Microbial Community Dynamics and the Correlation between Specific Bacterial Strains and Higher Alcohols Production in Tartary Buckwheat Huangjiu Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Tartary Buckwheat Huangjiu Fermentation and Samples Preparation
2.2. Microbial Community Analysis during Tartary Buckwheat Huangjiu Fermentation
2.2.1. DNA Isolation and Illumina MiSeq Sequencing
2.2.2. Sequencing Data Analysis
2.3. Higher Alcohols Determination during Tartary Buckwheat Huangjiu Fermentation
2.4. Correlation Analysis between Microbes and High Alcohols Production during Tartary Buckwheat Huangjiu Fermentation
2.5. Verification of the Correlation between the Wheat Qu Bacterial Strains and High Alcohols Production during Tartary Buckwheat Huangjiu Fermentation
3. Results
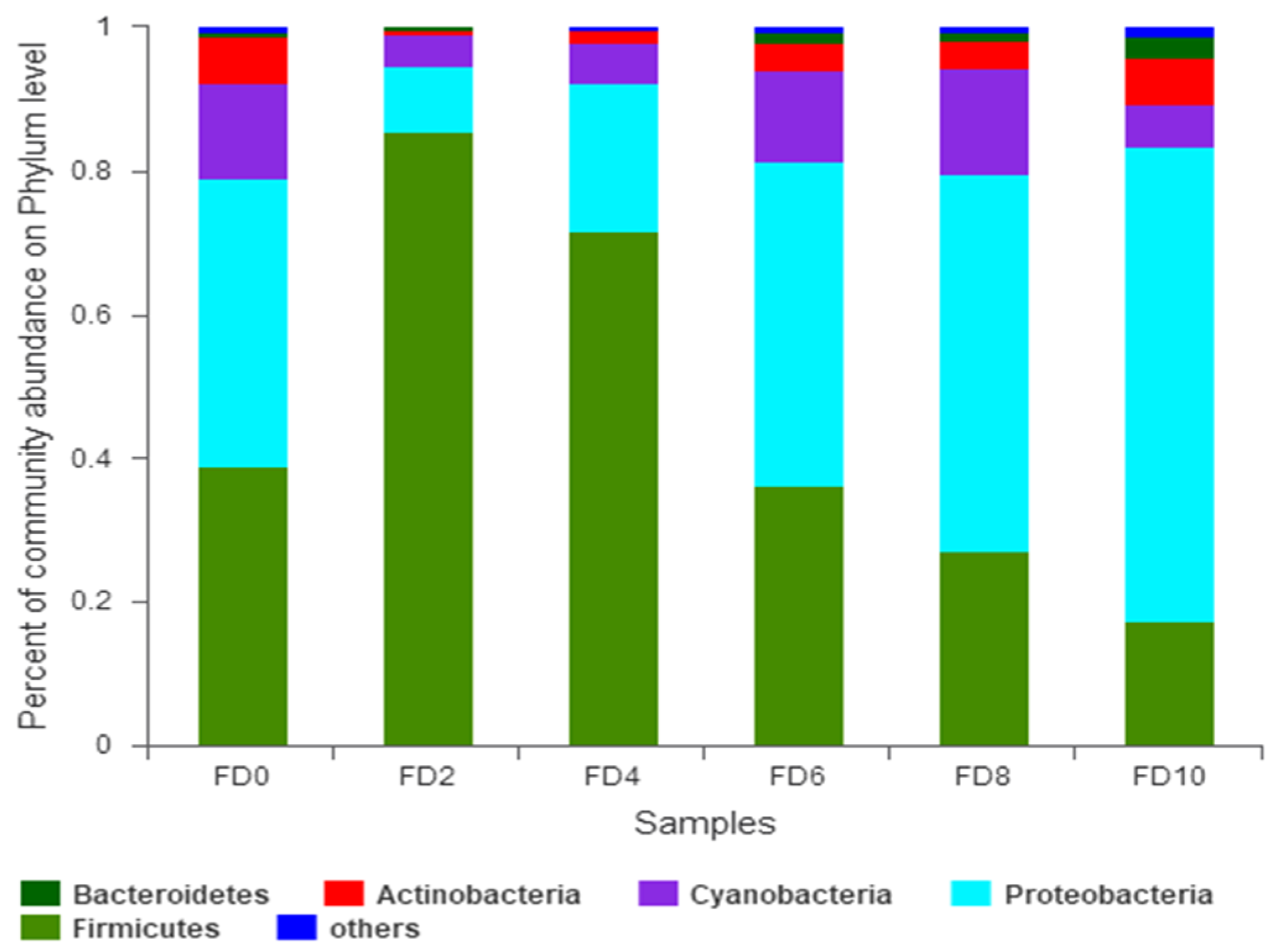
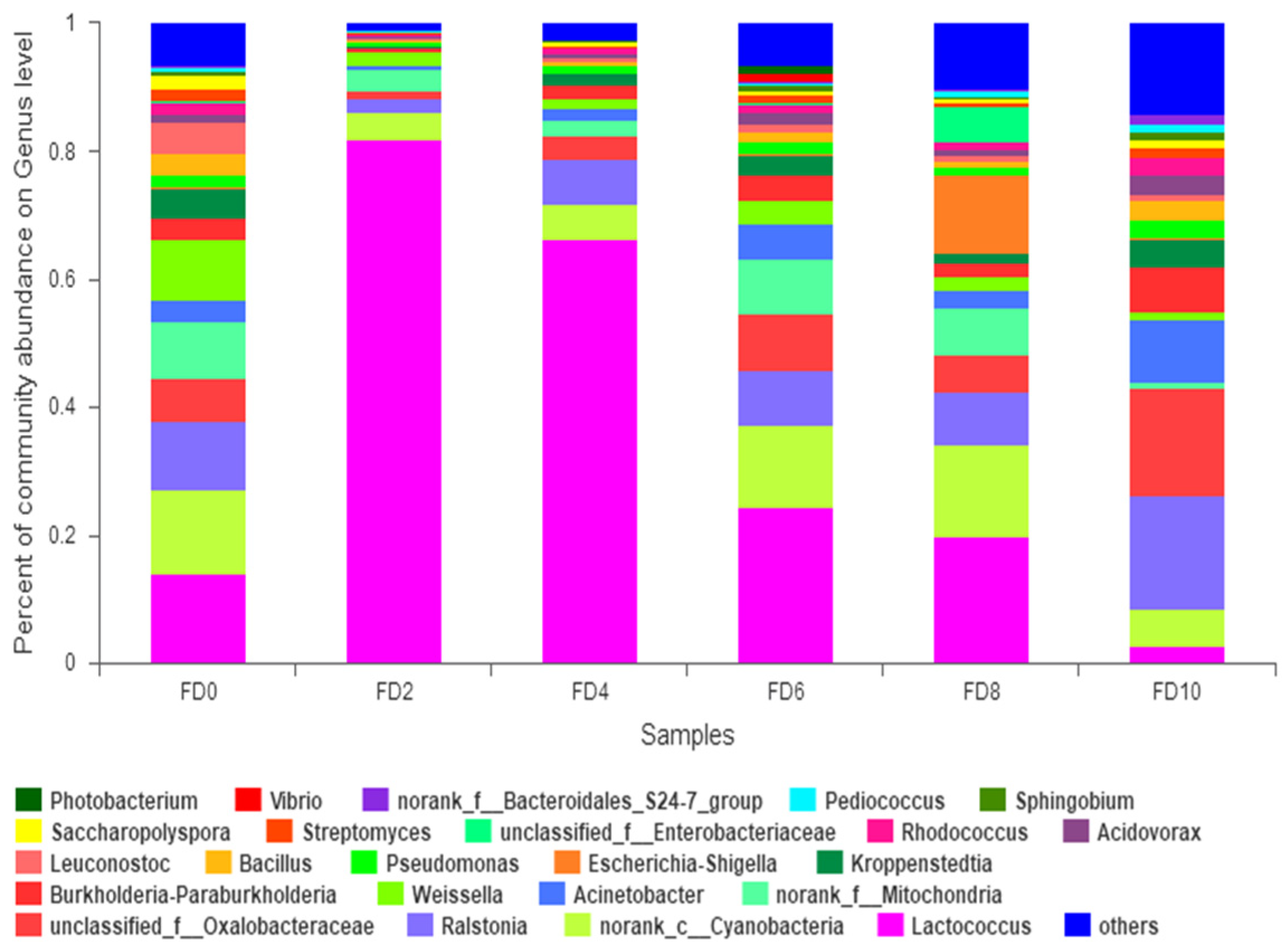
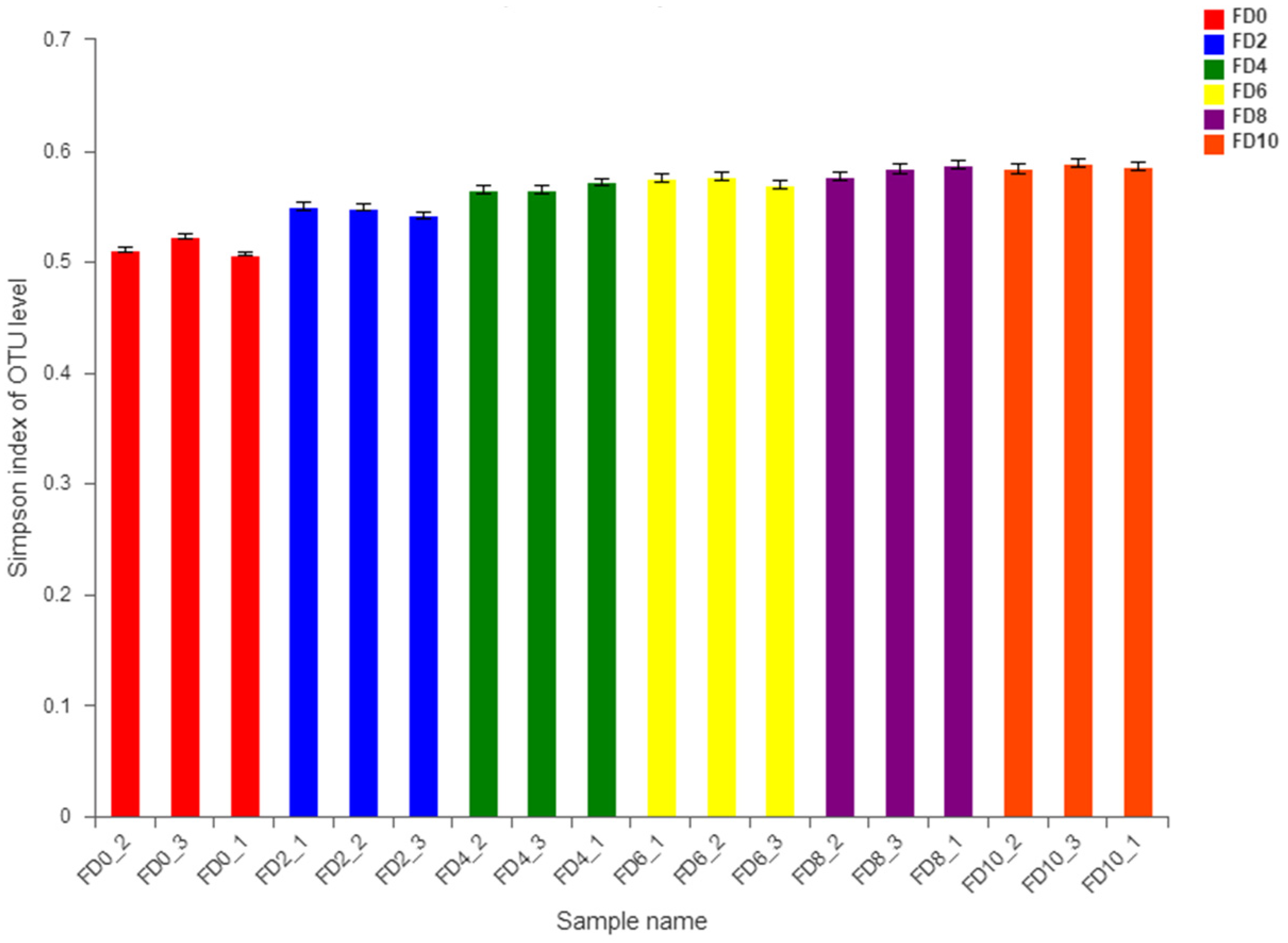
3.1. Dynamic Change in Bacterial Community during Tartary Buckwheat Huangjiu Fermentation
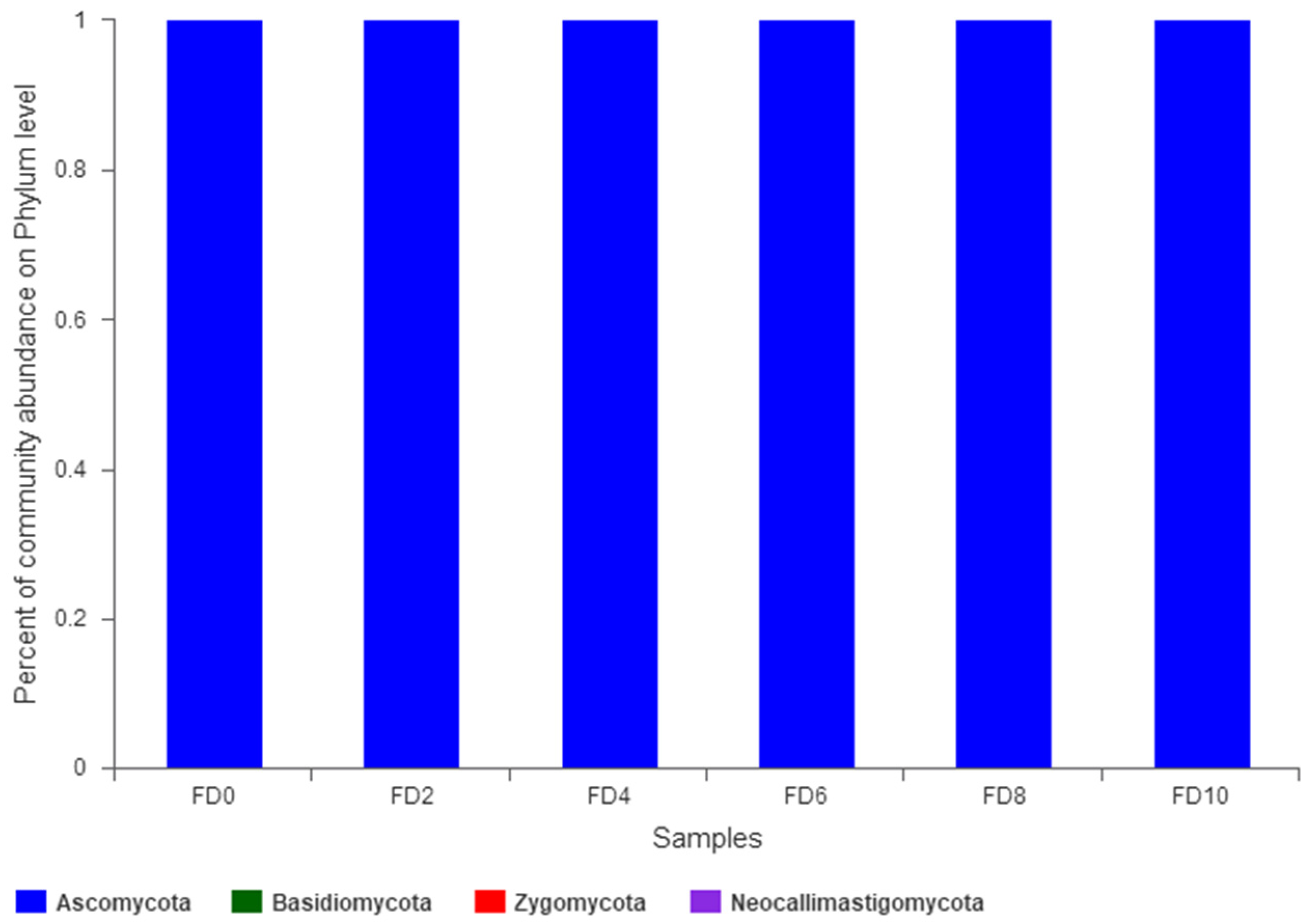
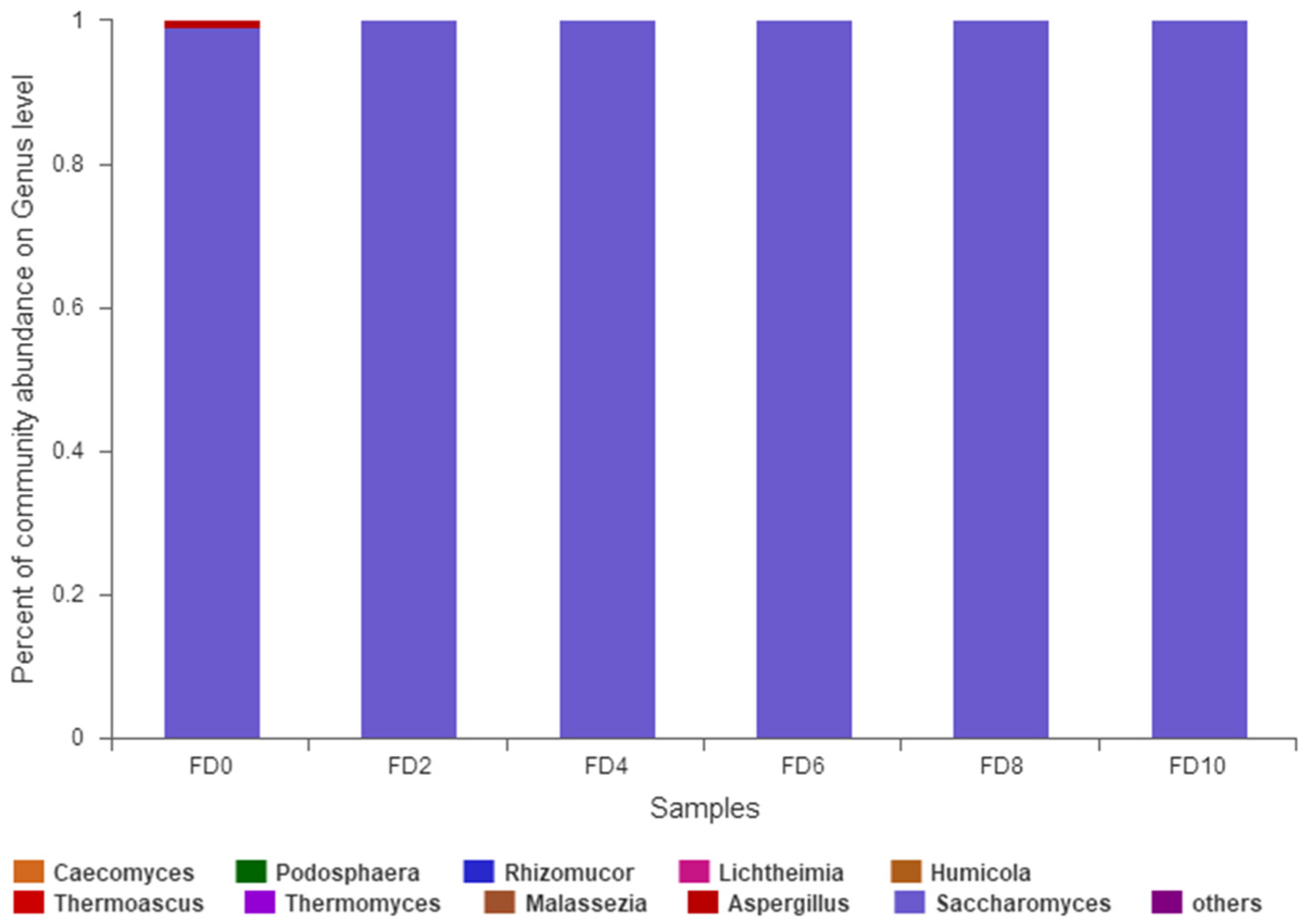
3.2. Dynamic Change in Fungal Community during Tartary Buckwheat Huangjiu Fermentation
3.3. Higher Alcohols Production during Tartary Buckwheat Huangjiu Fermentation
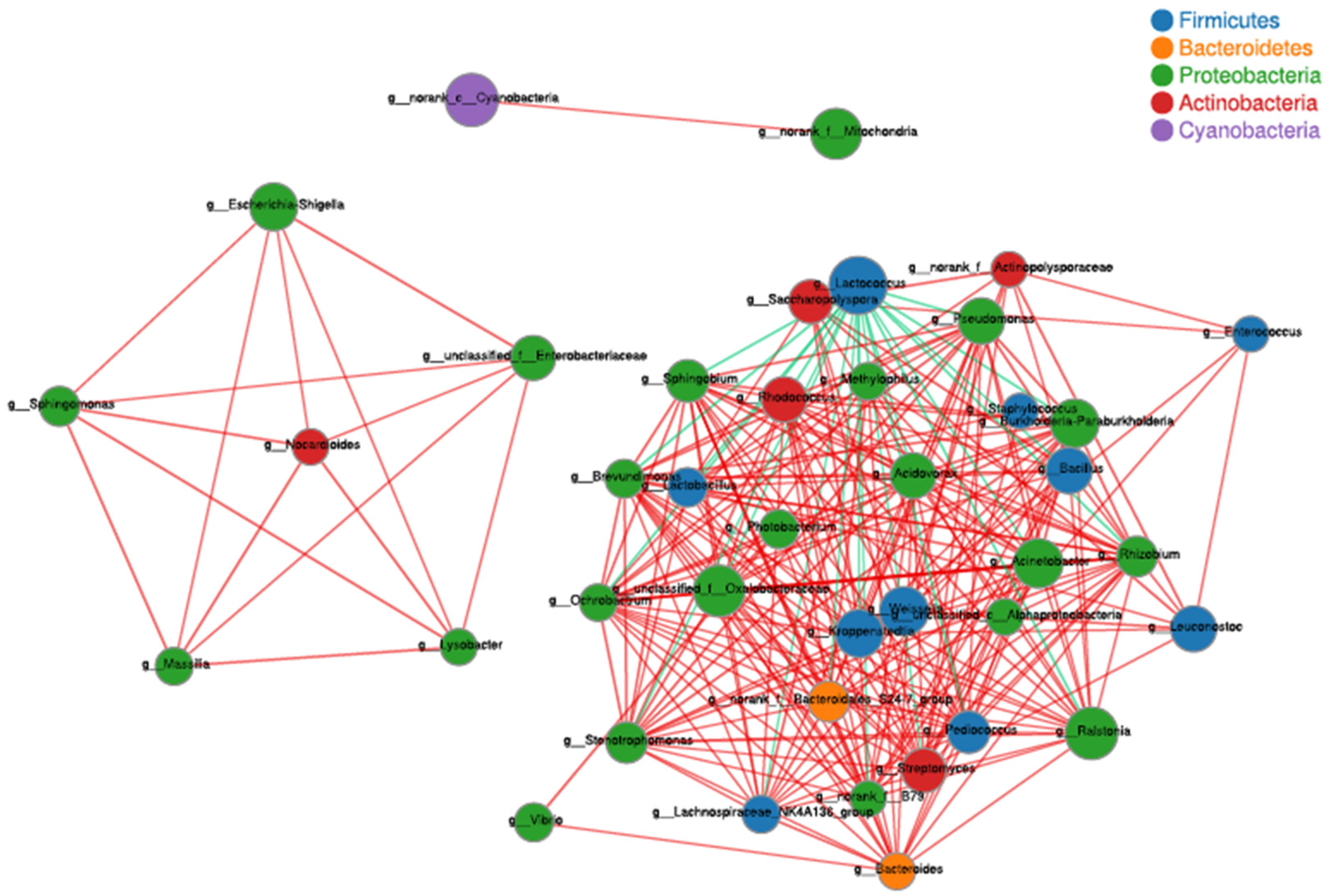
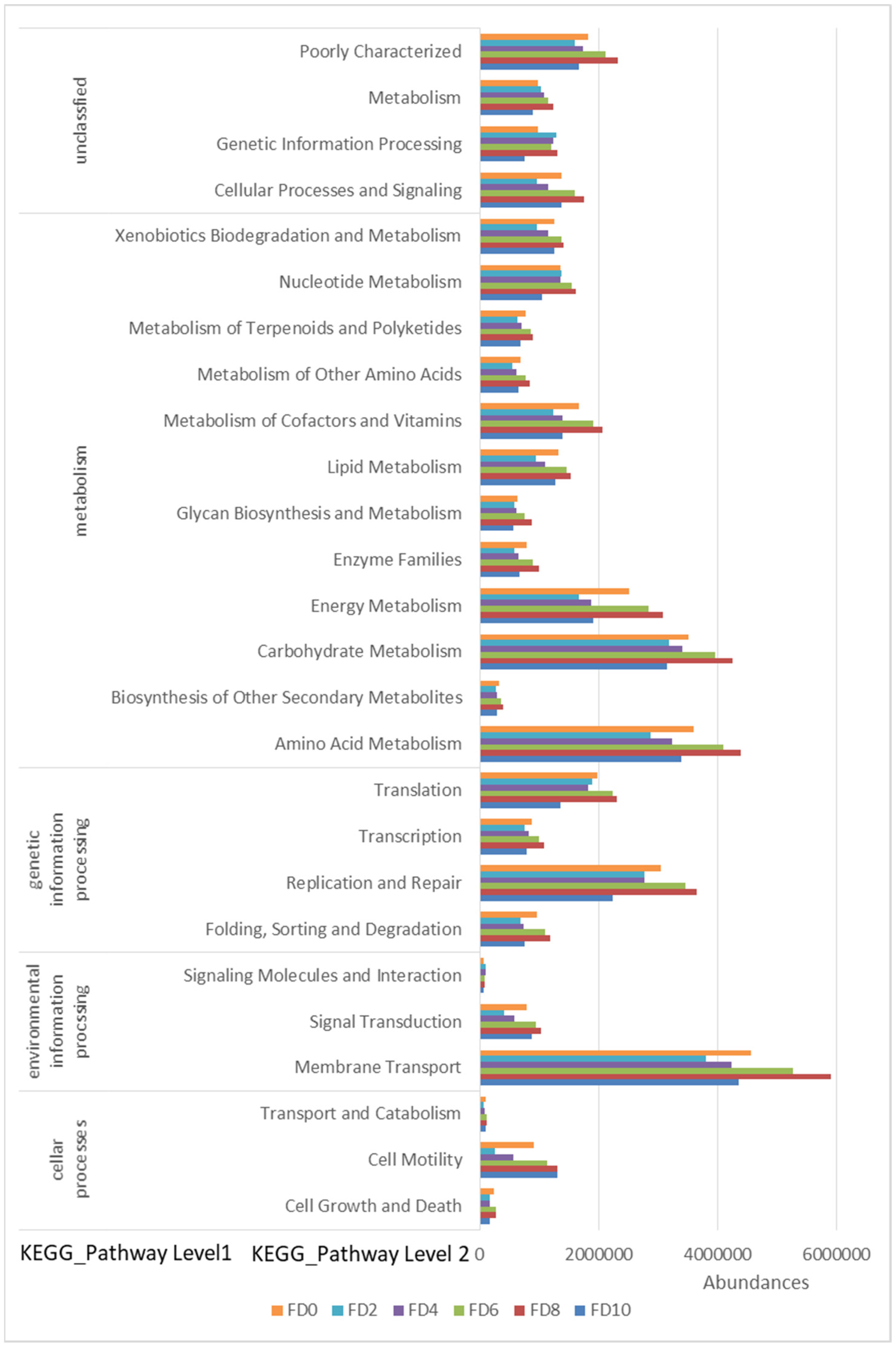
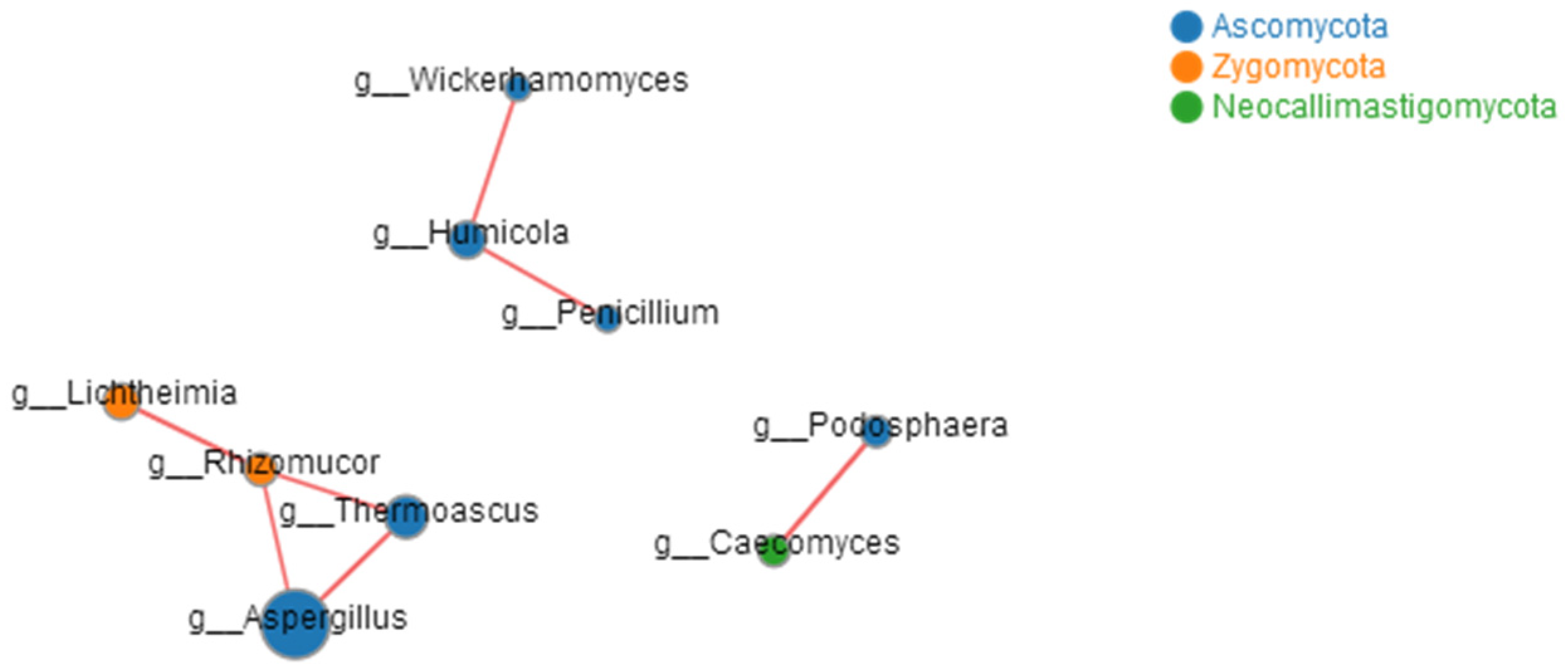
3.4. Correlation Analysis of Microbes and Higher Alcohols Production during Tartary Buckwheat Huangjiu Fermentation
3.5. Effects of Improved Wheat Qu Inoculated with Specific Bacterial Strains on Higher Alcohols Production in Tartary Buckwheat Huangjiu Fermentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lv, X.-C.; Jiang, Y.-J.; Liu, J.; Guo, W.-L.; Liu, Z.-B.; Zhang, W.; Rao, P.