Prevalence and Types of Extended-Spectrum β-Lactamase-Producing Bacteria in Retail Seafood
Abstract
1. Introduction
2. Materials and Methods
2.1. Review Protocol and Registration
2.2. Data Source and Literature Search Strategy
2.3. Selection Process and Inclusion/Exclusion Criteria
2.4. Data Extraction
2.5. Data Analysis
3. Results
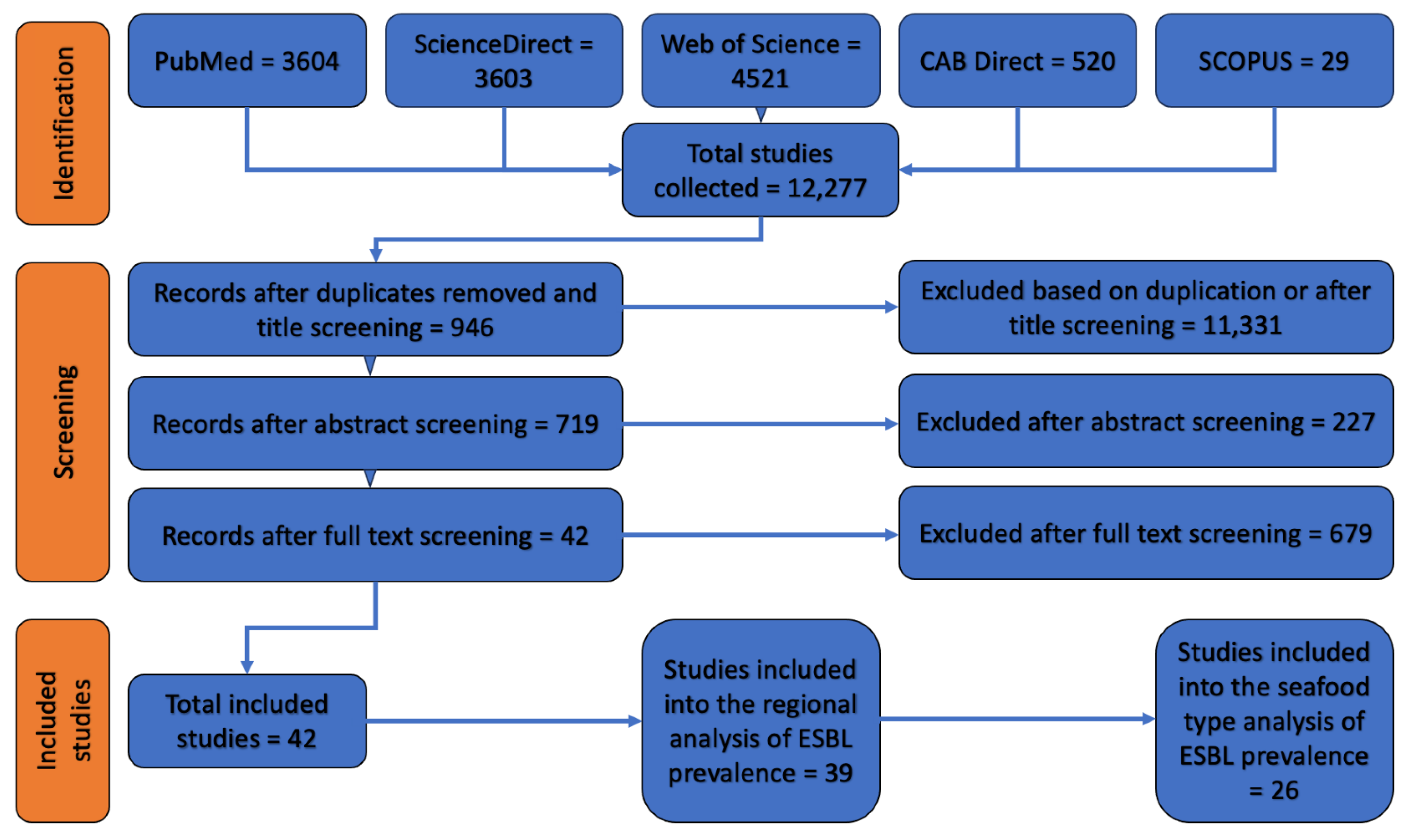
3.1. Literature Search Results
3.2. Prevalence of ESBL-Contaminated Products
3.3. Distribution of ESBL Types across Bacterial Hosts, Continents, and Seafood Types
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- European Commission. A European One Health Action Plan against Antimicrobial Resistance (AMR) CONTENTS. 2017. Available online: http://www.who.int/entity/drugresistance/documents/surveillancereport/en/index.html (accessed on 21 March 2022).
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 16 June 2022).
