The Screening and Isolation of Ethyl-Carbamate-Degrading Strains from Fermented Grains and Their Application in the Degradation of Ethyl Carbamate in Chinese Baijiu
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Collection
2.3. Screening of EC-Degrading Isolates
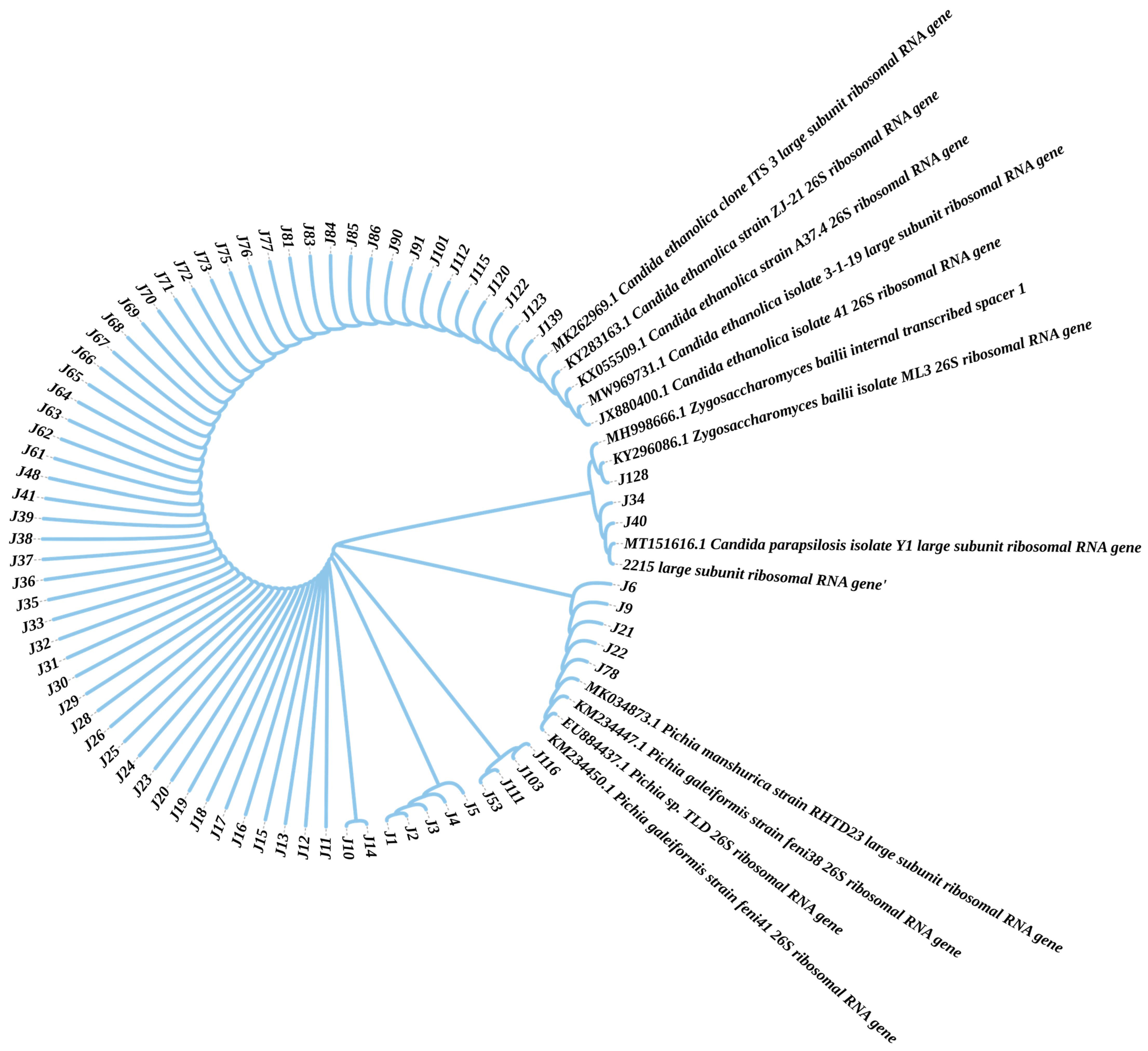
2.4. Phylogenetic Identification of the Isolates
2.5. Assimilation of Carbon Sources
2.6. Growth Curve Analysis of the Isolated Strains
2.7. Degradation of EC in Chinese Baijiu by the Immobilized Strain
2.8. Determination of VOC Content in Chinese Liquor
2.9. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Screening of EC-Degrading Strains from Fermented Grains
3.2. Carbon Source Assimilation by the Isolated Strains
3.3. Growth Curve of the Isolated Strains
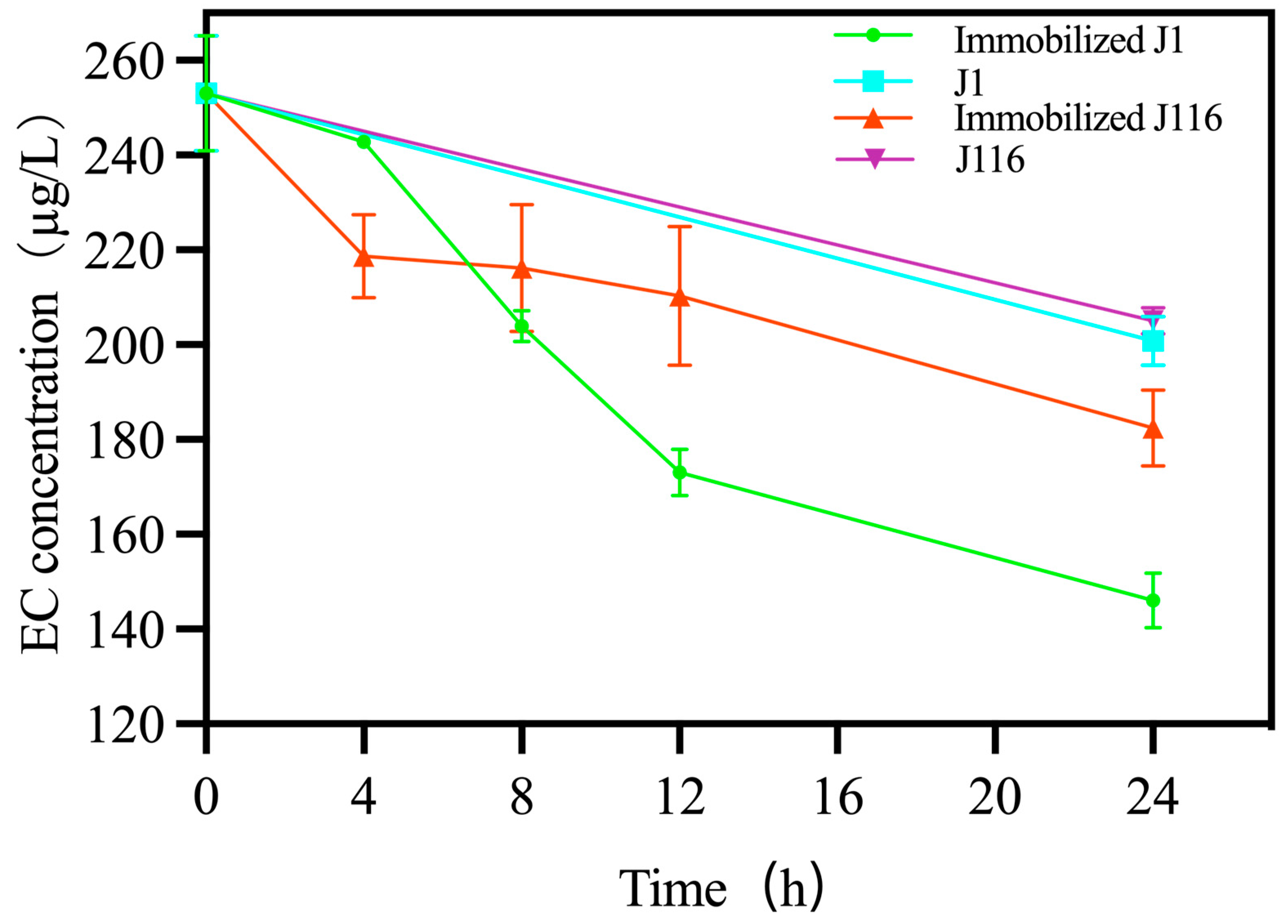
3.4. Degradation of EC in Chinese Wine by Strains and Immobilized Strains
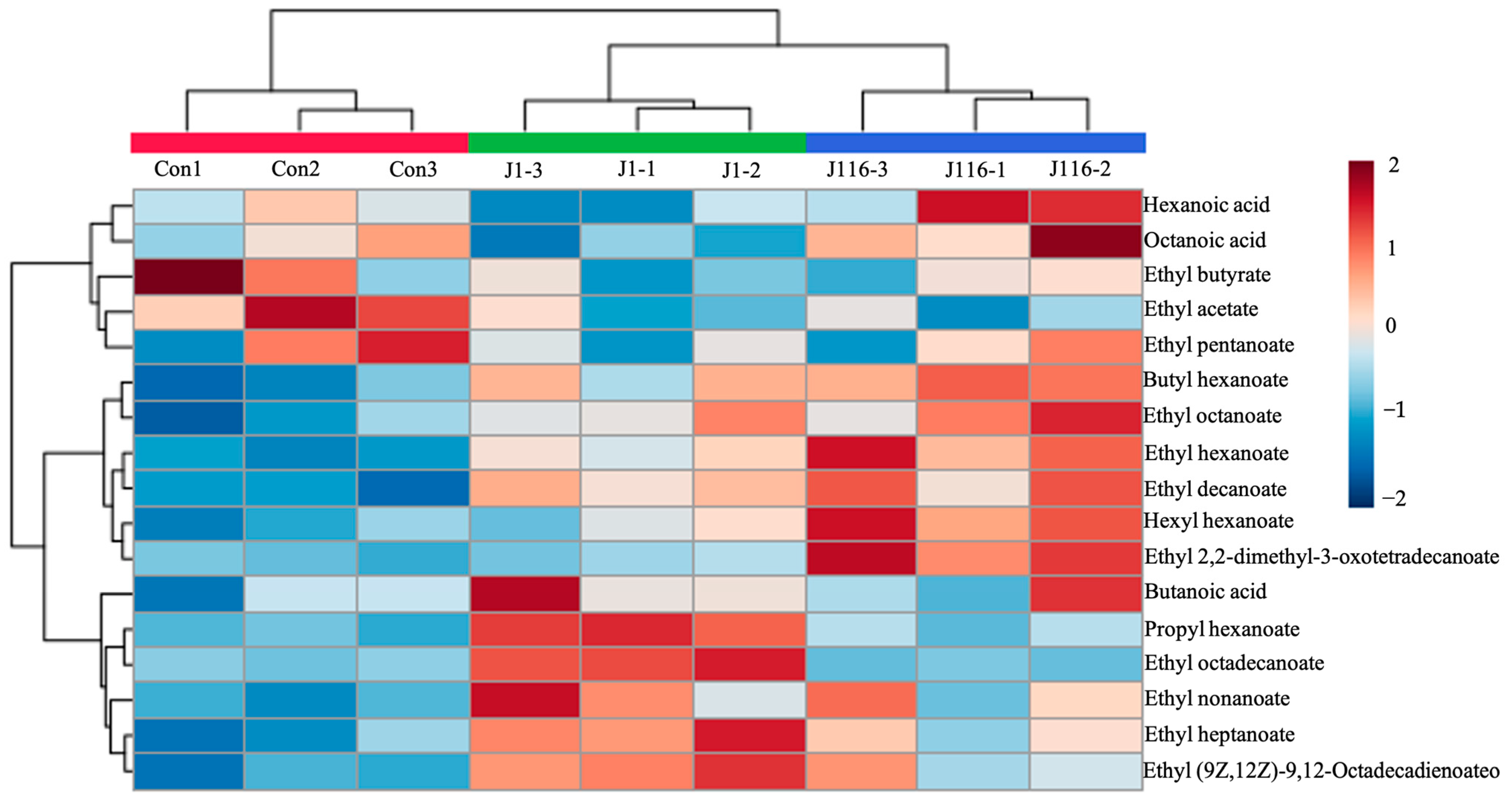
3.5. Effect of the Immobilized Strains on the VOC Content in Chinese Baijiu
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Le Kim, Y.K.; Koh, E.; Chung, H.J.; Kwon, H. Determination of Ethyl Carbamate in Some Fermented Korean Foods and Beverages. Food Addit. Contam. 2000, 17, 469–475. [Google Scholar] [CrossRef]
- Sakano, K.; Oikawa, S.; Hiraku, Y.; Kawanishi, S. Metabolism of Carcinogenic Urethane to Nitric Oxide Is Involved in Oxidative DNA Damage. Free Radic. Biol. Med. 2002, 33, 703–714. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization IARC Monographs on Evaluation of Carcinogenic Risk to Humans. Available online: https://web.archive.org/web/20170606065450/http://monographs.iarc.fr/ENG/Classification/index.php (accessed on 14 July 2023).
- Joint Fao/Who Expert Committee on Food Additives; International Program on Chemical Safety. Safety Evaluation of Certain Contaminants in Food; Food and Agriculture Organization: Rome, Italy, 2006; Volume 82, ISBN 9251054266. [Google Scholar]