-F.; Ni, L. Evaluation of different PCR primers for denaturing gradient gel electrophoresis (DGGE) analysis of fungal community structure in traditional fermentation starters used for Hong Qu glutinous rice wine. Int. J. Food Microbiol. 2017, 255, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xia, Y.; Wang, G.; Zhang, H.; Xiong, Z.; Yu, J.; Yu, H.; Ai, L. Comparison of oenological property, volatile profile, and sensory characteristic of Chinese rice wine fermented by different starters during brewing. Int. J. Food Prop. 2017, 20, S3195–S3211. [Google Scholar] [CrossRef]
- Ni, L.; Lv, X.; Huang, Z.; Cai, Q.; Liang, S.; Rao, P. A review of physiological functions and bioactives of rice wine. J. Chin. Inst. Food Sci. Technol. 2012, 12, 1–7. [Google Scholar] [CrossRef]
- Zhu, F. Chemical composition and health effects of Tartary buckwheat. Food Chem. 2016, 203, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Zeng, W.; Fang, F.; Zhou, J.; Du, G.-C. The microbiome of Chinese rice wine (Huangjiu). Curr. Res. Food Sci. 2022, 5, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Pires, E.J.; Teixeira, J.A.; Brányik, T.; Vicente, A.A. Yeast: The soul of beer’s aroma—A review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl. Microbiol. Biotechnol. 2014, 98, 1937–1949. [Google Scholar] [CrossRef]
- de-la-Fuente-Blanco, A.; Sáenz-Navajas, M.-P.; Ferreira, V. On the effects of higher alcohols on red wine aroma. Food Chem. 2016, 210, 107–114. [Google Scholar] [CrossRef]
- Lachenmeier, D.W.; Haupt, S.; Schulz, K. Defining maximum levels of higher alcohols in alcoholic beverages and surrogate alcohol products. Regul. Toxicol. Pharmacol. 2008, 50, 313–321. [Google Scholar] [CrossRef]
- Yuan, J.; Mishra, P.; Ching, C.B. Metabolically engineered Saccharomyces cerevisiae for branched-chain ester productions. J. Biotechnol. 2016, 239, 90–97. [Google Scholar] [CrossRef]
