Identification of Tartary Buckwheat (Fagopyrum tataricum (L.) Gaertn) and Common Buckwheat (Fagopyrum esculentum Moench) Using Gas Chromatography–Mass Spectroscopy-Based Untargeted Metabolomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction Methods
2.3. GC-MS Analysis
2.4. PCA Analysis
2.5. PLS-DA Analysis
2.6. Volcano Diagram Analysis
2.7. Heatmap Analysis
2.8. Differential Metabolite Annotation and Pathway Analysis
3. Results and Discussion
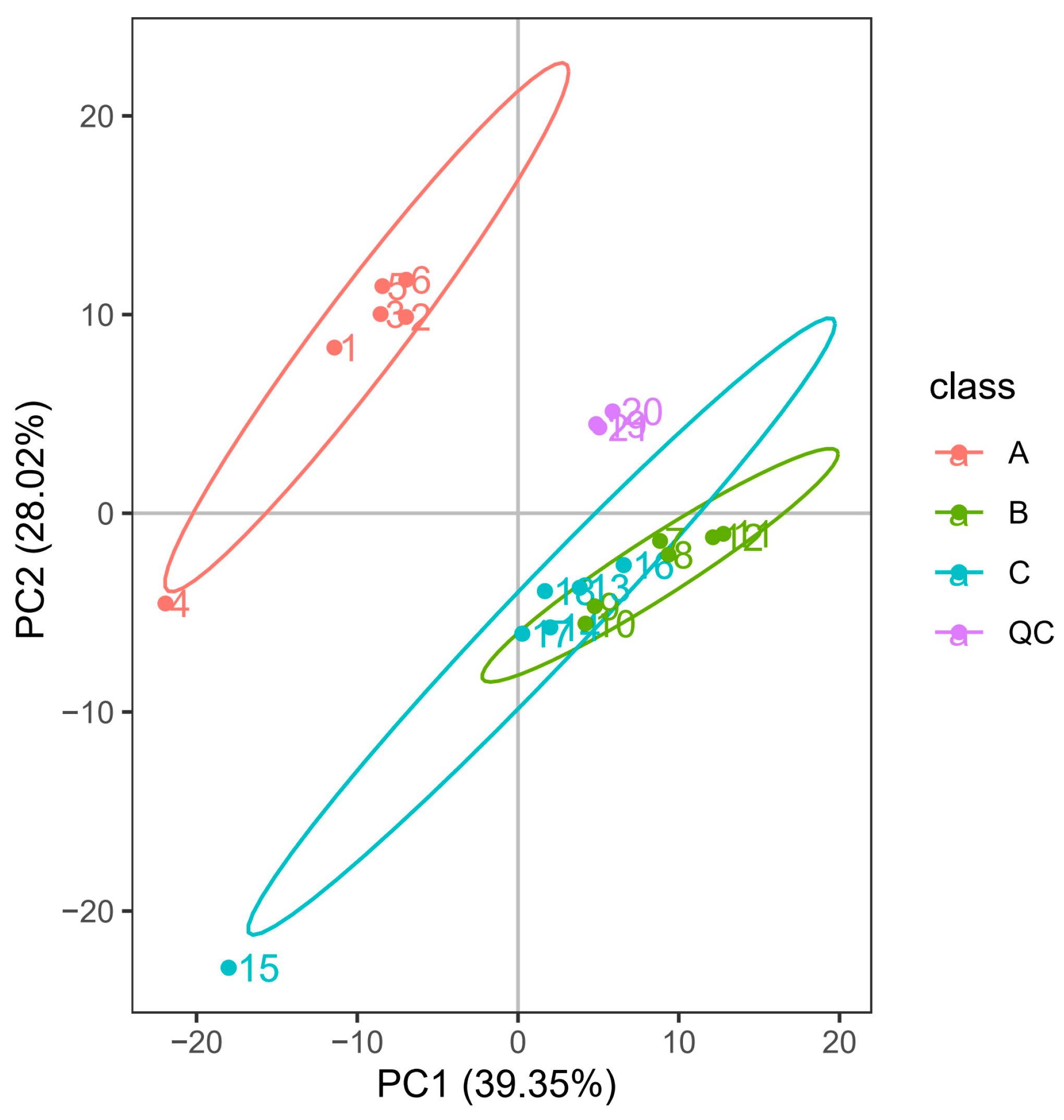
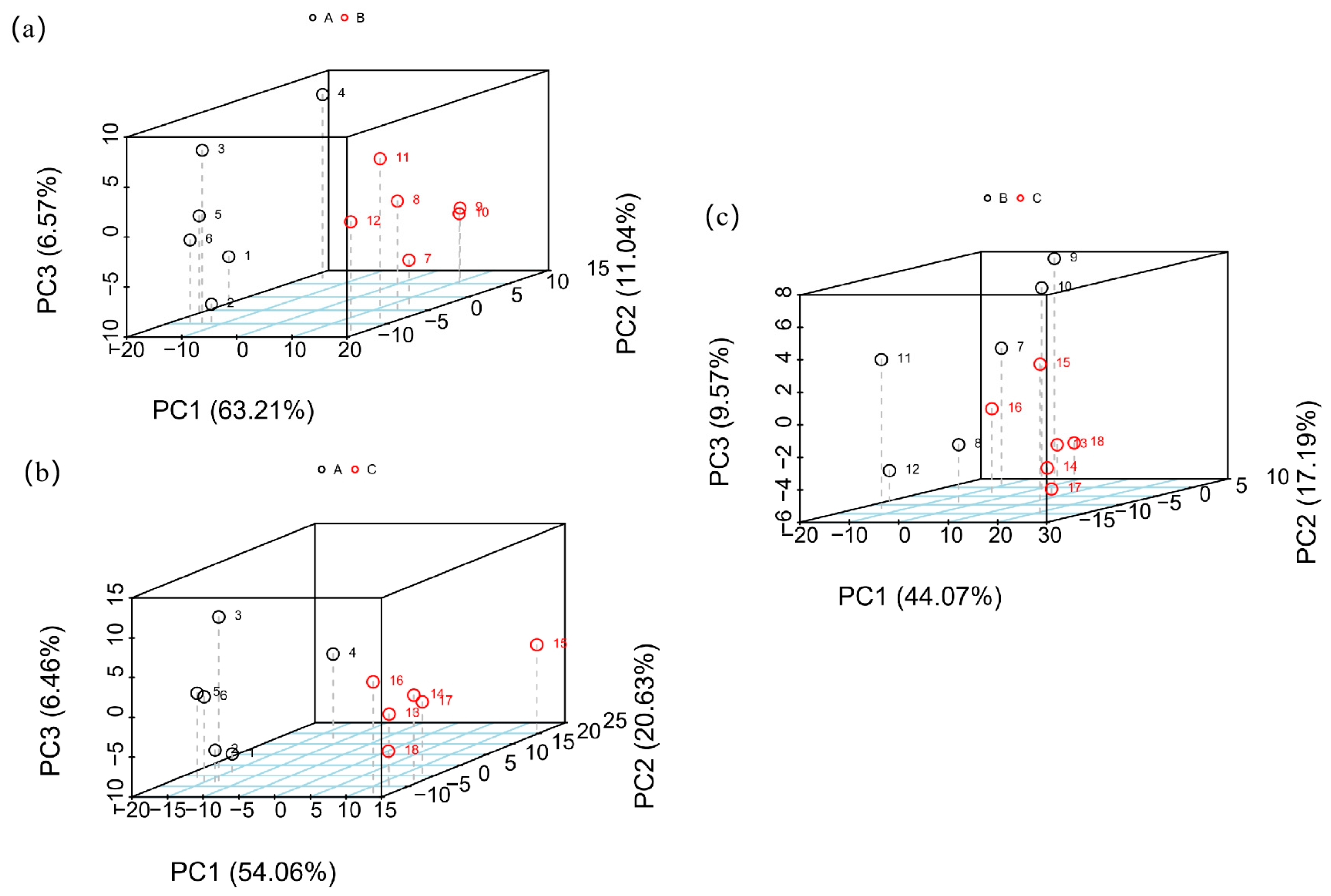
3.1. PCA
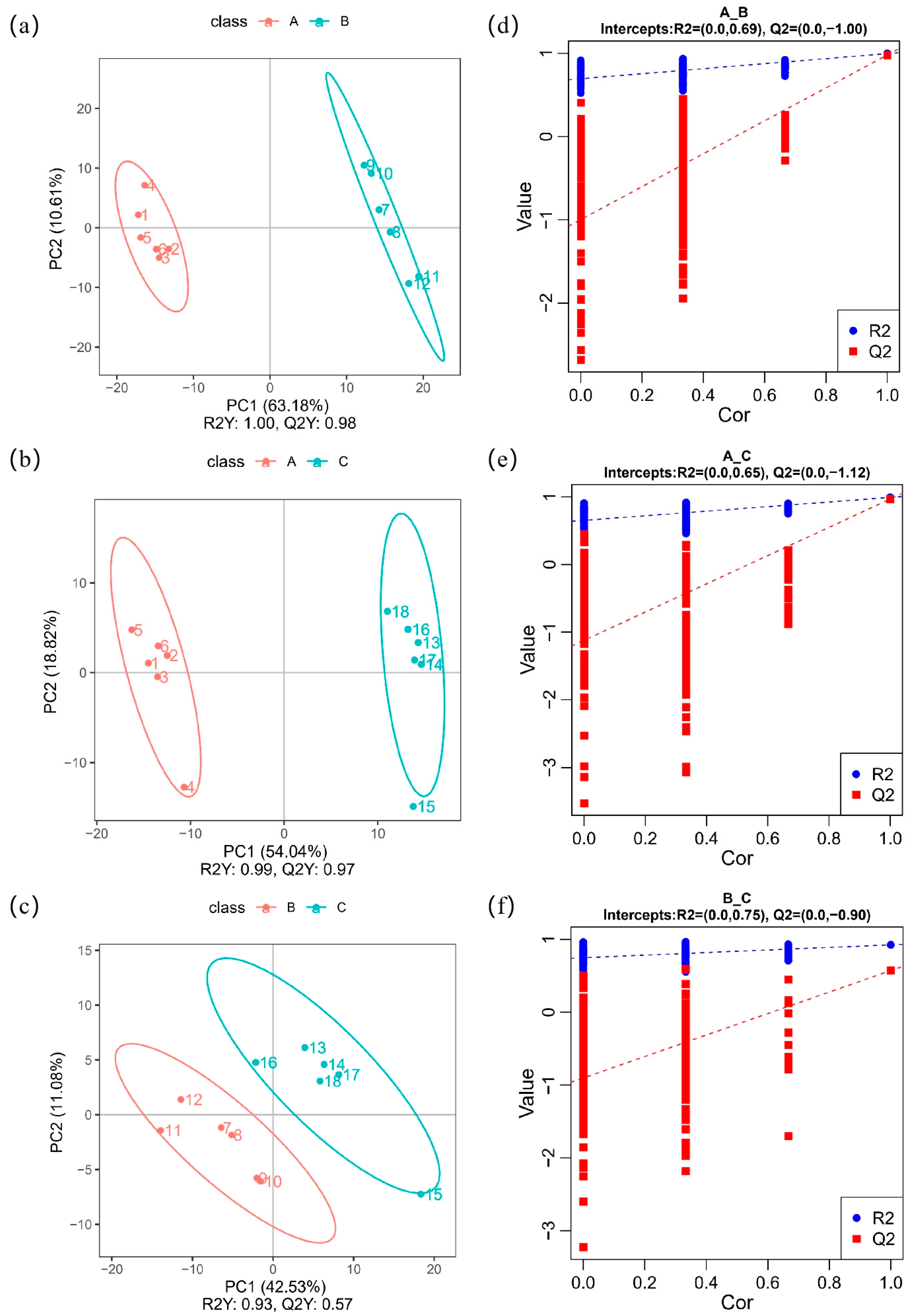
3.2. PLS-DA Analysis
3.3. Volcano Diagram Analysis
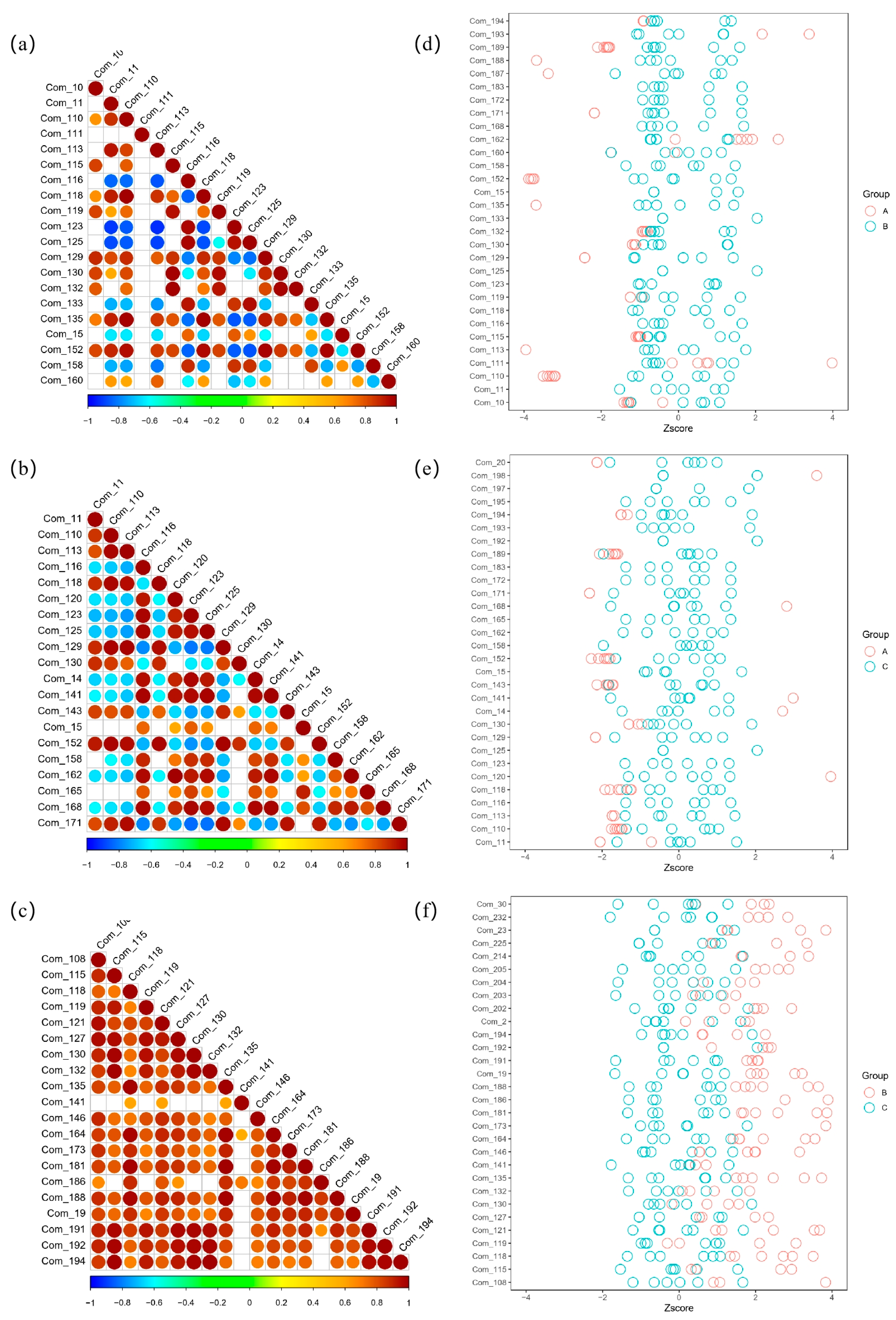
3.4. Heatmap Analysis
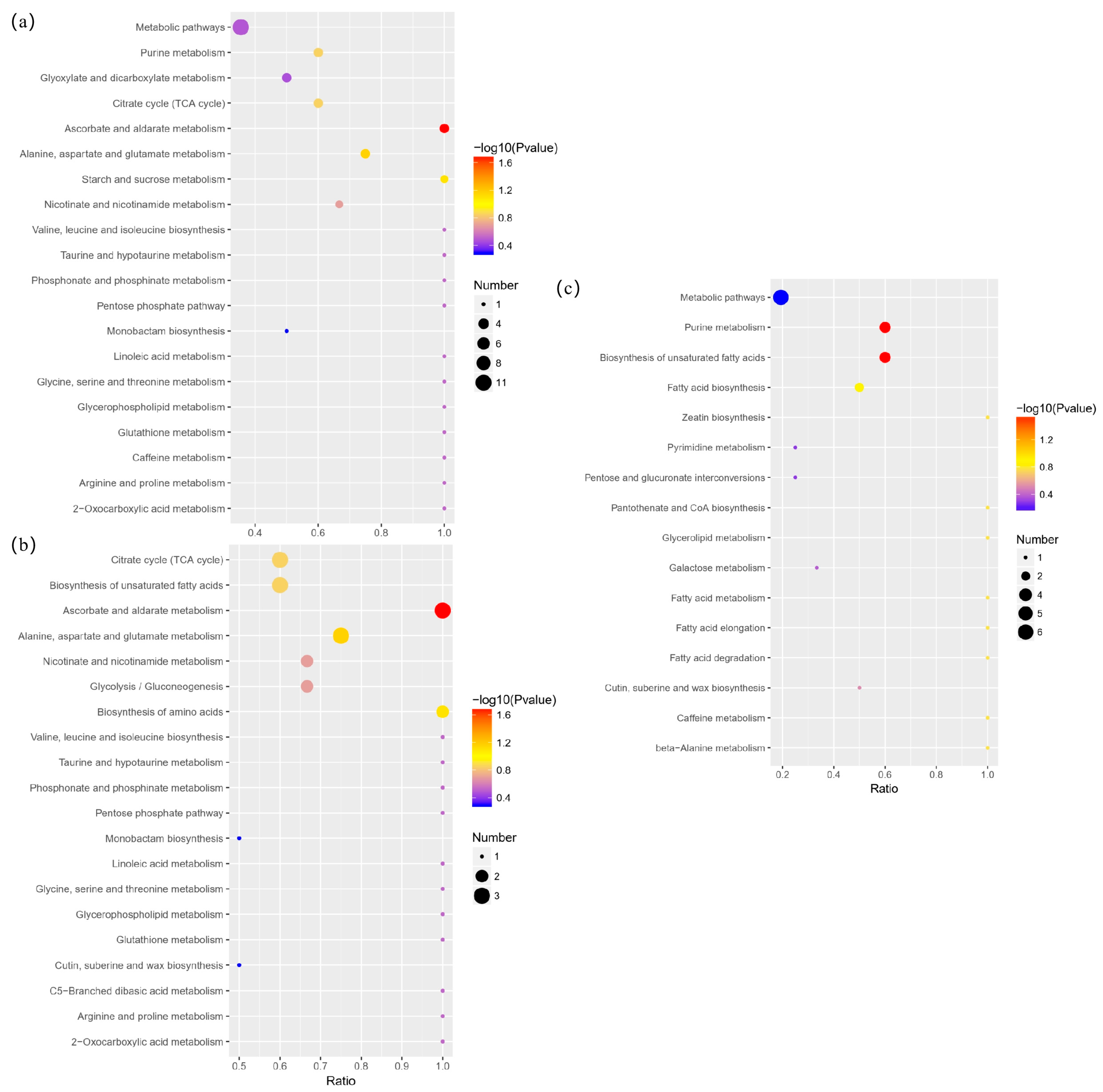
3.5. Differential Metabolite Annotation and Pathway Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sturza, A.; Paucean, A.; Chis, M.S.; Muresan, V.; Vodnar, D.C.; Man, S.M.; Urcan, A.C.; Rusu, I.E.; Fostoc, G.; Muste, S. Influence of buckwheat and buckwheat sprouts flours on the nutritional and textural parameters of wheat buns. Appl. Sci. 2020, 10, 7969. [Google Scholar] [CrossRef]
- Li, Z.; Ren, Z.; Zhao, L.; Chen, L.; Yu, Y.; Wang, D.; Mao, X.; Cao, G.; Zhao, Z.; Yang, H. Unique roles in health promotion of dietary flavonoids through gut microbiota regulation: Current understanding and future perspectives. Food Chem. 2022, 399, 133959. [Google Scholar] [CrossRef]