- Pang, X.-N.; Li, Z.-J.; Chen, J.-Y.; Gao, L.-J.; Han, B.-Z. A Comprehensive Review of Spirit Drink Safety Standards and Regulations from an International Perspective. J. Food Prot. 2017, 80, 431–442. [Google Scholar] [CrossRef]
- Ough, C.S.; Crowell, E.A.; Gutlove, B.R. Carbamyl Compound Reactions with Ethanol. Am. J. Enol. Vitic. 1988, 39, 239–242. [Google Scholar] [CrossRef]
- Abt, E.; Incorvati, V.; Robin, L.P.; Redan, B.W. Occurrence of Ethyl Carbamate in Foods and Beverages: Review of the Formation Mechanisms, Advances in Analytical Methods, and Mitigation Strategies. J. Food Prot. 2021, 84, 2195–2212. [Google Scholar] [CrossRef]
- Benito, Á.; Jeffares, D.; Palomero, F.; Calderón, F.; Bai, F.Y.; Bähler, J.; Benito, S. Selected Schizosaccharomyces Pombe Strains Have Characteristics That Are Beneficial for Winemaking. PLoS ONE 2016, 11, 151102. [Google Scholar] [CrossRef]
- Wu, Q.; Cui, K.; Lin, J.; Zhu, Y.; Xu, Y. Urea Production by Yeasts Other than Saccharomyces in Food Fermentation. FEMS Yeast Res. 2017, 17, fox072. [Google Scholar] [CrossRef] [Green Version]
- Jayakody, L.N.; Lane, S.; Kim, H.; Jin, Y.-S. Mitigating Health Risks Associated with Alcoholic Beverages through Metabolic Engineering. Curr. Opin. Biotechnol. 2016, 37, 173–181. [Google Scholar] [CrossRef]
- Fang, R.; Dong, Y.; Li, H.; Chen, Q. Ethyl Carbamate Formation Regulated by Saccharomyces Cerevisiae ZJU in the Processing of Chinese Yellow Rice Wine. Int. J. Food Sci. Technol. 2015, 50, 626–632. [Google Scholar] [CrossRef]
- Jia, Y.; Zhou, J.; Du, G.; Chen, J.; Fang, F. Identification of an Urethanase from Lysinibacillus Fusiformis for Degrading Ethyl Carbamate in Fermented Foods. Food Biosci. 2020, 36, 100666. [Google Scholar] [CrossRef]