- A Buijs, N.; Siewers, V.; Nielsen, J. Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr. Opin. Chem. Biol. 2013, 17, 480–488. [Google Scholar] [CrossRef]
- Hazelwood, L.A.; Daran, J.-M.; van Maris, A.J.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef]
- Vidal, E.E.; de Morais, M.A., Jr.; François, J.M.; de Billerbeck, G.M. Biosynthesis of higher alcohol flavour compounds by the yeast Saccharomyces cerevisiae: Impact of oxygen availability and responses to glucose pulse in minimal growth medium with leucine as sole nitrogen source. Yeast 2015, 32, 47–56. [Google Scholar] [CrossRef]
- Procopio, S.; Qian, F.; Becker, T. Function and regulation of yeast genes involved in higher alcohol and ester metabolism during beverage fermentation. Eur. Food Res. Technol. 2011, 233, 721–729. [Google Scholar] [CrossRef]
- Zhao, C.; Su, W.; Mu, Y.; Jiang, L.; Mu, Y. Correlations between microbiota with physicochemical properties and volatile flavor components in black glutinous rice wine fermentation. Food Res. Int. 2020, 138 Pt B, 109800. [Google Scholar] [CrossRef]
- Liu, S.P.; Mao, J.; Liu, Y.Y.; Meng, X.Y.; Ji, Z.W.; Zhou, Z.L.; Ai-Lati, A. Bacterial succession and the dynamics of volatile compounds during the fermentation of Chinese rice wine from Shaoxing region. World J. Microbiol. Biotechnol. 2015, 31, 1907–1921. [Google Scholar] [CrossRef]
- Wang, P.; Mao, J.; Meng, X.; Li, X.; Liu, Y.; Feng, H. Changes in flavor characteristics and bacterial diversity during the traditional fermentation of Chinese rice wines from Shaoxing region. Food Control 2014, 44, 58–63. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Tian, H.; Ai, L.; Yu, H. Metagenomic analysis reveals the impact of JIUYAO microbial diversity on fermentation and the volatile profile of Shaoxing-jiu. Food Microbiol. 2020, 86, 103326. [Google Scholar] [CrossRef]
