] [CrossRef]
- Duranti, A.; Sabbatucci, M.; Blasi, G.; Acciari, V.A.; Ancora, M.; Bella, A.; Busani, L.; Centorame, P.; Cammà, C.; Conti, F.; et al. A severe outbreak of listeriosis in central Italy with a rare pulsotype associated with processed pork products. J. Med. Microbiol. 2018, 67, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; Joseph, B.; Mertins, S.; Ecke, R.; Müller-Altrock, S.; Goebel, W. Overexpression of PrfA leads to growth inhibition of Listeria monocytogenes in glucose-containing culture media by interfering with glucose uptake. J. Bacteriol. 2006, 188, 3887–3901. [Google Scholar] [CrossRef]
- Polidoro, M.; De Biase, D.; Montagnini, B.; Guarrera, L.; Cavallo, S.; Valenti, P.; Stefanini, S.; Chiancone, E. The expression of the dodecameric ferritin in Listeria spp. is induced by iron limitation and stationary growth phase. Gene 2002, 296, 121–128. [Google Scholar] [CrossRef]
- Dussurget, O.; Dumas, E.; Archambaud, C.; Chafsey, I.; Chambon, C.; Hébraud, M.; Cossart, P. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol. Lett. 2005, 250, 253–261. [Google Scholar] [CrossRef]
- Olsen, K.N.; Larsen, M.H.; Gahan, C.G.M.; Kallipolitis, B.; Wolf, X.A.; Rea, R.; Hill, C.; Ingmer, H. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology 2005, 151, 925–933. [Google Scholar] [CrossRef]
- Bellapadrona, G.; Stefanini, S.; Zamparelli, C.; Theil, E.C.; Chiancone, E. Iron translocation into and out of Listeria innocua Dps and size distribution of the protein-enclosed nanomineral are modulated by the electrostatic gradient at the 3-fold “ferritin-like” pores. J. Biol. Chem. 2009, 284, 19101–19109. [Google Scholar] [CrossRef] [PubMed]
- Milecka, D.; Samluk, A.; Wasiak, K.; Krawczyk-Balska, A. An essential role of a ferritin-like protein in acid stress tolerance of Listeria monocytogenes. Arch. Microbiol. 2014, 197, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Theoret, J.R.; Cooper, K.K.; Zekarias, B.; Roland, K.L.; Law, B.F.; Curtiss, R., 3rd; Joens, L.A. The Campylobacter jejuni Dps homologue is important for in vitro biofilm formation and cecal colonization of poultry and may serve as a protective antigen for vaccination. Clin. Vaccine Immunol. 2012, 19, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Muchaamba, F.; Wambui, J.; Stephan, R.; Tasara, T. Cold shock proteins promote nisin tolerance in Listeria monocytogenes through modulation of cell envelope modification responses. Front. Microbiol. 2021, 12, 811939. [Google Scholar] [CrossRef]
- Thedieck, K.; Hain, T.; Mohamed, W.; Tindall, B.J.; Nimtz, M.; Chakraborty, T.; Wehland, J.; Jänsch, L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 2006, 62, 1325–1339. [Google Scholar] [CrossRef] [PubMed]
- Freitag, N.E.; Rong, L.; Portnoy, D.A. Regulation of the prfA transcriptional activator of Listeria monocytogenes: Multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 1993, 61, 2537–2544. [Google Scholar] [CrossRef]
- Thomsen, L.E.; Slutz, S.S.; Tan, M.W.; Ingmer, H. Caenorhabditis elegans is a model host for Listeria monocytogenes. Appl. Environ. Microbiol 2006, 72, 1700–1701. [Google Scholar] [CrossRef]
- Andersson, C.; Gripenland, J.; Johansson, J. Using the chicken embryo to assess virulence of Listeria monocytogenes and to model other microbial infections. Nat. Protoc. 2015, 10, 1155–1164. [Google Scholar] [CrossRef]
- Kulén, M.; Lindgren, M.; Hansen, S.; Cairns, A.G.; Grundström, C.; Begum, A.; van der Lingen, I.; Brännström, K.; Hall, M.; Sauer, U.H.; et al. Structure-based design of inhibitors targeting PrfA, the master virulence regulator of Listeria monocytogenes. J. Med. Chem. 2018, 61, 4165–4175. [Google Scholar] [CrossRef]
- Mains, D.R.; Eallonardo, S.J.; Freitag, N.E. Identification of Listeria monocytogenes genes contributing to oxidative stress resistance under conditions relevant to host infection. Infect. Immun. 2021, 89, e00700-20. [Google Scholar] [CrossRef]