- Esti, M.; Fidaleo, M.; Moresi, M.; Tamborra, P. Modeling of Urea Degradation in White and Rosé Wines by Acid Urease. J. Agric. Food Chem. 2007, 55, 2590–2596. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.-W.; Kang, W.-K.; Jang, H.W.; Turner, T.L.; Kim, H.J. CAR1 Deletion by CRISPR/Cas9 Reduces Formation of Ethyl Carbamate from Ethanol Fermentation by Saccharomyces Cerevisiae. J. Ind. Microbiol. Biotechnol. 2016, 43, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Husnik, J.I.; Van Vuuren, H.J.J. Functional Enhancement of Sake Yeast Strains to Minimize the Production of Ethyl Carbamate in Sake Wine. J. Appl. Microbiol. 2010, 109, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, X.; Shen, C.; Lu, J.; Chen, J.; Xie, G. Decreased Ethyl Carbamate Generation during Chinese Rice Wine Fermentation by Disruption of CAR1 in an Industrial Yeast Strain. Int. J. Food Microbiol. 2014, 180, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zeng, X.; Wang, L.; Yu, S.; Brennan, M.A. Ethyl Carbamate Control by Genomic Regulation of Arginase in Saccharomyces Cerevisiae EC1118 in Sugarcane Juice Fermentation. J. Food Process. Preserv. 2017, 41, e13261. [Google Scholar] [CrossRef]
- Wu, D.; Xie, W.; Li, X.; Cai, G.; Lu, J.; Xie, G. Metabolic Engineering of Saccharomyces Cerevisiae Using the CRISPR/Cas9 System to Minimize Ethyl Carbamate Accumulation during Chinese Rice Wine Fermentation. Appl. Microbiol. Biotechnol. 2020, 104, 4435–4444. [Google Scholar] [CrossRef]
- Masaki, K. Features and Application Potential of Microbial Urethanases. Appl. Microbiol. Biotechnol. 2022, 106, 3431–3438. [Google Scholar] [CrossRef]
- Dong, N.; Xue, S.; Guo, H.; Xiong, K.; Lin, X.; Liang, H.; Ji, C.; Huang, Z.; Zhang, S. Genetic Engineering Production of Ethyl Carbamate Hydrolase and Its Application in Degrading Ethyl Carbamate in Chinese Liquor. Foods 2022, 11, 937. [Google Scholar] [CrossRef]