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Shamsrizi, P.; Gladstone, B.P.; Carrara, E.; Luise, D.; Cona, A.; Bovo, C.; Tacconelli, E. Variation of effect estimates in the analysis of mortality and length of hospital stay in patients with infections caused by bacteria-producing extended-spectrum beta-lactamases: A systematic review and meta-analysis. BMJ Open 2020, 10, e030266. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, J.; Guedes, C.; Silva, C.; Sá, S.; Oliveira, M.; Accioly, G.; Baylina, P.; Barata, P.; Pereira, C.; Fernandes, R. New CTX-M Group Conferring β-Lactam Resistance: A Compendium of Phylogenetic Insights from Biochemical, Molecular, and Structural Biology. Biology 2022, 11, 256. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Perestrelo, S.; Carreira, G.C.; Valentin, L.; Fischer, J.; Pfeifer, Y.; Werner, G.; Schmiedel, J.; Falgenhauer, L.; Imirzalioglu, C.; Chakraborty, T.; et al. Comparison of approaches for source attribution of ESBL-producing Escherichia coli in Germany. PLoS ONE 2022, 17, e0271317. [Google Scholar] [CrossRef]
- Martak, D.; Guther, J.; Verschuuren, T.D.; Valot, B.; Conzelmann, N.; Bunk, S.; Riccio, M.E.; Salamanca, E.; Meunier, A.; Henriot, C.P.; et al. Populations of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae are different in human-polluted environment and food items: A multicentre European study. Clin. Microbiol. Infect. 2021, 28, 447.e7–447.e14. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Dorado-García, A.; Van Duijkeren, E.; Van Den Bunt, G.; Dierikx, C.M.; Bonten, M.J.M.; Bootsma, M.C.J.; Schmitt, H.; Hald, T.; Evers, E.G.; et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet. Health 2019, 3, e357–e369. [Google Scholar] [CrossRef]
- Higgins, J.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.; Welch, V. Cochrane Handbook for Systematic Reviews of Interventions; Cochrane Training (6.3); John Wiley & Sons: Hoboken, NJ, USA, 2022; Available online: https://training.cochrane.org/handbook (accessed on 28 September 2022).
- PROSPERO. International Prospective Register of Systematic Reviews 2022. Available online: https://www.crd.york.ac.uk/prospero/ (accessed on 28 September 2022).
- Wickramanayake, M.V.K.S.; Dahanayake, P.S.; Hossain, S.; De Zoysa, M.; Heo, G.-J. Aeromonas spp. Isolated from Pacific Abalone (Haliotis discus hannai) Marketed in Korea: Antimicrobial and Heavy-Metal Resistance Properties. Curr. Microbiol. 2020, 77, 1707–1715. [Google Scholar] [CrossRef]
- Dahanayake, P.; Hossain, S.; Wickramanayake, M.; Heo, G. Prevalence of virulence and extended-spectrum β-lactamase (ESBL) genes harbouring Vibrio spp. isolated from cockles (Tegillarca granosa) marketed in Korea. Lett. Appl. Microbiol. 2019, 71, 61–69. [Google Scholar] [CrossRef]
- Wickramanayake, M.V.K.S.; Dahanayake, P.S.; Hossain, S.; De Silva, B.C.J.; Heo, G.J. Characterisation of pathogenic Vibrio spp. isolated from live pacific abalone (Haliotis discus hannai ino, 1953) marketed in South Korea. Indian J. Fish. 2020, 67, 105–113. [Google Scholar] [CrossRef]
- Le, Q.P.; Ueda, S.; Nguyen, T.N.H.; Dao, T.V.K.; Van Hoang, T.A.; Tran, T.T.N.; Hirai, I.; Nakayama, T.; Kawahara, R.; Do, T.H.; et al. Characteristics of Extended-Spectrum β-Lactamase–Producing Escherichia coli in Retail Meats and Shrimp at a Local Market in Vietnam. Foodborne Pathog. Dis. 2015, 12, 719–725. [Google Scholar] [CrossRef]
- Beshiru, A.; Okareh, O.T.; Okoh, A.I.; Igbinosa, E.O. Detection of antibiotic resistance and virulence genes of Vibrio strains isolated from ready-to-eat shrimps in Delta and Edo States, Nigeria. J. Appl. Microbiol. 2020, 129, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Hagiya, H.; Akeda, Y.; Aye, M.M.; Win, H.P.M.; Sakamoto, N.; Shanmugakani, R.K.; Takeuchi, D.; Nishi, I.; Ueda, A.; et al. Dissemination of carbapenemase-producing Enterobacteriaceae harbouring blaNDM or blaIMI in local market foods of Yangon, Myanmar. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Pongsilp, N.; Nimnoi, P. Diversity and antibiotic resistance patterns of enterobacteria isolated from seafood in Thailand. CyTA-J. Food 2018, 16, 793–800. [Google Scholar] [CrossRef]
- Singh, A.S.; Nayak, B.B.; Kumar, S.H. High Prevalence of Multiple Antibiotic-Resistant, Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli in Fresh Seafood Sold in Retail Markets of Mumbai, India. Vet. Sci. 2020, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.T.T.; Alter, T.; Roesler, U.; Roschanski, N.; Huehn, S. Investigation of Extended-Spectrum and AmpC β-Lactamase–Producing Enterobacteriaceae from Retail Seafood in Berlin, Germany. J. Food Prot. 2018, 81, 1079–1086. [Google Scholar] [CrossRef]
- Singh, A.S.; Lekshmi, M.; Prakasan, S.; Nayak, B.B.; Kumar, S. Multiple Antibiotic-Resistant, Extended Spectrum-β-Lactamase (ESBL)-Producing Enterobacteria in Fresh Seafood. Microorganisms 2017, 5, 53. [Google Scholar] [CrossRef]