- Xu, J.; Wu, H.; Wang, Z.; Zheng, F.; Lu, X.; Li, Z.; Ren, Q. Microbial dynamics and metabolite changes in Chinese Rice Wine fermentation from sorghum with different tannin content. Sci. Rep. 2018, 8, 4639. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef]
- Chen, G.-M.; Huang, Z.-R.; Wu, L.; Wu, Q.; Guo, W.-L.; Zhao, W.-H.; Liu, B.; Zhang, W.; Rao, P.-F.; Lv, X.-C.; et al. Microbial diversity and flavor of Chinese rice wine (Huangjiu): An overview of current research and future prospects. Curr. Opin. Food Sci. 2021, 42, 37–50. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Tian, H.; Yu, H. Research progress on the microbial community and their relationship with flavor formation in the brewing process of Chinese rice wine. J. Food Saf. Qual. 2019, 10, 4856–4863. [Google Scholar] [CrossRef]
- Zhang, K.; Li, Q.; Wu, W.; Yang, J.; Zou, W. Wheat Qu and its production technology, microbiota, flavor, and metabolites. J. Food Sci. 2019, 84, 2373–2386. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Xie, X.; Li, X.; Deng, B.; Xiao, Y.; Wu, D.; Zhai, S. Research progress on microorganisms and their metabolites of Huangjiu Qu. Amino Acids Biotic. Resour. 2019, 41, 104–111. [Google Scholar] [CrossRef]
- Li, P.; Aflakpui, F.W.K.; Yu, H.; Luo, L.; Lin, W.-T. Characterization of activity and microbial diversity of typical types of Daqu for traditional Chinese vinegar. Ann. Microbiol. 2015, 65, 2019–2027. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Yan, J.; Zhou, M.; Woo, S.-H.; Weng, W.; Cheng, J.; Zhang, K. Tartary Buckwheat: An under-utilized edible and medicinal herb for food and nutritional security. Food Rev. Int. 2022, 38, 440–454. [Google Scholar] [CrossRef]