- Cui, K.; Wu, Q.; Xu, Y. Biodegradation of Ethyl Carbamate and Urea with Lysinibacillus Sphaericus MT33 in Chinese Liquor Fermentation. J. Agric. Food Chem. 2018, 66, 1583–1590. [Google Scholar] [CrossRef]
- Aylott, R.I.; McNeish, A.S.; Walker, D.A. Determination of Ethyl Carbamate in Distilled Spirits Using Nitrogen Specific and Mass Spectrometric Detection. J. Inst. Brew. 1987, 93, 382–386. [Google Scholar] [CrossRef]
- Lachenmeier, D.W.; Sarsh, B.; Rehm, J. The Composition of Alcohol Products from Markets in Lithuania and Hungary, and Potential Health Consequences: A Pilot Study. Alcohol Alcohol. 2009, 44, 93–102. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S.; Hu, P. A Survey of Levels of Ethyl Carbamate in Alcoholic Beverages in 2009–2012, Hebei Province, China. Food Addit. Contam. Part B 2013, 6, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Madrera, R.R.; Valles, B.S. Determination of Ethyl Carbamate in Cider Spirits by HPLC-FLD. Food Control 2009, 20, 139–143. [Google Scholar] [CrossRef]
- Liu, Y.P.; Dong, B.; Qin, Z.S.; Yang, N.J.; Lu, Y.; Yang, L.X.; Chang, F.Q.; Wu, Y.N. Ethyl Carbamate Levels in Wine and Spirits from Markets in Hebei Province, China. Food Addit. Contam. Part B Surveill 2011, 4, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Ji, L.; Han, X.; Wu, T.; Han, B.; Li, C.; Zhan, J.; Huang, W.; You, Y. Research Progress on the Application of Different Controlling Strategies to Minimizing Ethyl Carbamate in Grape Wine. Compr. Rev. Food Sci. Food Saf. 2023, 22, 1495–1516. [Google Scholar] [CrossRef] [PubMed]
- Shoeb, E.; Ahmed, N.; Akhter, J.; Badar, U.; Siddiqui, K.; ANSARI, F.; Waqar, M.; Imtiaz, S.; Akhtar, N.; Shaikh, Q.U.A. Screening and Characterization of Biosurfactant-Producing Bacteria Isolatedfrom the Arabian Sea Coast of Karachi. Turk. J. Biol. 2015, 39, 210–216. [Google Scholar] [CrossRef]