- Fabjan, N.; Rode, J.; Košir, I.J.; Wang, Z.; Zhang, Z.; Kreft, I. Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J. Agric. Food Chem. 2003, 51, 6452–6455. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.L.; Wang, L.; Li, W.X.; Xing, Y.G.; Zhang, P.; Wang, Q.; Li, H.; Liu, H.; Yang, H.; Liu, X.C.; et al. Scented Tartary buckwheat tea: Aroma components and antioxidant activity. Molecules 2019, 24, 4368. [Google Scholar] [CrossRef]
- Noda, T.; Ishiguro, K.; Suzuki, T.; Morishita, T. Roasted Tartary buckwheat bran as a material for producing rutin-rich tea beverages. Plants 2021, 10, 2662. [Google Scholar] [CrossRef]
- Li, Y.Q.; Zhou, F.C.; Gao, F.; Bian, J.S.; Shan, F. Comparative evaluation of quercetin, isoquercetin and rutin as inhibitors of α-glucosidase. J. Agric. Food Chem. 2009, 57, 11463–11468. [Google Scholar] [CrossRef]
- Li, H.Y.; Lv, Q.Y.; Liu, A.; Wang, J.R.; Sun, X.Q.; Deng, J.; Chen, Q.F.; Wu, Q. Comparative metabolomics study of Tartary (Fagopyrum tataricum (L.) Gaertn) and common (Fagopyrum esculentum Moench) buckwheat seeds. Food Chem. 2022, 371, 131125. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kang, H.B.; Lee, Y.M.; Gwak, Y.S.; Kim, H.Y. Development and validation of a multiplex real-time PCR assay for accurate authentication of common buckwheat (Fagopyrum esculentum) and tartary buckwheat (Fagopyrum tataricum) in food. Food Control 2023, 145, 109442. [Google Scholar] [CrossRef]