- Hossain, S.; Wickramanayake, M.V.K.S.; Dahanayake, P.S.; Heo, G.-J. Occurrence of Virulence and Extended-Spectrum β-Lactamase Determinants in Vibrio spp. Isolated from Marketed Hard-Shelled Mussel (Mytilus coruscus). Microb. Drug Resist. 2020, 26, 391–401. [Google Scholar] [CrossRef]
- Beshiru, A.; Igbinosa, I.H.; Igbinosa, E.O. Prevalence of Antimicrobial Resistance and Virulence Gene Elements of Salmonella Serovars From Ready-to-Eat (RTE) Shrimps. Front. Microbiol. 2019, 10, 1613. [Google Scholar] [CrossRef] [PubMed]
- Elhadi, N. Prevalence of extended-spectrum-β-lactamase-producing Escherichia coli in imported frozen freshwater fish in Eastern Province of Saudi Arabia. Saudi J. Med. Med. Sci. 2016, 4, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Dahanayake, P.S.; Hossain, S.; Wickramanayake, M.; Heo, G. Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam (Ruditapes philippinarum) in Korea. J. Appl. Microbiol. 2019, 127, 941–952. [Google Scholar] [CrossRef]
- Dahanayake, P.; Hossain, S.; Wickramanayake, M.; Heo, G. Prevalence of virulence and antimicrobial resistance genes in Aeromonas species isolated from marketed cockles (Tegillarca granosa) in Korea. Lett. Appl. Microbiol. 2020, 71, 94–101. [Google Scholar] [CrossRef]
- Nguyen, D.P.; Nguyen, T.A.D.; Le, T.H.; Tran, N.M.D.; Ngo, T.P.; Dang, V.C.; Kawai, T.; Kanki, M.; Kawahara, R.; Jinnai, M.; et al. Dissemination of Extended-Spectrumβ-Lactamase- and AmpCβ-Lactamase-ProducingEscherichia coliwithin the Food Distribution System of Ho Chi Minh City, Vietnam. BioMed Res. Int. 2016, 2016, 8182096. [Google Scholar] [CrossRef]
- de Greef, S.; Mounton, J. NethMap 2017: Consumption of Antimicrobial Agents and Antimi-Crobial Resistance among Medically Important Bacteria in the Netherlands/MARAN 2017: Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2016|RIVM. 2017. Available online: https://www.rivm.nl/publicaties/nethmap-2017-consumption-of-antimicrobial-agents-and-antimicrobial-resistance-among (accessed on 6 April 2021).
- de greeff, S.; Mounton, J. NethMap 2018: Consumption of Antimicrobial Agents and Antimi-Crobial Resistance among Medically Important Bacteria in the Netherlands/MARAN 2018: Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2017|RIVM. 2018. Available online: https://www.rivm.nl/publicaties/nethmap-2018-consumption-of-antimicrobial-agents-and-antimicrobial-resistance-among (accessed on 6 April 2021).
- de Greeff, S.; Mounton, J.; Heoing, A.; Verduin, C. NethMap 2019: Consumption of Antimi-Crobial Agents and Antimicrobial Resistance among Medically Important Bacteria in the Netherlands/MARAN 2019: Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2018|RIVM. 2019. Available online: https://www.rivm.nl/publicaties/nethmap-2019-consumption-of-antimicrobial-agents-and-antimicrobial-resistance-among (accessed on 6 April 2021).
- Danmap. Danmap, Danish Programme for Surveillance of Antimicrobial Consumption and Resistance in Bacteria from Food Animals, Food and Humans. 2018. Available online: https://www.danmap.org/ (accessed on 6 April 2021).
- Cerdeira, L.; Monte, D.F.; Fuga, B.; Sellera, F.P.; Neves, I.; Rodrigues, L.; Landgraf, M.; Lincopan, N. Genomic insights of Klebsiella pneumoniae isolated from a native Amazonian fish reveal wide resistome against heavy metals, disinfectants, and clinically relevant antibiotics. Genomics 2020, 112, 5143–5146. [Google Scholar] [CrossRef]
- Nadimpalli, M.; Fabre, L.; Yith, V.; Sem, N.; Gouali, M.; Delarocque-Astagneau, E.; Sreng, N.; Le Hello, S.; Raheliarivao, B.T.; Randrianirina, F.; et al. CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J. Antimicrob. Chemother. 2018, 74, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, F.; Zheng, Y.; Jiao, X.; Guo, L. Isolation, Molecular Characterization and Antibiotic Susceptibility Pattern of Vibrio parahaemolyticus from Aquatic Products in the Southern Fujian Coast, China. J. Microbiol. Biotechnol. 2020, 30, 856–867. [Google Scholar] [CrossRef]
- Nguyen, D.T.A.; Kanki, M.; Nguyen, P.D.; Le, H.T.; Ngo, P.T.; Tran, D.N.M.; Le, N.H.; Van Dang, C.; Kawai, T.; Kawahara, R.; et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016, 236, 115–122. [Google Scholar] [CrossRef]
- Osman, K.M.; da Silva Pires, Á.; Franco, O.L.; Saad, A.; Hamed, M.; Naim, H.; Ali, A.H.; Elbehiry, A. Nile tilapia (Oreochromis niloticus) as an aquatic vector for Pseudomonas species of medical importance: Antibiotic Resistance Association with Biofilm Formation, Quorum Sensing and Virulence. Aquaculture 2020, 532, 736068. [Google Scholar] [CrossRef]
- Nakayama, T.; Hoa, T.T.T.; Huyen, H.M.; Yamaguchi, T.; Jinnai, M.; Minh, D.T.N.; Hoang, O.N.; Le Thi, H.; Thanh, P.N.; Hoai, P.H.; et al. Isolation of carbapenem-resistant Enterobacteriaceae harbouring NDM-1, 4, 5, OXA48 and KPC from river fish in Vietnam. Food Control 2021, 133, 108594. [Google Scholar] [CrossRef]