- Yu, H.; Xie, T.; Xie, J.; Ai, L.; Tian, H. Characterization of key aroma compounds in Chinese rice wine using gas chromatography-mass spectrometry and gas chromatography-olfactometry. Food Chem. 2019, 293, 8–14. [Google Scholar] [CrossRef]
- Hua, D.; Xu, P. Recent advances in biotechnological production of 2-phenylethanol. Biotechnol. Adv. 2011, 29, 654–660. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Lu, X.; Zong, H.; Zhuge, B. Advances in 2-phenylethanol production from engineered microorganisms. Biotechnol. Adv. 2019, 37, 403–409. [Google Scholar] [CrossRef]
- Etschmann, M.; Bluemke, W.; Sell, D.; Schrader, J. Biotechnological production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 2002, 59, 1–8. [Google Scholar] [CrossRef]
- Etschmann, M.M.W.; Sell, D.; Schrader, J. Screening of yeasts for the production of the aroma compound 2-phenylethanol in a molasses-based medium. Biotechnol. Lett. 2003, 25, 531–536. [Google Scholar] [CrossRef]
- Fabre, C.E.; Duviau, V.J.; Blanc, P.J.; Goma, G. Identification of flavour volatile compounds obtained in culture of Kluyveromyces marxianus. Biotechnol. Lett. 1995, 17, 1207–1212. [Google Scholar] [CrossRef]
- Forti, L.; Di Mauro, S.; Cramarossa, M.R.; Filippucci, S.; Turchetti, B.; Buzzini, P. Non-conventional yeasts whole cells as efficient biocatalysts for the production of flavors and fragrances. Molecules 2015, 20, 10377–10398. [Google Scholar] [CrossRef]
- Gao, F.; Daugulis, A.J. Bioproduction of the aroma compound 2-phenylethanol in a solid-liquid two-phase partitioning bioreactor system by Kluyveromyces marxianus. Biotechnol. Bioeng. 2009, 104, 332–339. [Google Scholar] [CrossRef]