- Sinkovič, L.; Ogrinc, N.; Potočnik, D.; Meglič, V. Isotope Fingerprints of Common and Tartary Buckwheat Grains and Milling Fractions: A Preliminary Study. Foods 2022, 11, 1414. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhuang, L.; Zhi, X.; Du, B. Application of NIR analysis technology in quality control of tartary buckwheat products. In Advances in Graphic Communication, Printing and Packaging: Proceedings of 2018 9th China Academic Conference on Printing and Packaging; Springer: Singapore, 2019; pp. 1022–1030. [Google Scholar]
- Lankadurai, B.P.; Nagato, E.G.; Simpson, M.J. Environmental metabolomics: An emerging approach to study organism responses to environmental stressor. Environ. Rev. 2013, 21, 180–205. [Google Scholar] [CrossRef]
- Zhao, Q.; Xi, J.; Xu, D.; Jin, Y.; Wu, F.; Tong, Q.; Yin, Y.; Xu, X. A comparative HS-SPME/GC-MS-based metabolomics approach for discriminating selected japonica rice varieties from different regions of China in raw and cooked form. Food Chem. 2022, 385, 132701. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Chen, S.; Zhou, Z. Age-dependent characterization of volatile organic compounds and age discrimination in Chinese rice wine using an untargeted GC/MS-based metabolomic approach. Food Chem. 2020, 385, 126900. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tan, M.; Deng, H.; Yang, X.; Yu, Y.; Zhou, D.; Dong, H. Geographical origin differentiation of rice by LC–MS-based non-targeted metabolomics. Foods 2022, 11, 3318. [Google Scholar] [CrossRef] [PubMed]
- Janes, D.; Prosen, H.; Kreft, S. Identification and quantification of aroma compounds of tartary buckwheat (fagopyrum tataricum gaertn.) and some of its milling fractions. J. Food Sci. 2012, 77, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Pang, X.L.; Liu, H.H.; Zou, H.; Liao, X.J. Volatiles identification of pomelo based on GC-MS-O and Multivariate Statistical Analysis. J. Chin. Inst. Food Sci. Technol. 2020, 20, 283–292. [Google Scholar]
- Dong, H.; Zhao, X.; Cai, M.; Gu, H.; E, H.; Li, X.; Zhang, Y.; Lu, H.; Zhou, C. Metabolomics analysis of Morchella sp. from different geographical origins of China using UPLC-Q-TOF-MS. Front. Nutr. 2022, 9, 865531. [Google Scholar] [CrossRef]
- Gad, H.A.; Mamadalieva, N.Z.; Bohmdorfer, S.; Rosenau, T.; Zengin, G.; Mamadalieva, R.Z.; Al Musayeib, N.M.; Ashour, M.L. GC-MS Based identification of the volatile components of six astragalus species from uzbekistan and their biological activity. Plants 2021, 10, 124. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.C.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.Y.; Lee, B.L.; et al. HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Res. 2022, 50, 622–631. [Google Scholar] [CrossRef]