- Dahanayake, P.; Hossain, S.; Wickramanayake, M.; Wimalasena, S.; Heo, G. Manila clam (Ruditapes philippinarum) marketed in Korea as a source of vibrios harbouring virulence and β-lactam resistance genes. Lett. Appl. Microbiol. 2019, 71, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Yen, N.T.P.; Nhung, N.T.; Van, N.T.B.; Van Cuong, N.; Chau, L.T.T.; Trinh, H.N.; Van Tuat, C.; Tu, N.D.; Lan, N.P.H.; Campbell, J.; et al. Antimicrobial residues, non-typhoidal Salmonella, Vibrio spp. and associated microbiological hazards in retail shrimps purchased in Ho Chi Minh city (Vietnam). Food Control 2019, 107, 106756. [Google Scholar] [CrossRef] [PubMed]
- Dib, A.L.; Agabou, A.; Chahed, A.; Kurekci, C.; Moreno, E.; Espigares, M.; Espigares, E. Isolation, molecular characterization and antimicrobial resistance of enterobacteriaceae isolated from fish and seafood. Food Control 2018, 88, 54–60. [Google Scholar] [CrossRef]
- Silva, V.; Nunes, J.; Gomes, A.; Capita, R.; Alonso-Calleja, C.; Pereira, J.E.; Torres, C.; Igrejas, G.; Poeta, P. Detection of Antibiotic Resistance in Escherichia coli Strains: Can Fish Commonly Used in Raw Preparations such as Sushi and Sashimi Constitute a Public Health Problem? J. Food Prot. 2019, 82, 1130–1134. [Google Scholar] [CrossRef]
- Gawish, M.F.; Ahmed, A.M.; Torky, H.A.; Shimamoto, T. Prevalence of extended-spectrum β-lactamase (ESBL)-producing Salmonella enterica from retail fishes in Egypt: A major threat to public health. Int. J. Food Microbiol. 2021, 351, 109268. [Google Scholar] [CrossRef]
- Sivaraman, G.; Sudha, S.; Muneeb, K.; Shome, B.; Holmes, M.; Cole, J. Molecular assessment of antimicrobial resistance and virulence in multi drug resistant ESBL-producing Escherichia coli and Klebsiella pneumoniae from food fishes, Assam, India. Microb. Pathog. 2020, 149, 104581. [Google Scholar] [CrossRef] [PubMed]
- Vitas, A.I.; Naik, D.; Pérez-Etayo, L.; González, D. Increased exposure to extended-spectrum β-lactamase-producing multidrug-resistant Enterobacteriaceae through the consumption of chicken and sushi products. Int. J. Food Microbiol. 2018, 269, 80–86. [Google Scholar] [CrossRef]
- Moremi, N.; Manda, E.V.; Falgenhauer, L.; Ghosh, H.; Imirzalioglu, C.; Matee, M.; Chakraborty, T.; Mshana, S.E. Predominance of CTX-M-15 among ESBL Producers from Environment and Fish Gut from the Shores of Lake Victoria in Mwanza, Tanzania. Front. Microbiol. 2016, 7, 1862. [Google Scholar] [CrossRef]
- Le, H.V.; Kawahara, R.; Khong, D.T.; Tran, H.T.; Nguyen, T.N.; Pham, K.N.; Jinnai, M.; Kumeda, Y.; Nakayama, T.; Ueda, S.; et al. Widespread dissemination of extended-spectrum β-lactamase-producing, multidrug-resistant Escherichia coli in livestock and fishery products in Vietnam. Int. J. Food Contam. 2015, 2, 1–6. [Google Scholar] [CrossRef]
- Sapugahawatte, D.N.; Li, C.; Zhu, C.; Dharmaratne, P.; Wong, K.T.; Lo, N.; Ip, M. Prevalence and Characteristics of Extended-Spectrum-β-Lactamase-Producing and Carbapenemase-Producing Enterobacteriaceae from Freshwater Fish and Pork in Wet Markets of Hong Kong. mSphere 2020, 5, e00107-20. [Google Scholar] [CrossRef] [PubMed]
- Sola, M.; Mani, Y.; Saras, E.; Drapeau, A.; Grami, R.; Aouni, M.; Madec, J.-Y.; Haenni, M.; Mansour, W. Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Enterobacterales from Tunisian Seafood. Microorganisms 2022, 10, 1364. [Google Scholar] [CrossRef]
- Kavinesan, K.; Sugumar, G.; Chrisolite, B.; Muthiahsethupathy, A.; Sudarshan, S.; Parthiban, F.; Mansoor, M. Phenotypic and genotypic characterization of pathogenic Escherichia coli identified in resistance mapping of β-lactam drug-resistant isolates from seafood along Tuticorin coast. Environ. Sci. Pollut. Res. 2023, 30, 68111–68128. [Google Scholar] [CrossRef]
- Nakayama, T.; Yamamoto, S.; Ohata, N.; Yamaguchi, T.; Jinnai, M.; Minh, D.T.N.; Hoang, O.N.; Le Thi, H.; Thanh, P.N.; Hoai, P.H.; et al. Common presence of plasmid encoding blaCTX-M-55 in extended-spectrum β-lactamase-producing Salmonella enterica and Escherichia coli isolates from the same edible river fish. J. Microorg. Control 2023, 28, 49–56. [Google Scholar] [CrossRef]
- Thongkao, K.; Sudjaroen, Y. Screening of antibiotic resistance genes in pathogenic bacteria isolated from tiny freshwater shrimp (Macrobrachium lanchesteri) and “Kung Ten”, the uncooked Thai food. J. Adv. Vet.-Anim. Res. 2020, 7, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.-J.; Woo, G.-J. Characterization of Antimicrobial Resistance of Escherichia coli Recovered from Foods of Animal and Fish Origin in Korea. J. Food Prot. 2012, 75, 966–972. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of World Fisheries and Aquaculture 2022; Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Directorate-General for Maritime Affairs and Fisheries. EU Consumer Habits Regarding Fishery and Aquaculture Products: Report; European Commission; 2021; Issue January. Available online: https://data.europa.eu/doi/10.2771/87688 (accessed on 21 July 2023).