- Xu, W.; Wu, R.; Li, C.; Lü, J.; Ji, C.; Liang, H.; Sun, Z.; Lin, X. Separation and Identification of Yeast from Traditional Shrimp Paste and Their Carbon Utilization Characteristics. J. Chin. Inst. Food Sci. Technol. 2021, 21, 303–309. [Google Scholar] [CrossRef]
- Artíguez, M.L.; Martínez de Marañón, I. Effect of Pulsed Light Treatment on the Germination of Bacillus Subtilis Spores. Food Bioproc. Technol. 2015, 8, 478–485. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, Y.; Wang, D.; Xu, Y. Immobilized Rhodotorula Mucilaginosa: A Novel Urethanase-Producing Strain for Degrading Ethyl Carbamate. Appl. Biochem. Biotechnol. 2013, 171, 2220–2232. [Google Scholar] [CrossRef]
- Xia, Q.; Yuan, H.; Wu, C.; Zheng, J.; Zhang, S.; Shen, C.; Yi, B.; Zhou, R. An Improved and Validated Sample Cleanup Method for Analysis of Ethyl Carbamate in Chinese Liquor. J. Food Sci. 2014, 79, T1854–T1860. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; He, Z.; Wang, X.; Song, G.; Chen, H.; Lin, X.; Ji, C.; Zhang, S. Bacterial Profiles and Volatile Flavor Compounds in Commercial Suancai with Varying Salt Concentration from Northeastern China. Food Res. Int. 2020, 137, 109384. [Google Scholar] [CrossRef]
- Rivas, B.; Torre, P.; Domínguez, J.M.; Perego, P.; Converti, A.; Parajó, J.C. Carbon Material and Bioenergetic Balances of Xylitol Production from Corncobs by Debaryomyces Hansenii. Biotechnol. Prog. 2003, 19, 706–713. [Google Scholar] [CrossRef]
- Koganti, S.; Ju, L.K. Debaryomyces Hansenii Fermentation for Arabitol Production. Biochem. Eng. J. 2013, 79, 112–119. [Google Scholar] [CrossRef]