- Bowman, J.P.; Chang, K.J.L.; Pinfold, T.; Ross, T. Transcriptomic and phenotypic responses of Listeria monocytogenes strains possessing different growth efficiencies under acidic conditions. Appl. Environ. Microbiol. 2010, 76, 4836–4850. [Google Scholar] [CrossRef] [PubMed]
- Gründling, A.; Burrack, L.S.; Bouwer, H.G.A.; Higgins, D.E. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 2004, 101, 12318–12323. [Google Scholar] [CrossRef]
- Alves, Â.; Magalhães, R.; Brandão, T.R.S.; Pimentel, L.; Rodríguez-Alcalá, L.M.; Teixeira, P.; Ferreira, V. Impact of exposure to cold and cold-osmotic stresses on virulence-associated characteristics of Listeria monocytogenes strains. Food Microbiol. 2020, 87, 103351. [Google Scholar] [CrossRef] [PubMed]
- van der Veen, S.; Abee, T. Contribution of Listeria monocytogenes RecA to acid and bile survival and invasion of human intestinal Caco-2 cells. Int. J. Med. Microbiol. 2011, 301, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Lenhart, J.S.; Schroeder, J.W.; Walsh, B.W.; Simmons, L.A. DNA repair and genome maintenance in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 2012, 76, 530–564. [Google Scholar] [CrossRef]
- Ojha, D.; Greeshma, M.V.; Patil, K.N. Expression, purification and biochemical characterization of Listeria monocytogenes single stranded DNA binding protein 1. Protein Expr. Purif. 2019, 161, 63–69. [Google Scholar] [CrossRef]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef]
- Mérino, D.; Réglier-Poupet, H.; Berche, P.; The European Listeria Genome Consortium; Charbit, A. A hypermutator phenotype attenuates the virulence of Listeria monocytogenes in a mouse model. Mol. Microbiol. 2002, 44, 877–887. [Google Scholar] [CrossRef]
- Markkula, A.; Mattila, M.; Lindström, M.; Korkeala, H. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3°C. Env. Microbiol. Rep. 2012, 14, 2223–2232. [Google Scholar] [CrossRef]
- Bäreclev, C.; Vaitkevicius, K.; Netterling, S.; Johansson, J. DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression. RNA Biol. 2014, 11, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.C.; Raengpradub, S.; Boor, K.J.; Wiedmann, M. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 2007, 73, 6484–6498. [Google Scholar] [CrossRef]
- van der Veen, S.; Abee, T. Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl. Environ. Microbiol. 2010, 76, 7854–7860. [Google Scholar] [CrossRef] [PubMed]
- Duche, O.; Trémoulet, F.; Glaser, P.; Labadie, J. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 2002, 68, 1491–1498. [Google Scholar] [CrossRef]
- Duche, O.; Trémoulet, F.; Namane, A.; Labadie, J. A proteomic analysis of the salt stress response of Listeria monocytogenes. FEMS Microbiol. Lett. 2002, 215, 183–188. [Google Scholar] [CrossRef]
- Chastanet, A.; Derre, I.; Nair, S.; Msadek, T. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 2004, 186, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Wonderling, L.D.; Wilkinson, B.J.; Bayles, D.O. The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 2004, 70, 1935–1943. [Google Scholar] [CrossRef]
- van der Veen, S.; Hain, T.; Wouters, J.A.; Hossain, H.; de Vos, W.M.; Abee, T.; Chakraborty, T.; Wells-Bennik, M.H. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 2007, 153, 3593–3607. [Google Scholar] [CrossRef]
- Chassaing, D.; Auvray, F. The lmo1078 gene encoding a putative UDP-glucose pyrophosphorylase is involved in growth of Listeria monocytogenes at low temperature. FEMS Microbiol. Lett. 2007, 275, 31–37. [Google Scholar] [CrossRef]
- Balogh, D.; Eckel, K.; Fetzer, C.; Sieber, S.A. Listeria monocytogenes utilizes the ClpP1/2 proteolytic machinery for fine-tuned substrate degradation at elevated temperatures. RSC Chem. Biol. 2022, 3, 955–971. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, Y.; Qiu, X.; Wang, G.; Hu, Z.; Chen, S.; Wu, Z.; Yuan, N.; Gao, H.; Wang, J.; et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat. Immunol. 2019, 20, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, I.; Favini-Stabile, S.; Dessen, A. Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci. 2014, 23, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Manso, B.; Melero, B.; Stessl, B.; Jaime, I.; Wagner, M.; Rovira, J.; Rodríguez-Lázaro, D. The response to oxidative stress in Listeria monocytogenes is temperature dependent. Microorganisms 2020, 8, 521. [Google Scholar] [CrossRef]