- FAO. The State of World Fisheries and Aquaculture 2020. In Sustainability in Action; FAO: Rome, Italy, 2020; Volume 32, p. 244. [Google Scholar] [CrossRef]
- Weissfeld, A.S. Infections from Eating Raw or Undercooked Seafood. Clin. Microbiol. Newsl. 2014, 36, 17–21. [Google Scholar] [CrossRef]
- Golden, O.; Caldeira, A.J.; Santos, M.J. Raw fish consumption in Portugal: A survey on trends in consumption and consumer characteristics. Food Control 2022, 135, 108810. [Google Scholar] [CrossRef]
- Schar, D.; Klein, E.Y.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in aquaculture. Sci. Rep. 2020, 10, 21878. [Google Scholar] [CrossRef]
- Woerther, P.-L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in Human Fecal Carriage of Extended-Spectrum β-Lactamases in the Community: Toward the Globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef]
- Campos, C.J.A.; Kershaw, S.R.; Lee, R.J. Environmental Influences on Faecal Indicator Organisms in Coastal Waters and Their Accumulation in Bivalve Shellfish. Estuaries Coasts 2013, 36, 834–853. [Google Scholar] [CrossRef]
- Reverter, M.; Sarter, S.; Caruso, D.; Avarre, J.-C.; Combe, M.; Pepey, E.; Pouyaud, L.; Vega-Heredía, S.; de Verdal, H.; Gozlan, R.E. Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 2020, 11, 1870. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef]
- Fischer, J.; Rodríguez, I.; Baumann, B.; Guiral, E.; Beutin, L.; Schroeter, A.; Kaesbohrer, A.; Pfeifer, Y.; Helmuth, R.; Guerra, B. bla CTX-M-15-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J. Antimicrob. Chemother. 2014, 69, 2951–2958. [Google Scholar] [CrossRef] [PubMed]
- Zamudio, R.; Boerlin, P.; Beyrouthy, R.; Madec, J.-Y.; Schwarz, S.; Mulvey, M.R.; Zhanel, G.G.; Cormier, A.; Chalmers, G.; Bonnet, R.; et al. Dynamics of extended-spectrum cephalosporin resistance genes in Escherichia coli from Europe and North America. Nat. Commun. 2022, 13, 7490. [Google Scholar] [CrossRef]
- Sargeant, J.; Amezcua, M.; Waddell, L. A Guide to Conducting Systematic Reviews in Agri-Food Public Health; Public Health Agency of Canada: Hamilton, ON, Canada, 2005. [Google Scholar]
- EFSA. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2011. EFSA J. 2013, 11, 359. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef]


| Study Type [Reference] | Data Collection/Publication Year | Study Country | Seafood Origin (N Samples) | Seafood Type(s) (N Samples) | Sample Size (N Samples) | ESBL Prevalence (%) |
|---|---|---|---|---|---|---|
| Original article [15] | 2018/2020 | Korea | Korea (120) | Snail | 120 | 29 (24.2) |
| Original article [16] | 2018/2019 | Korea | Korea (145) | Clam | 145 | 36 (24.8) |
| Original article [17] | 2018/2020 | Korea | Korea (120) | Snail | 120 | 32 (26.7) |
| Original article [18] | 2013/2015 | Vietnam | Vietnam (60) | Shrimp | 60 | 11 (18.3) |
| Original article [19] | 2016/2020 | Nigeria | Unknown (1440) | Shrimp | 1440 | 120 (8.3) |
| Original article [20] | 2017/2019 | Myanmar | Unknown (21) | Clam | 5 | 1 (20.0) |
| Shrimp | 16 | 1 (6.3) | ||||
| Original article [21] | 2014/2018 | Thailand | Unknown (35) | Undefined seafood | 35 | 31 (88.6) |
| Original article [22] | 2016/2020 | India | Unknown (50) | Sardine | 5 | 1 (20.0) |
| Perch | 5 | 1 (20.0) | ||||
| Croaker | 3 | 1 (33.3) | ||||
| Sea Bass | 2 | 1 (50.0) | ||||
| Sea Bream | 2 | 1 (50.0) | ||||
| Moon fish | 2 | 1 (50.0) | ||||
| Anchovy | 3 | 1 (33.3) | ||||
| Lizard fish | 4 | 1 (25.0) | ||||
| Belt fish | 2 | 1 (50.0) | ||||
| Moon tail | 2 | 1 (50.0) | ||||
| Mackerel | 2 | 1 (50.0) | ||||
| Gizzard | 3 | 1 (33.3.0) | ||||
| Pomfret | 2 | 1 (50.0) | ||||
| Shrimp | 5 | 1 (20.0) | ||||
| Shrimp | 3 | 1 (33.3.0) | ||||
| Clam | 3 | 1 (33.3.0) | ||||
| Squid | 2 | 0 | ||||
| Original article [23] | 2015/2018 | Germany | Bangladesh (14), Denmark (14), Ecuador (12), France (7), Germany (8), India (10), Ireland (12), Italy (17), Netherlands (12), Spain (1), Vietnam (4), Unknown (49) | Shrimp, mussel, clam, cockle | 160 | 31 (19.4) |
| Original article [24] | 2013/2017 | India | Unknown (19) | Undefined seafood | 19 | 19 (100.0) |
| Original article [25] | 2018/2019 | Korea | Korea (275) | Mussel | 275 | 32 (11.6) |
| Original article [26] | 2016/2019 | Nigeria | Unknown (1440) | Shrimp | 1440 | 0 |
| Original article [27] | 2012/2016 | Saudi Arabia | Thailand (260), India (75), Vietnam (35), Myanmar (35) | Catfish | 65 | 32 (49.2) |
