A Self-Supervised Anomaly Detector of Fruits Based on Hyperspectral Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.1.1. Strawberry Data Set
2.1.2. Blueberry Data Set
2.2. Hyperspectral Data Measurement
2.3. Data Preprocessing
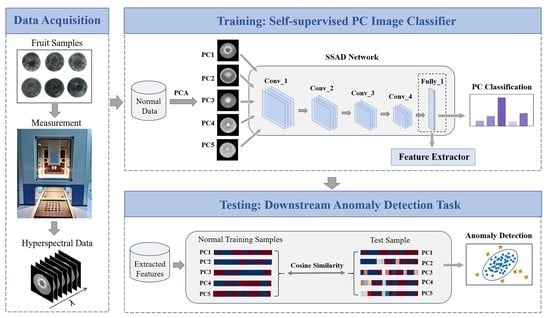
2.4. The Proposed Method
2.4.1. Architecture of SSAD
2.4.2. Detailed Description of Training Procedure
2.4.3. Detailed Description of Testing Procedure
2.4.4. Hyperparameter Settings and Training Configurations
2.5. Metrics for Model Evaluation
2.6. Methods for Comparison
2.7. Software and Hardware Environment
3. Results
3.1. Spectra and PC Images
3.2. Comparison of Anomaly Detection Performance
4. Discussion
4.1. Effect of the Principal Components
4.2. Effect of the Layers for Feature Extraction
4.3. Effect of Data Pollution
5. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, D.; Sun, D.W. Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review-Part I: Fundamentals. Innov. Food Sci. Emerg. 2013, 19, 1–14. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, J.; Lin, T.; Ying, Y. Food and agro-product quality evaluation based on spectroscopy and deep learning: A review. Trends Food Sci. Technol. 2021, 112, 431–441. [Google Scholar] [CrossRef]
- Tian, Y.; Sun, J.; Zhou, X.; Yao, K.; Tang, N. Detection of soluble solid content in apples based on hyperspectral technology combined with deep learning algorithm. J. Food Process. Pres. 2022, 46, e16414. [Google Scholar] [CrossRef]
- Teerachaichayut, S.; Ho, H.T. Non-destructive prediction of total soluble solids, titratable acidity and maturity index of limes by near infrared hyperspectral imaging. Postharvest Biol. Technol. 2017, 133, 20–25. [Google Scholar] [CrossRef]
- Hu, M.H.; Dong, Q.L.; Liu, B.L.; Opara, U.L.; Chen, L. Estimating blueberry mechanical properties based on random frog selected hyperspectral data. Postharvest Biol. Technol. 2015, 106, 1–10. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, Y.; Xuan, G.; Gao, Z.; Hu, Z.; Gao, C.; Wang, K. Assessment of strawberry ripeness using hyperspectral imaging. Anal. Lett. 2020, 54, 1547–1560. [Google Scholar] [CrossRef]
- Baranowski, P.; Mazurek, W.; Wozniak, J.; Majewska, U. Detection of early bruises in apples using hyperspectral data and thermal imaging. J. Food. Eng. 2012, 110, 345–355. [Google Scholar] [CrossRef]
- Mehl, P.M.; Chen, Y.R.; Kim, M.S.; Chan, D.E. Development of hyperspectral imaging technique for the detection of apple surface defects and contaminations. J. Food. Eng. 2004, 61, 67–81. [Google Scholar] [CrossRef]
- Babellahi, F.; Paliwal, J.; Erkinbaev, C.; Amodio, M.L.; Chaudhry, M.M.A.; Colelli, G. Early detection of chilling injury in green bell peppers by hyperspectral imaging and chemometrics. Postharvest Biol. Technol. 2022, 162, 111100. [Google Scholar] [CrossRef]
- Li, J.; Huang, W.; Tian, X.; Wang, C.; Fan, S.; Zhao, C. Fast detection and visualization of early decay in citrus using Vis-NIR hyperspectral imaging. Comput. Electron Agric. 2016, 127, 582–592. [Google Scholar] [CrossRef]
- Yu, K.; Zhao, Y.; Li, X.; Shao, Y.; Zhu, F.; He, Y. Identification of crack features in fresh jujube using Vis/NIR hyperspectral imaging combined with image processing. Comput. Electron Agric. 2014, 103, 1–10. [Google Scholar] [CrossRef]