- Masuo, S.; Osada, L.; Zhou, S.; Fujita, T.; Takaya, N. Aspergillus oryzae pathways that convert phenylalanine into the flavor volatile 2-phenylethanol. Fungal Genet. Biol. 2015, 77, 22–30. [Google Scholar] [CrossRef]
- Chung, H., Jr.; Lee, S.L.; Chou, C.C. Production and molar yield of 2-phenylethanol by Pichia fermentans L-5 as affected by some medium components. J. Biosci. Bioeng. 2000, 90, 142–147. [Google Scholar] [CrossRef]
- Lu, X.; Wang, Y.; Zong, H.; Ji, H.; Zhuge, B.; Dong, Z. Bioconversion of L-phenylalanine to 2-phenylethanol by the novel stress-tolerant yeast Candida glycerinogenes WL2002-5. Bioengineered 2016, 7, 418–423. [Google Scholar] [CrossRef]
- Qian, X.; Yan, W.; Zhang, W.; Dong, W.; Ma, J.; Ochsenreither, K.; Jiang, M.; Xin, F. Current status and perspectives of 2-phenylethanol production through biological processes. Crit. Rev. Biotechnol. 2019, 39, 235–248. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, K.; Cao, W.; Zhang, G.; Chen, G.; Yang, H.; Wang, Q.; Liu, H.; Xian, M.; Zhang, H. Genome mining of 2-phenylethanol biosynthetic genes from Enterobacter sp. CGMCC 5087 and heterologous overproduction in Escherichia coli. Biotechnol. Biofuels 2018, 11, 305. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, J.; Bai, Y.; Fan, T.-P.; Zhao, Y.; Zheng, X.; Cai, Y. Mimicking a new 2-phenylethanol production pathway from Proteus mirabilis JN458 in Escherichia coli. J. Agric. Food Chem. 2018, 66, 3498–3504. [Google Scholar] [CrossRef]
- Ziosi, P.; Tabanelli, T.; Fornasari, G.; Cocchi, S.; Cavani, F.; Righi, P. Carbonates as reactants for the production of fine chemicals: The synthesis of 2-phenoxyethanol. Catal. Sci. Technol. 2014, 4, 4386–4395. [Google Scholar] [CrossRef]