- Pinilla, C.M.B.; Stincone, P.; Brandelli, A. Proteomic dataset of Listeria monocytogenes exposed to sublethal concentrations of free and nanoencapsulated nisin. Data Brief 2022, 43, 108343. [Google Scholar] [CrossRef]
- Yan, M.Y.; Yan, H.Q.; Ren, G.X.; Zhao, J.P.; Guo, X.P.; Sun, Y.C. CRISPR-Cas12a-assisted recombineering in bacteria. Appl. Environ. Microbiol. 2017, 83, e00947-17. [Google Scholar] [CrossRef] [PubMed]

| Gene | Entry | Protein Name | Length (AA) | Condition |
|---|---|---|---|---|
| Pathogenesis | ||||
| eno | P64074 | Enolase | 430 | C1, C2, C3, C4 |
| dps | Q8Y8G1 | DNA protection during starvation protein | 156 | C1, C2, C3, C4 |
| dltA | Q8Y8D4 | D-alanine--D-alanyl carrier protein ligase | 510 | C2, C3, C4 |
| prfA | P22262 | Listeriolysin regulatory protein | 237 | C2 |
| mogR | P0DJO8 | Motility gene repressor MogR | 306 | C3 |
| inlA | P0DJM0 | Internalin A | 800 | C4 |
| Response to stress | ||||
| ssb1 | Q8YAR8 | Single-stranded DNA-binding protein | 178 | C1, C2, C3, C4 |
| radA | Q48761 | DNA repair protein | 457 | C1, C2, C3, C4 |
| mutS2 | Q8Y7P1 | Endonuclease MutS2 | 785 | C1, C2, C3, C4 |
| recA | P0DJP0 | Protein RecA | 348 | C1, C2, C3, C4 |
| mutS | Q8Y789 | DNA mismatch repair protein MutS | 860 | C1, C2, C3, C4 |
| mutL | Q8Y788 | DNA mismatch repair protein MutL | 601 | C1, C2, C3, C4 |
| cshB | Q8Y755 | DEAD-box ATP-dependent RNA helicase CshB | 435 | C1, C2, C3, C4 |
| dnaJ | P0DJM1 | Chaperone protein DnaJ | 377 | C1, C2, C3, C4 |
| dnaK | P0DJM2 | Chaperone protein DnaK | 613 | C1, C2, C3, C4 |
| ruvB | Q8Y6Z8 | Holliday junction ATP-dependent DNA helicase RuvB | 335 | C1, C2, C3, C4 |
| clpB | Q8Y570 | Chaperone protein ClpB | 866 | C1, C2, C3, C4 |
| addA | Q8Y511 | ATP-dependent helicase/nuclease subunit A | 1235 | C1, C2, C3, C4 |
| addB | Q8Y510 | ATP-dependent helicase/deoxyribonuclease subunit B | 1157 | C1, C2, C3, C4 |
| trxB | O32823 | Thioredoxin reductase | 319 | C1, C2, C3, C4 |
| uvrA | Q8Y4F6 | UvrABC system protein A | 956 | C1, C2, C3, C4 |
| uvrB | Q8Y4F5 | UvrABC system protein A | 658 | C1, C2, C3, C4 |
| ruvA | Q8Y6Z7 | Holliday junction ATP-dependent DNA helicase RuvA | 201 | C1, C2, C3 |
| recF | Q8YAV8 | DNA replication and repair protein RecF | 370 | C2, C3, C4 |
| uvrC | Q8Y7P0 | UvrABC system protein C | 603 | C2, C3, C4 |
| lexA | Q8Y7H7 | LexA repressor | 204 | C1, C3, C4 |
| msrB | Q8Y641 | Peptide methionine sulfoxide reductase MsrB | 145 | C2 |
| ppaX | Q8Y4G3 | Pyrophosphatase PpaX | 217 | C3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Onofrio, F.; Schirone, M.; Paparella, A.; Krasteva, I.; Tittarelli, M.; Pomilio, F.; Iannetti, L.; D’Alterio, N.; Luciani, M. Stress Adaptation Responses of a Listeria monocytogenes 1/2a Strain via Proteome Profiling. Foods 2023, 12, 2166. https://doi.org/10.3390/foods12112166
D’Onofrio F, Schirone M, Paparella A, Krasteva I, Tittarelli M, Pomilio F, Iannetti L, D’Alterio N, Luciani M. Stress Adaptation Responses of a Listeria monocytogenes 1/2a Strain via Proteome Profiling. Foods. 2023; 12(11):2166. https://doi.org/10.3390/foods12112166
Chicago/Turabian StyleD’Onofrio, Federica, Maria Schirone, Antonello Paparella, Ivanka Krasteva, Manuela Tittarelli, Francesco Pomilio, Luigi Iannetti, Nicola D’Alterio, and Mirella Luciani. 2023. "Stress Adaptation Responses of a Listeria monocytogenes 1/2a Strain via Proteome Profiling" Foods 12, no. 11: 2166. https://doi.org/10.3390/foods12112166
APA StyleD’Onofrio, F., Schirone, M., Paparella, A., Krasteva, I., Tittarelli, M., Pomilio, F., Iannetti, L., D’Alterio, N., & Luciani, M. (2023). Stress Adaptation Responses of a Listeria monocytogenes 1/2a Strain via Proteome Profiling. Foods, 12(11), 2166. https://doi.org/10.3390/foods12112166