- Liu, Q.; Sun, K.; Peng, J.; Xing, M.; Pan, L.; Tu, K. Identification of bruise and fungi contamination in strawberries using hyperspectral imaging technology and multivariate analysis. Food Anal. Methods 2018, 11, 1518–1527. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, C.; Li, J.; Fan, S.; Yang, Y.; Huang, W. Detection of early decay on citrus using LW-NIR hyperspectral reflectance imaging coupled with two-band ratio and improved watershed segmentation algorithm. Food Chem. 2021, 360, 130077. [Google Scholar] [CrossRef] [PubMed]
- ElMasry, G.; Wang, N.; Vigneault, C. Detecting chilling injury in Red Delicious apple using hyperspectral imaging and neural networks. Postharvest Biol. Technol. 2009, 52, 1–8. [Google Scholar] [CrossRef]
- Zhang, M.; Jiang, Y.; Li, C.; Yang, F. Fully convolutional networks for blueberry bruising and calyx segmentation using hyperspectral transmittance imaging. Biosyst. Eng. 2020, 192, 159–175. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, S.; Han, W.; Liu, W.; Qiu, Z.; Li, C. Convolutional neural network for hyperspectral data analysis and effective wavelengths selection. Anal. Chim. Acta. 2019, 1086, 46–54. [Google Scholar] [CrossRef]
- Chandola, V.; Banerjee, A.; Kumar, V. Anomaly detection: A survey. ACM Comput. Surv. 2009, 41, 1–58. [Google Scholar] [CrossRef]
- Chalapathy, R.; Chawla, S. Deep learning for anomaly detection: A survey. arXiv 2019, arXiv:1901.03407. [Google Scholar]
- An, J.; Cho, S. Variational autoencoder based anomaly detection using reconstruction probability. Spec. Lect. IE 2015, 2, 1–18. [Google Scholar]
- Breunig, M.M.; Kriegel, H.P.; Ng, R.T.; Sander, J. LOF: Identifying density-based local outliers. In Proceedings of the 2000 ACM SIGMOD International Conference on Management of Data, Dalles, TX, USA, 15–18 May 2000; pp. 93–104. [Google Scholar]
- Zong, B.; Song, Q.; Min, M.R.; Cheng, W.; Lumezanu, C.; Cho, D.; Chen, H. Deep autoencoding gaussian mixture model for unsupervised anomaly detection. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Schölkopf, B.; Williamson, R.C.; Smola, A.; Shawe-Taylor, J.; Platt, J. Support vector method for novelty detection. In Proceedings of the Advances in Neural Information Processing Systems 12, NIPS Conference, Denver, CO, USA, 29 November–4 December 1999. [Google Scholar]
- Bergman, L.; Hoshen, Y. Classification-based anomaly detection for general data. arXiv 2020, arXiv:2005.02359. [Google Scholar]
- Mohseni, S.; Pitale, M.; Yadawa, J.B.S.; Wang, Z. Self-supervised learning for generalizable out-of-distribution detection. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 5216–5223. [Google Scholar]
- Golan, I.; El-Yaniv, R. Deep anomaly detection using geometric transformations. arXiv 2018, arXiv:1805.10917. [Google Scholar]
- Zhang, X.; Mu, J.; Zhang, X.; Liu, H.; Zong, L.; Li, Y. Deep anomaly detection with self-supervised learning and adversarial training. Pattern Recogn. 2022, 121, 108234. [Google Scholar] [CrossRef]
- Tack, J.; Mo, S.; Jeong, J.; Shin, J. CSI: Novelty detection via contrastive learning on distributionally shifted instances. Adv. Neural Inf. Process. Syst. 2020, 33, 11839–11852. [Google Scholar]
- Xu, L.; Yan, S.M.; Cai, C.B.; Yu, X.P. One-class partial least squares (OCPLS) classifier. Chemometr. Intell. Lab. 2013, 126, 1–5. [Google Scholar] [CrossRef]
- Ranzan, L.; Trierweiler, L.F.; Hitzmann, B.; Trierweiler, J.O. Avoiding misleading predictions in fluorescence-based soft sensors using autoencoders. Chemometr. Intell. Lab. 2022, 223, 104527. [Google Scholar] [CrossRef]