- Liu, Z.; Lan, X.; Li, X.; Zhao, H.; Gan, J.; Li, R.; Chen, B. A plant-derived alkanol induces teliospore germination in Sporisorium scitamineum. J. Fungi 2022, 8, 209. [Google Scholar] [CrossRef] [PubMed]

| Day 0 | Day 2 | Day 4 | Day 6 | Day 8 | Day 10 | |
|---|---|---|---|---|---|---|
| 2-phenylethanol | 14.56 ± 1.35 | 312.76 ± 2.47 | 811.16 ± 1.14 | 1551.68 ± 0.82 | 1728.42 ± 0.91 | 1596.30 ± 1.00 |
| isobutanol | ND | 26.37 ± 1.24 | 37.42 ± 1.07 | 317.34 ± 1.10 | ND | ND |
| isopentanol | ND | 237.82 ± 1.72 | 396.22 ± 0.87 | 2089.44 ± 2.22 | ND | 2513.07 ± 3.21 |
| pentanol | ND | ND | ND | ND | 2102.71 ± 1.41 | ND |
| isooctanol | ND | 5.48 ± 2.85 | 57.53 ± 0.81 | ND | ND | ND |
| methionol | ND | 3.91 ± 0.66 | 10.02 ± 0.55 | ND | ND | ND |
| 2-phenoxyethanol | 14.54 ± 1.07 | 15.89 ± 0.56 | 161.50 ± 1.04 | ND | 908.45 ± 0.94 | 1760.51 ± 1.08 |
| 2-ethyl-2-methyl-tridecanol | 79.56 ± 1.98 | ND | ND | 24439.67 ± 4.88 | 9051.63 ± 0.66 | ND |
| 1-hendecanol | ND | ND | ND | ND | 350.83 ± 23.54 | ND |
| 1-dodecanol | 40.42 ± 1.58 | ND | ND | ND | ND | ND |
| 1-hexadecanol | 27.60 ± 0.29 | 12.57 ± 1.53 | 62.29 ± 1.19 | ND | 4343.59 ± 0.41 | ND |
| linalool | ND | ND | 46.32 ± 1.05 | ND | ND | ND |
| lavandulol | ND | ND | 17.66 ± 1.33 | ND | ND | ND |
| linoleny alcohol | ND | ND | 150.80 ± 53.36 | ND | ND | ND |
| 2-(2-ethylhexyloxy) ethanol | ND | ND | ND | ND | 517.87 ± 0.55 | ND |
| 3,3-dimethyl-2-butanol | ND | ND | ND | 465.31 ± 1.94 | ND | ND |
| 2-(2-butoxyethoxy) ethanol | ND | 29.45 ± 1.06 | 70.19 ± 0.89 | ND | ND | ND |
| 2-isopropyl-5-methyl-1-heptanol | 13.53 ± 0.54 | ND | 72.11 ± 1.05 | ND | ND | ND |
| Higher Alcohols | Qu | QT1 | QT2 | QT3 | QT4 |
|---|---|---|---|---|---|
| isopentanol | 2.65 ± 0.08 a | 2.81 ± 0.12 a | 2.57 ± 0.10 a | 2.53 ± 0.09 a | 2.84 ± 0.12 b |
| 2-phenylethanol | 1.83 ± 0.11 a | 1.77 ± 0.13 a | 1.79 ± 0.12 a | 2.26 ± 0.14 b | 1.81 ± 0.09 a |