- Mitchell, J.M.; Flight, R.M.; Moseley, H.N.B. Deriving lipid classification based on molecular formulas. Metabolites 2020, 10, 122. [Google Scholar] [CrossRef]
- Cotter, D.; Maer, A.; Guda, C.; Saunders, B.; Subramaniam, S. LMPD: LIPID MAPS proteome database. Nucleic Acids Res. 2006, 34, 507–510. [Google Scholar] [CrossRef]
- Merrill, A.H.; Dennis, E.A.; McDonald, J.G.; Fahy, E. Lipidomics technologies at the end of the first decade and the beginning of the next. Adv. Nutr. 2013, 4, 565–567. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Boulesteix, A.L.; Strimmer, K. Partial least squares: A versatile tool for the analysis of high-dimensional genomic data. Brief. Bioinform. 2007, 8, 32–44. [Google Scholar] [CrossRef]
- Kieffer, D.A.; Piccolo, B.D.; Vaziri, N.D.; Liu, S.M.; Lau, W.L.; Khazaeli, M.; Nazertehrani, S.; Moore, M.E.; Marco, M.L.; Martin, R.J.; et al. Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats. Am. J. Physiol. Ren. Physiol. 2016, 310, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Heischmann, S.; Quinn, K.; Cruickshank-Quinn, C.; Liang, L.P.; Reisdorph, R.; Reisdorph, N.; Patel, M. Exploratory metabolomics profiling in the kainic acid rat model reveals depletion of 25-hydroxyvitamin D3 during epileptogenesis. Sci. Rep. 2016, 6, 31424. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Song, J.; Ma, Y.; Yu, Y.; He, X.; Guo, Y.; Dou, J.; Dong, H. Identification of aged-rice adulteration based on near-infrared spectroscopy combined with partial least squares regression and characteristic wavelength variables. Food Chem. X 2023, 17, 100539. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.; dos Santos, A.M.P.; Ferreira, M.M.C.; Ferreira, S.L.C.; Nepomuceno, A.F.S.F. The use of ANOVA-PCA and DD-SIMCA in the development of corn flour laboratory reference materials. Food Chem. 2022, 367, 130748. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Wang, J.; Ma, H.; Wang, S.; Luo, L.; Wang, S. Effect of microwave radiation on antioxidant capacities of Tartary buckwheat sprouts. J. Food Sci. Technol. 2020, 57, 3913–3919. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, Q.; Li, J.; Xiong, J.; Zhou, L.; He, S.; Zhang, J.; Chen, Z.; He, S.; Liu, H. Effects of exogenous sulfur on alleviating cadmium stress in tartary buckwheat. Sci. Rep. 2019, 9, 7397. [Google Scholar] [CrossRef]
- Ma, H.; Xu, X.; Wang, S.; Wang, J.; Wang, S. Effects of microwave irradiation of Fagopyrum tataricum seeds on the physicochemical and functional attributes of sprouts. LWT 2022, 165, 113738. [Google Scholar] [CrossRef]