| Tilapia | 60 | 18 (30.0) | ||||
| Carfoo | 50 | 2 (4.0) | ||||
| Tilapia | 75 | 28 (37.3) | ||||
| Mirgal | 45 | 3 (6.7) | ||||
| Rohu | 40 | 5 (12.5) | ||||
| Milkfish | 35 | 19 (54.3) | ||||
| Rohu | 35 | 0 | ||||
| Original article [28] | 2018/2019 | Korea | Korea (120) | Cockle | 120 | 32 (26.7) |
| Original article [29] | 2018/2019 | Korea | Korea (120) | Cockle | 120 | 32 (26.7) |
| Original article [30] | 2012/2016 | Vietnam | Vietnam (154) | Undefined seafood | 101 | 1 (1.0) |
| Shrimp | 53 | 0 | ||||
| Surveillance report [31] | 2017/2018 | Netherlands | Asia (56) | Undefined seafood | 56 | 7 (12.5) |
| Surveillance report [32] | 2018/2019 | Netherlands | Asia (304) | Undefined seafood | 304 | 5 (1.7) |
| Surveillance report [33] | 2019/2020 | Netherlands | Asia (304) | Undefined seafood | 304 | 3 (1.0) |
| Surveillance report [34] | 2017/2018 | Denmark | Asia (300) | Undefined seafood | 300 | 1 (0.3) |
| Original article [35] | 2019/2020 | Brazil | Brazil (1) | Catfish | 1 | 1 (100) |
| Original article [36] | 2016/2018 | Cambodia | Unknown (60) | Undefined seafood | 60 | 10 (16.7) |
| Original article [37] | 2018/2020 | China | China (200) | Seafood | 200 | 27 (13.5) |
| Original article [38] | 2015/2016 | Vietnam | Vietnam (82) | Seafood | 82 | 24 (29.3) |
| Original article [39] | 2017/2020 | Egypt | Egypt (100) | Tilapia | 100 | 29 (29.0) |
| Original article [40] | 2019/2022 | Vietnam | Unknown (103) | Undefined seafood | 103 | 10 (9.7) |
| Original article [41] | 2018/2019 | Korea | Korea (120) | Clam | 120 | 27 (22.5) |
| Original article [42] | 2018/2020 | Vietnam | Unknown (40) | Shrimp | 40 | 24 (60.0) |
| Original article [30] | 2012/2016 | Vietnam | Vietnam (82) | Undefined seafood | 82 | 37 (45.1) |
| Original article [43] | 2015/2018 | Algeria | Algeria (14) | Sardines | 10 | 4 (40.0) |
| Shrimp | 4 | 0 | ||||
| Original article [44] | 2017/2019 | Portugal | Unknown (150) | Tuna (sushi) | 30 | 1 (3.3) |
| Seabass (sushi) | 30 | 1 (3.3) | ||||
| Salmon (sushi) | 30 | 1 (3.3) | ||||
| Snapper (sushi) | 30 | 0 | ||||
| Bramble shark (sushi) | 30 | 0 | ||||
| Original article [45] | 2019/2021 | Egypt | Egypt (200) | Tilapia | 100 | 14 (14) |
| Mullet | 100 | 5 (5.0) | ||||
| Original article [46] | 2019/2020 | India | Unknown (79) | Piranha, Catfish, shrimp, carp, Snakehead, Eel, Puria, Barb, Potasi, Loach, perch, Bulla machi, Karkaria, featherback, Narva | 79 | 65 (82.3) |
| Original article [47] | 2016/2018 | Spain | Unknown (97) | Undefined seafood | 97 | 16 (16.5) |
| Original article [48] | 2015/2016 | Tanzania | Tanzania (196) | Tilapia | 196 | 26 (13.3) |
| Original article [49] | 2014/2015 | Vietnam | Unknown (124) | Shrimp | 60 | 45 (75.0) |
| Undefined seafood | 64 | 40 (62.5) | ||||
| Original article [50] | 2018/2020 | China | China (411) | Snakehead | 213 | 2 (1.0) |
| Carp | 198 | 1 (0.5) | ||||
| Original article [51] | 2016/2022 | Tunisia | Tunisia (1716) | Sea bream | 485 | 8 (1.7) |
| Sea Bass | 156 | 1 (0.7) | ||||
| Clam | 1075 | 18 (1.7) | ||||
| Original article [52] | 2021/2023 | India | India (17) | Undefined seafood | 17 | 7 (41.2) |
| Original article [53] | 2022/2023 | Vietnam | Unknown (80) | Snakehead, tilapia, carp, catfish, anabas | 80 | 3 (3.8) |
| Original article [54] | 2019/2019 | Thailand | Thailand (120) | Shrimp | 120 | N/A |
| Original article [55] | 2004/2011 | Korea | Unknown (N/A) | Undefined seafood | N/A | N/A |
| Reference | Bacterial Isolation | Detection of Presumptive ESBL | ESBL Confirmation and Typing |
|---|---|---|---|
| [15] | Sample homogenate enriched in peptone broth and plated onto Aeromonas base (RYAN) agar | Double-disk synergy test | PCR at class level |
| [16] | Sample homogenate enriched in peptone broth and plated onto Aeromonas base (RYAN) agar | Double-disk synergy test | PCR at class level |
| [17] | Sample homogenate enriched in peptone broth and plated onto thiosulphate citrate bile salts sucrose (TCBS) agar | Double-disk synergy test | PCR at class level |
| [18] | Sample homogenate enriched in peptone broth and plated onto tryptone bile X-glucuronide (TBX) agar containing 2 μg/mL of cefotaxime | Double-disk synergy test | PCR at group level |
| [19] | Sample homogenate enriched in peptone broth and plated onto thiosulphate citrate bile salts sucrose (TCBS) agar | Double-disk synergy test | PCR at class level |