- Tian, S.; Zeng, W.; Zhou, J.; Du, G. Correlation between the Microbial Community and Ethyl Carbamate Generated during Huzhou Rice Wine Fermentation. Food Res. Int. 2022, 154, 111001. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Xie, G.; Wu, D.; Li, X.; Lu, J. A Lactobacillus Brevis Strain with Citrulline Re-Uptake Activity for Citrulline and Ethyl Carbamate Control during Chinese Rice Wine Fermentation. Food Biosci. 2020, 36, 100612. [Google Scholar] [CrossRef]
- Masaki, K.; Fujihara, K.; Kakizono, D.; Mizukure, T.; Okuda, M.; Mukai, N. Aspergillus Oryzae Acetamidase Catalyzes Degradation of Ethyl Carbamate. J. Biosci. Bioeng. 2020, 130, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Masaki, K.; Mizukure, T.; Kakizono, D.; Fujihara, K.; Fujii, T.; Mukai, N. New Urethanase from the Yeast Candida Parapsilosis. J. Biosci. Bioeng. 2020, 130, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.; Lin, J.; Yang, L.; Wu, M. Expression, Isolation, and Identification of an Ethanol-Resistant Ethyl Carbamate-Degrading Amidase from Agrobacterium Tumefaciens D3. J. Biosci. Bioeng. 2021, 132, 220–225. [Google Scholar] [CrossRef]
- Liu, X.; Fang, F.; Xia, X.; Du, G.; Chen, J. Stability Enhancement of Urethanase from Lysinibacillus Fusiformis by Site-Directed Mutagenesis. Shengwu Gongcheng Xuebao/Chin. J. Biotechnol. 2016, 32, 1233–1242. [Google Scholar] [CrossRef]
- Liu, Q.; Yao, X.; Liang, Q.; Li, J.; Fang, F.; Du, G.; Kang, Z. Molecular Engineering of Bacillus Paralicheniformis Acid Urease to Degrade Urea and Ethyl Carbamate in Model Chinese Rice Wine. J. Agric. Food Chem. 2018, 66, 13011–13019. [Google Scholar] [CrossRef]
- Van Gemert, L.J. Compilations of Odour Threshold Values in Air, Water and Other Media; Oliemans Punter: Houten, The Netherlands, 2003. [Google Scholar]
- Liu, S.Q.; Holland, R.; Crow, V.L. Esters and Their Biosynthesis in Fermented Dairy Products: A Review. Int. Dairy J. 2004, 14, 923–945. [Google Scholar] [CrossRef]