- Ma, W.; Kim, J.K.; Jia, C.; Yin, F.; Kim, H.J.; Akram, W.; Hu, X.; Li, X. Comparative transcriptome and metabolic profiling analysis of buckwheat (Fagopyrum Tataricum (L.) Gaertn.) under salinity stress. Metabolites 2019, 9, 225. [Google Scholar] [CrossRef] [PubMed]



| Sample Number | Group Name | Buckwheat Type | Region |
|---|---|---|---|
| 1 | A | Common buckwheat | Kunming, Yunnan |
| 2 | Common buckwheat | Bijie, Guizhou | |
| 3 | Common buckwheat | Bijie, Guizhou | |
| 4 | Common buckwheat | Yilan County, Harbin, Heilongjiang | |
| 5 | Common buckwheat | Zhaodong City, Suihua, Heilongjiang | |
| 6 | Common buckwheat | Jining, Shandong | |
| 7 | B | Tartary buckwheat | Zhaojue County, Liangshan, Sichuan |
| 8 | Tartary buckwheat | Zhaojue County, Liangshan, Sichuan | |
| 9 | Tartary buckwheat | Zhaojue County, Liangshan, Sichuan | |
| 10 | Tartary buckwheat | Zhaojue County, Liangshan, Sichuan | |
| 11 | Tartary buckwheat | Xichang County, Liangshan, Sichuan | |
| 12 | Tartary buckwheat | Xichang County, Liangshan, Sichuan | |
| 13 | C | Tartary buckwheat | Qujing, Yunnan |
| 14 | Tartary buckwheat | Qujing, Yunnan | |
| 15 | Tartary buckwheat | Qujing, Yunnan | |
| 16 | Tartary buckwheat | Qujing, Yunnan | |
| 17 | Tartary buckwheat | Zhaotong, Yunnan | |
| 18 | Tartary buckwheat | Zhaotong, Yunnan |
| Group | Number of Differential Metabolites | Up | Down |
|---|---|---|---|
| A vs. B | 63 | 25 | 38 |