| [20] | Sample homogenate enriched in peptone broth and plated onto chromogenic ECC (CHROMagar) agar containing 0.25 μg/mL meropenem | Double-disk synergy test | WGS |
| [21] | Sample homogenate enriched in peptone broth and plated onto violet-red bile glucose (VRBG) agar | Double-disk synergy test | PCR at class level |
| [22] | Sample homogenate enriched in tryptone broth and plated onto MacConkey agar | Double-disk synergy test | PCR at class level |
| [23] | Non-homogenized sample enriched in peptone broth and plated onto MacConkey agar containing 1 μg/mL cefotaxime | Double-disk synergy test | PCR at group level and WGS |
| [24] | Sample enriched in enterobacteria enrichment (EE) broth and plated onto MacConkey agar | Double-disk synergy test | PCR at class level |
| [25] | Sample homogenate enriched in peptone broth and plated onto thiosulphate citrate bile salts sucrose (TCBS) agar | Double-disk synergy test | PCR at class level |
| [26] | Sample homogenate enriched in tryptone soy broth (TSB) and plated onto xylose lysine deoxycholate (XLD) agar and hektoen enteric agar | Double-disk synergy test | PCR at class level |
| [27] | Sample homogenate enriched in EC broth and plated onto ESBL chromogenic (CHROMagar) agar | Double-disk synergy test | PCR at class level |
| [28] | Sample homogenate enriched in peptone broth and plated onto Aeromonas base (RYAN) agar | Double-disk synergy test | PCR at class level |
| [29] | Sample homogenate enriched in peptone broth and plated onto thiosulphate citrate bile salts sucrose (TCBS) agar | Double-disk synergy test | PCR at class level |
| [30] | Sample homogenate enriched in peptone broth and plated onto xylose lysine deoxycholate agar (XLD) and CHROMagar Salmonella (CHROMagar) | Double-disk synergy test | PCR at group level |
| [31] | Unspecified | MIC determination using broth microdilution | PCR at variant level |
| [32] | Unspecified | MIC determination using broth microdilution | PCR at variant level |
| [33] | Unspecified | MIC determination using broth microdilution | PCR at variant level |
| [34] | Unspecified | MIC determination using broth microdilution | WGS |
| [35] | Sample rinsed in MacConkey broth and rinse directly plated onto MacConkey agar containing 2 μg/mL cefotaxime | Double-disk synergy test | WGS |
| [36] | Sample homogenate enriched in peptone broth and plated onto xylose lysine deoxycholate (XLD) agar containing 2 μg/mL cefotaxime | Double-disk synergy test | PCR at variant level |
| [37] | Sample streaked onto thiosulphate citrate bile salts sucrose (TCBS) agar | Double-disk synergy test | PCR at class level |
| [38] | Sample homogenate enriched in peptone broth and plated onto chromogenic ECC (CHROMagar) agar | Double-disk synergy test | PCR at class level |
| [39] | Sample streaked onto Columbia Blood (CBA) Agar | Double-disk synergy test | PCR at variant level |
| [40] | Sample homogenate enriched in peptone broth and plated onto chromogenic ECC (CHROMagar) agar | Double-disk synergy test | PCR at group level |
| [41] | Sample homogenate enriched in peptone broth and plated onto chromogenic Rambach agar (CHROMagar) | Double-disk synergy test | PCR at class level |
| [42] | Sample homogenate enriched in peptone broth and plated onto thiosulphate citrate bile salts sucrose (TCBS) agar | Double-disk synergy test | PCR at group level |
| [30] | Sample homogenate enriched in peptone broth and plated onto chromogenic ECC (CHROMagar) agar containing 1 μg/mL cefotaxime | Double-disk synergy test | PCR at class level |
| [43] | Sample homogenate enriched in peptone broth and plated onto xylose lysine deoxycholate (XLD) agar | Double-disk synergy test | PCR at variant level |
| [44] | Sample homogenate enriched in peptone broth and plated onto Levine (EMB) agar with and without cefotaxime (2 μg/mL) | Double-disk synergy test | PCR at class level |
| [45] | Sample homogenate enriched in peptone broth and plated onto xylose lysine deoxycholate (XLD) agar | Double-disk synergy test | PCR at variant level |
| [46] | Sample homogenate enriched in Brilliant Green Bile Lactose (BGBLB) Broth and plated onto MacConkey agar containing 1 μg/mL cefotaxime | Double-disk synergy test | PCR at variant level |
| [47] | Sample homogenate enriched in peptone Broth and plated onto chromogenic ESBL (ChromID) agar containing 1 μg/mL cefotaxime | MIC determination using broth microdilution | PCR at variant level |