| 2-phenoxyethanol | 1.76 ± 0.10 a | 1.68 ± 0.09 a | 1.80 ± 0.13 a | 1.83 ± 0.12 a | 1.79 ± 0.12 a |
| pentanol | 0.62 ± 0.07 a | 0.70 ± 0.05 a | 0.57 ± 0.06 a | 0.83 ± 0.08 b | 0.58 ± 0.06 a |
| isooctanol | 0.04 ± 0.001 a | 0.03 ± 0.001 a | 0.03 ± 0.001 a | 0.03 ± 0.001 a | 0.04 ± 0.002 a |
| isobutanol | 0.03 ± 0.002 a | 0.04 ± 0.001 a | 0.04 ± 0.002 a | 0.04 ± 0.001 a | 0.72 ± 0.011 b |
| others | 0.81 ± 0.07 a | 0.72 ± 0.08 a | 0.74 ± 0.05 a | 1.25 ± 0.11 b | 1.18 ± 0.12 b |
| total content | 7.65 ± 0.14 a | 7.75 ± 0.16 a | 7.54 ± 0.13 a | 8.77 ± 0.16 b | 8.96 ± 0.15 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, S.; Huang, M.; Wang, J.; Liu, B.; Ren, Q. Microbial Community Dynamics and the Correlation between Specific Bacterial Strains and Higher Alcohols Production in Tartary Buckwheat Huangjiu Fermentation. Foods 2023, 12, 2664. https://doi.org/10.3390/foods12142664
Yin S, Huang M, Wang J, Liu B, Ren Q. Microbial Community Dynamics and the Correlation between Specific Bacterial Strains and Higher Alcohols Production in Tartary Buckwheat Huangjiu Fermentation. Foods. 2023; 12(14):2664. https://doi.org/10.3390/foods12142664
Chicago/Turabian StyleYin, Sheng, Mingquan Huang, Jiaxuan Wang, Bo Liu, and Qing Ren. 2023. "Microbial Community Dynamics and the Correlation between Specific Bacterial Strains and Higher Alcohols Production in Tartary Buckwheat Huangjiu Fermentation" Foods 12, no. 14: 2664. https://doi.org/10.3390/foods12142664
APA StyleYin, S., Huang, M., Wang, J., Liu, B., & Ren, Q. (2023). Microbial Community Dynamics and the Correlation between Specific Bacterial Strains and Higher Alcohols Production in Tartary Buckwheat Huangjiu Fermentation. Foods, 12(14), 2664. https://doi.org/10.3390/foods12142664