- Takahashi, T.; Ohara, Y.; Sueno, K. Breeding of a Sake Yeast Mutant with Enhanced Ethyl Caproate Productivity in Sake Brewing Using Rice Milled at a High Polishing Ratio. J. Biosci. Bioeng. 2017, 123, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, F.; Guo, J.; Liu, G.; Guo, X.; Xiao, D. Enhanced Ethyl Caproate Production of Chinese Liquor Yeast by Overexpressing EHT1 with Deleted FAA1. J. Ind. Microbiol. Biotechnol. 2014, 41, 563–572. [Google Scholar] [CrossRef] [PubMed]

| Glucose | Sucrose | Sorbitol | Glycerin | Mannitol | EC | |
|---|---|---|---|---|---|---|
| J1 | +++ | + | ++ | + | ++ | ++ |
| J10 | +++ | +++ | +++ | + | ++ | + |
| J14 | +++ | + | ++ | + | + | + |
| J34 | + | + | + | ++ | + | + |
| J53 | ++ | + | ++ | − | ++ | + |
| J78 | + | + | ++ | + | + | + |
| J116 | ++ | +++ | ++ | + | + | ++ |
| J128 | ++ | ++ | ++ | + | + | + |
| Compounds | Concentration (mg/L) | ||
|---|---|---|---|
| Control | J1 | J116 | |
| Ethyl acetate | 13.99 ± 1.48 a | 9.78 ± 1.24 b | 9.74 ± 1.22 b |
| Ethyl butyrate | 31.95 ± 4.49 a | 26.03 ± 1.98 a | 27.47 ± 2.04 a |
| Ethyl pentanoate | 15.81 ± 1.88 a | 14.46 ± 0.81 a | 15.15 ± 1.36 a |
| Ethyl hexanoate | 451.94 ± 10.56 c | 554.32 ± 15.28 b | 638.95 ± 37.72 a |
| Propyl hexanoate | 1.22 ± 0.18 b | 5.10 ± 0.27 a | 1.86 ± 0.40 b |
| Ethyl heptanoate | 20.30 ± 2.36 c | 32.11 ± 1.90 a | 26.09 ± 2.22 b |
| Butyl hexanoate | 11.29 ± 1.03 b | 15.11 ± 1.23 a | 16.91 ± 0.62 a |
| Ethyl octanoate | 46.05 ± 3.09 b | 54.54 ± 2.81 ab | 57.88 ± 4.01 a |
| Ethyl nonanoate | 1.77 ± 0.18 b | 3.63 ± 0.75 a | 3.00 ± 0.77 ab |
| Hexyl hexanoate | 23.02 ± 2.25 b | 27.45 ± 2.38 b | 36.17 ± 2.42 a |
| Ethyl decanoate | 3.77 ± 0.42 b | 6.69 ± 0.41 a | 7.45 ± 0.96 a |
| Ethyl 2-phenylacetate | 4.86 ± 0.23 b | 6.06 ± 0.53 a | 5.15 ± 0.51 ab |
| Ethyl 2,2-dimethyl-3-oxotetradecanoate | 8.04 ± 0.48 b | 9.29 ± 0.67 b | 17.57 ± 1.55 a |