- Vasafi, P.S.; Hinrichs, J.; Hitzmann, B. Establishing a novel procedure to detect deviations from standard milk processing by using online Raman spectroscopy. Food Control 2022, 131, 108442. [Google Scholar] [CrossRef]
- Vasafi, P.S.; Paquet-Durand, O.; Brettschneider, K.; Hinrichs, J.; Hitzmann, B. Anomaly detection during milk processing by autoencoder neural network based on near-infrared spectroscopy. J. Food. Eng. 2021, 299, 110510. [Google Scholar] [CrossRef]
- Shen, G.; Fan, X.; Yang, Z.; Han, L. A feasibility study of non-targeted adulterant screening based on NIRM spectral library of soybean meal to guarantee quality: The example of non-protein nitrogen. Food Chem. 2016, 210, 35–42. [Google Scholar] [CrossRef]
- Zhang, M.; Li, C.; Yang, F. Optical properties of blueberry flesh and skin and monte carlo multi-layered simulation of light interaction with fruit tissues. Postharvest Biol. Technol. 2019, 150, 28–41. [Google Scholar] [CrossRef]
- Sioma, A. Geometry and resolution in triangulation vision systems. In Proceedings of the Photonics Applications in Astronomy, Communications, Industry, and High Energy Physics Experiments, Wilga, Poland, 31 August–2 September 2020. [Google Scholar]
- Bieniek, A.; Moga, A. An efficient watershed algorithm based on connected components. Pattern Recognit. 2000, 33, 907–916. [Google Scholar] [CrossRef]
- Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 1, 2579–2605. [Google Scholar]
- Siedliska, A.; Baranowski, P.; Zubik, M.; Mazurek, W.; Sosnowska, B. Detection of fungal infections in strawberry fruit by VNIR/SWIR hyperspectral imaging. Postharvest Biol. Technol. 2018, 139, 115–126. [Google Scholar] [CrossRef]
- Liu, Q.; Wei, K.; Xiao, H.; Tu, S.; Sun, K.; Sun, Y.; Pan, L.; Tu, K. Near-infrared hyperspectral imaging rapidly detects the decay of postharvest strawberry based on water-soluble sugar analysis. Food Anal. Methods 2018, 139, 115–126. [Google Scholar] [CrossRef]
- Hu, M.; Zhao, Y.; Zhai, G. Active learning algorithm can establish classifier of blueberry damage with very small training dataset using hyperspectral transmittance data. Chemometr. Intell. Lab. 2018, 172, 52–57. [Google Scholar] [CrossRef]










| Strawberry | |||||||
| Total | Normal | Anomaly | |||||
| Training | Test | Bruised | Infected | Chilling | Contaminated | ||
| Number of samples | 1045 | 300 | 301 | 139 | 100 | 105 | 100 |
| Blueberry | |||||||
| Total | Normal | Anomaly | |||||
| Training | Test | Bruised | Infected | Chilling | Wrinkled | ||
| Number of samples | 1335 | 404 | 404 | 120 | 120 | 150 | 137 |
| Strawberry Data Set | ||
|---|---|---|
| Name of Layers | Network Parameters | Output Feature Size |
| Convolution_1 | Filters = 16, Filter_size = 3 × 3, Activation = ReLu | 120 × 120 × 16 |
| Average_pooling_1 | Filter_size = 2, Stride =2 | 60 × 60 × 16 |
| Convolution_2 | Filters = 16, Filter_size = 3 × 3, Activation = ReLu | 60 × 60 × 16 |
| Average_pooling_2 | Filter_size = 2, Stride =2 | 30 × 30 × 16 |
| Convolution_3 | Filters = 16, Filter_size = 3 × 3, Activation = ReLu | 30 × 30 × 16 |
| Average_pooling_3 | Filter_size = 2, Stride =2 | 15 × 15 × 16 |
| Convolution_4 | Filters = 4, Filter_size = 3 × 3, Activation = ReLu | 15 × 15 × 4 |
| Fully_connected_1 | Nodes = 16, Activation = Tanh | 16 |
| Output | Nodes = 5, Activation = Softmax | 5 |