| [48] | Sample homogenized in 0.9% saline before plating onto MacConkey agar containing 2 μg/mL cefotaxime | Double-disk synergy test | WGS |
| [49] | Sample homogenate enriched in peptone broth and plated onto chromogenic ECC (CHROMagar) agar containing 1 μg/mL cefotaxime | Double-disk synergy test | PCR at group level |
| [50] | Sample homogenized in saline before plating onto chromogenic ESBL (ChromID) agar | Double-disk synergy test | PCR at group level |
| [51] | Sample enriched in peptone broth and plated onto MacConkey agar containing 2 μg/mL cefotaxime | Double-disk synergy test | WGS |
| [52] | Sample homogenate enriched in EC broth and plated onto sorbitol (MUG) agar with tellurite-cefixime supplement | Double-disk synergy test | PCR at class level |
| [53] | Sample homogenate enriched in peptone broth and plated onto CHROMagar Salmonella (CHROMagar) | Double-disk synergy test | PCR at class level |
| [54] | Sample streaked onto blood, MacConkey, and chocolate agar | Double-disk synergy test | PCR at class level |
| [55] | Unspecified | Double-disk synergy test | PCR at class level |
| Bacteria isolated | No. ESBL-Positive Isolates | CTX-M Total | Undefined CTX-M Class | CTX-M-1 Group | CTX-M-9 Group | CTX-M Group 2 | SHV | TEM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Undefined CTX-M-1 Group | CTX-M-15 | CTX-M-55 | CTX-M-3 | CTX-M-130 | Undefined CTX-M-9 Group | CTX-M-14 | CTX-M-27 | CTX-M-32 | |||||||
| E. coli | 521 (59.5) | 481 (92.3) | 174 (33.4) | 79 (15.2) | 81 (15.5) | 11 (2.1) | 0 | 0 | 126 (24.2) | 6 (1.2) | 3 (0.6) | 1 (0.2) | 0 | 10 (1.9) | 36 (6.9) |
| Salmonella spp. | 5 (4.2) | 5 (100) | 1 (20) | 0 | 0 | 4 (80) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S. enterica | 24 (16.7) | 23 (95.8) | 1 (4.2) | 3 (12.5) | 11 (45.8) | 0 | 1 (4.2) | 1 (4.2) | 3 (12.5) | 3 (12.5) | 0 | 0 | 0 | 3 (12.5) | 0 |
| Klebsiella spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K. pneumoniae | 57 (76) | 50 (87.7) | 19 (33.3) | 1 (1.8) | 26 (45.6) | 0 | 0 | 0 | 3 (5.3) | 1 (1.8) | 0 | 0 | 0 | 9 (15.8) | 4 (7) |
| K. oxytoca | 41 (100) | 35 (85.4) | 35 (85.4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 (14.6) | 13 (31.7) |
| Enterobacter spp. | 5 (11.4) | 4 (80) | 3 (60) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (20) | 0 | 0 | 1 (20) | 0 |
| E. cloacae | 7 (31.8) | 2 (28.6) | 0 | 0 | 2 (28.6) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (71.4) | 0 |
| Citrobacter spp. | 21 (48.8) | 17 (81) | 15 (71.4) | 0 | 2 (9.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (14.3) | 4 (19) |
| Proteus spp. | 18 (64.3) | 18 (100) | 18 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Providencia spp. | 8 (38.1) | 6 (75) | 6 (75) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (12.5) | 2 (25) |
| Shigella sonnie | 2 (100) | 2 (100) | 2 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterobacterales total | 709 (49) | 643 (90.7) | 274 (38.6) | 83 (11.7) | 122 (17.2) | 15 (2.1) | 1 (0.1) | 1 (0.1) | 132 (18.6) | 10 (1.4) | 4 (0.6) | 1 (0.1) | 0 | 38 (5.4) | 59 (8.3) |
| Vibrio spp. | 151 (34.2) | 151 (100) | 106 (70.2) | 18 (11.9) | 0 | 0 | 0 | 0 | 27 (17.9) | 0 | 0 | 0 | 0 | 0 | 0 |
| Aeromonas spp. | 26 (25.2) | 26 (100) | 26 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other spp. a | 34 (56.7) | 27 (79.4) | 8 (23.5) | 13 (38.2) | 0 | 0 | 3 (8.8) | 0 | 0 | 0 | 0 | 0 | 3 (8.8) | 7 (20.6) | 1 (2.9) |
| Total Isolates | 920 (44.8) | 847 (92.1) | 414 (45) | 114 (12.4) | 122 (13.3) | 15 (1.6) | 4 (0.4) | 1 (0.1) | 159 (17.3) | 10 (1.1) | 4 (0.4) | 1 (0.1) | 3 (0.3) | 45 (4.9) | 60 (6.5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pearce, R.; Conrady, B.; Guardabassi, L. Prevalence and Types of Extended-Spectrum β-Lactamase-Producing Bacteria in Retail Seafood. Foods 2023, 12, 3033. https://doi.org/10.3390/foods12163033
Pearce R, Conrady B, Guardabassi L. Prevalence and Types of Extended-Spectrum β-Lactamase-Producing Bacteria in Retail Seafood. Foods. 2023; 12(16):3033. https://doi.org/10.3390/foods12163033
Chicago/Turabian StylePearce, Ryan, Beate Conrady, and Luca Guardabassi. 2023. "Prevalence and Types of Extended-Spectrum β-Lactamase-Producing Bacteria in Retail Seafood" Foods 12, no. 16: 3033. https://doi.org/10.3390/foods12163033
APA StylePearce, R., Conrady, B., & Guardabassi, L. (2023). Prevalence and Types of Extended-Spectrum β-Lactamase-Producing Bacteria in Retail Seafood. Foods, 12(16), 3033. https://doi.org/10.3390/foods12163033