| Ethyl hexadecanoate | 44.98 ± 2.03 b | 46.35 ± 2.74 b | 62.82 ± 7.32 a |
| Ethyl octadecanoate | 0.63 ± 0.16 b | 5.34 ± 0.33 a | 0.37 ± 0.12 b |
| Ethyl (E)-octadec-9-enoate | 15.71 ± 1.73 b | 44.84 ± 1.75 a | 14.58 ± 0.93 b |
| Ethyl (9Z,12Z)-9,12-Octadecadienoateo | 16.42 ± 1.86 c | 31.00 ± 1.86 a | 24.36 ± 3.65 b |
| Hexadecyl octanoate | 1.59 ± 0.29 a | 0.28 ± 0.22 b | 0.37 ± 0.03 b |
| Butanoic acid | 2.31 ± 0.19 a | 2.74 ± 0.28 a | 2.56 ± 0.34 a |
| Pentanoic acid | 1.38 ± 0.21 ab | 1.23 ± 0.15 b | 1.73 ± 0.18 a |
| Hexanoic acid | 43.33 ± 1.22 ab | 39.58 ± 2.00 b | 47.21 ± 3.69 a |
| Heptanoic acid | 1.53 ± 0.17 a | 1.41 ± 0.21 a | 1.81 ± 0.11 a |
| Octanoic acid | 2.51 ± 0.16 ab | 2.16 ± 0.12 b | 2.77 ± 0.25 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, S.; Dong, N.; Xiong, K.; Guo, H.; Dai, Y.; Liang, H.; Chen, Y.; Lin, X.; Zhu, B.; Zhang, S. The Screening and Isolation of Ethyl-Carbamate-Degrading Strains from Fermented Grains and Their Application in the Degradation of Ethyl Carbamate in Chinese Baijiu. Foods 2023, 12, 2843. https://doi.org/10.3390/foods12152843
Xue S, Dong N, Xiong K, Guo H, Dai Y, Liang H, Chen Y, Lin X, Zhu B, Zhang S. The Screening and Isolation of Ethyl-Carbamate-Degrading Strains from Fermented Grains and Their Application in the Degradation of Ethyl Carbamate in Chinese Baijiu. Foods. 2023; 12(15):2843. https://doi.org/10.3390/foods12152843
Chicago/Turabian StyleXue, Siyu, Naihui Dong, Kexin Xiong, Hui Guo, Yiwei Dai, Huipeng Liang, Yingxi Chen, Xinping Lin, Beiwei Zhu, and Sufang Zhang. 2023. "The Screening and Isolation of Ethyl-Carbamate-Degrading Strains from Fermented Grains and Their Application in the Degradation of Ethyl Carbamate in Chinese Baijiu" Foods 12, no. 15: 2843. https://doi.org/10.3390/foods12152843