| Blueberry Data Set | ||
|---|---|---|
| Name of Layers | Network Parameters | Output Feature Size |
| Convolution_1 | Filters = 16, Filter_size = 3 × 3, Activation = ReLu | 60 × 60 × 16 |
| Average_pooling_1 | Filter_size = 2, Stride =2 | 30 × 30 × 16 |
| Convolution_2 | Filters = 16, Filter_size = 3 × 3, Activation = ReLu | 30 × 30 × 16 |
| Average_pooling_2 | Filter_size = 2, Stride =2 | 15 × 15 × 16 |
| Convolution_3 | Filters = 16, Filter_size = 3 × 3, Activation = ReLu | 15 × 15 × 16 |
| Convolution_4 | Filters = 4, Filter_size = 3 × 3, Activation = ReLu | 15 × 15 × 4 |
| Fully_connected_1 | Nodes = 16, Activation = Tanh | 16 |
| Output | Nodes = 5, Activation = Softmax | 5 |
| Methods | AUC | F1 Score | Acc_Normal | Acc_Bruised | Acc_Infected | Acc_Chilling | Acc_Contaminated |
|---|---|---|---|---|---|---|---|
| OCSVM | 0.773 ± 0.009 | 0.758 ± 0.007 | 0.643 ± 0.009 | 0.788 ± 0.020 | 0.594 ± 0.015 | 0.904 ± 0.005 | 0.776 ± 0.003 |
| AE-1D | 0.748 ± 0.005 | 0.727 ± 0.005 | 0.597 ± 0.007 | 0.684 ± 0.007 | 0.552 ± 0.009 | 0.995 ± 0.001 | 0.902 ± 0.005 |
| VAE-1D | 0.829 ± 0.004 | 0.784 ± 0.005 | 0.681 ± 0.008 | 0.742 ± 0.012 | 0.496 ± 0.016 | 0.753 ± 0.018 | 0.869 ± 0.015 |
| AE-2D | 0.690 ± 0.024 | 0.690 ± 0.017 | 0.543 ± 0.026 | 0.460 ± 0.051 | 0.764 ± 0.013 | 0.949 ± 0.025 | 0.866 ± 0.021 |
| VAE-2D | 0.659 ± 0.021 | 0.704 ± 0.014 | 0.542 ± 0.022 | 0.475 ± 0.014 | 0.767 ± 0.021 | 0.914 ± 0.003 | 0.717 ± 0.015 |
| SSAD | 0.913 ± 0.006 | 0.869 ± 0.005 | 0.807 ± 0.007 | 0.835 ± 0.031 | 0.834 ± 0.025 | 0.876 ± 0.022 | 0.945 ± 0.018 |
| Methods | AUC | F1 Score | Acc_Normal | Acc_Bruised | Acc_Infected | Acc_Chilling | Acc_Wrinkled |
|---|---|---|---|---|---|---|---|
| OCSVM | 0.744 ± 0.005 | 0.710 ± 0.005 | 0.622 ± 0.007 | 0.789 ± 0.010 | 0.582 ± 0.013 | 0.900 ± 0.006 | 0.614 ± 0.013 |
| AE-1D | 0.818 ± 0.028 | 0.779 ± 0.025 | 0.712 ± 0.033 | 0.838 ± 0.044 | 0.769 ± 0.057 | 0.743 ± 0.035 | 0.772 ± 0.027 |
| VAE-1D | 0.794 ± 0.008 | 0.754 ± 0.009 | 0.678 ± 0.011 | 0.910 ± 0.014 | 0.630 ± 0.020 | 0.823 ± 0.005 | 0.691 ± 0.013 |
| AE-2D | 0.655 ± 0.016 | 0.643 ± 0.011 | 0.534 ± 0.015 | 0.699 ± 0.037 | 0.991 ± 0.002 | 0.602 ± 0.031 | 0.249 ± 0.023 |
| VAE-2D | 0.803 ± 0.005 | 0.768 ± 0.003 | 0.697 ± 0.004 | 0.864 ± 0.006 | 0.963 ± 0.003 | 0.420 ± 0.006 | 0.774 ± 0.007 |
| SSAD | 0.932 ± 0.015 | 0.875 ± 0.012 | 0.837 ± 0.016 | 0.944 ± 0.018 | 0.909 ± 0.037 | 0.838 ± 0.021 | 0.810 ± 0.027 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhou, S.; Wan, Z.; Qiu, Z.; Zhao, L.; Pang, K.; Li, C.; Yin, Z. A Self-Supervised Anomaly Detector of Fruits Based on Hyperspectral Imaging. Foods 2023, 12, 2669. https://doi.org/10.3390/foods12142669
Liu Y, Zhou S, Wan Z, Qiu Z, Zhao L, Pang K, Li C, Yin Z. A Self-Supervised Anomaly Detector of Fruits Based on Hyperspectral Imaging. Foods. 2023; 12(14):2669. https://doi.org/10.3390/foods12142669
Chicago/Turabian StyleLiu, Yisen, Songbin Zhou, Zhiyong Wan, Zefan Qiu, Lulu Zhao, Kunkun Pang, Chang Li, and Zexuan Yin. 2023. "A Self-Supervised Anomaly Detector of Fruits Based on Hyperspectral Imaging" Foods 12, no. 14: 2669. https://doi.org/10.3390/foods12142669
APA StyleLiu, Y., Zhou, S., Wan, Z., Qiu, Z., Zhao, L., Pang, K., Li, C., & Yin, Z. (2023). A Self-Supervised Anomaly Detector of Fruits Based on Hyperspectral Imaging. Foods, 12(14), 2669. https://doi.org/10.3390/foods12142669