| A vs. C | 61 | 37 | 24 |
| B vs. C | 37 | 36 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Li, Z.; Zhu, H.; Zi, R.; Xue, F.; Yu, Y. Identification of Tartary Buckwheat (Fagopyrum tataricum (L.) Gaertn) and Common Buckwheat (Fagopyrum esculentum Moench) Using Gas Chromatography–Mass Spectroscopy-Based Untargeted Metabolomics. Foods 2023, 12, 2578. https://doi.org/10.3390/foods12132578
Wu Y, Li Z, Zhu H, Zi R, Xue F, Yu Y. Identification of Tartary Buckwheat (Fagopyrum tataricum (L.) Gaertn) and Common Buckwheat (Fagopyrum esculentum Moench) Using Gas Chromatography–Mass Spectroscopy-Based Untargeted Metabolomics. Foods. 2023; 12(13):2578. https://doi.org/10.3390/foods12132578
Chicago/Turabian StyleWu, Yuling, Zhanming Li, Hui Zhu, Run Zi, Fang Xue, and Yue Yu. 2023. "Identification of Tartary Buckwheat (Fagopyrum tataricum (L.) Gaertn) and Common Buckwheat (Fagopyrum esculentum Moench) Using Gas Chromatography–Mass Spectroscopy-Based Untargeted Metabolomics" Foods 12, no. 13: 2578. https://doi.org/10.3390/foods12132578
APA StyleWu, Y., Li, Z., Zhu, H., Zi, R., Xue, F., & Yu, Y. (2023). Identification of Tartary Buckwheat (Fagopyrum tataricum (L.) Gaertn) and Common Buckwheat (Fagopyrum esculentum Moench) Using Gas Chromatography–Mass Spectroscopy-Based Untargeted Metabolomics. Foods, 12(13), 2578. https://doi.org/10.3390/foods12132578






