A Multi-Enzyme Cascade Response for the Colorimetric Recognition of Organophosphorus Pesticides Utilizing Core-Shell Pd@Pt Nanoparticles with High Peroxidase-like Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Nanozyme Synthesis
2.2.2. Peroxidase Activity Test for Nanozymes
2.2.3. POD Kinetic Test of Nanozymes
2.2.4. Effect of pH on Nanozymes Activity
2.2.5. Effect of pH on Nanozymes Activity
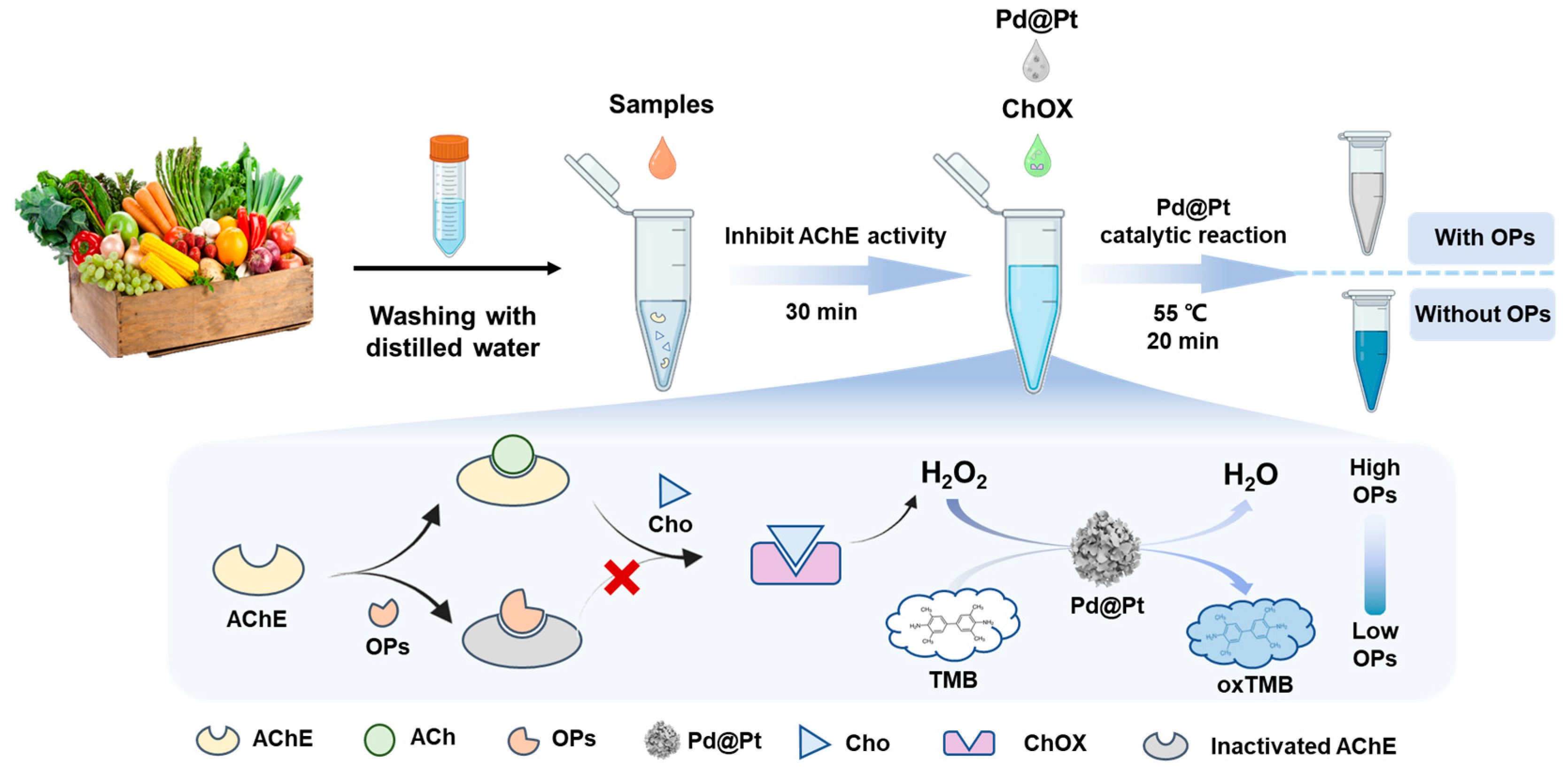
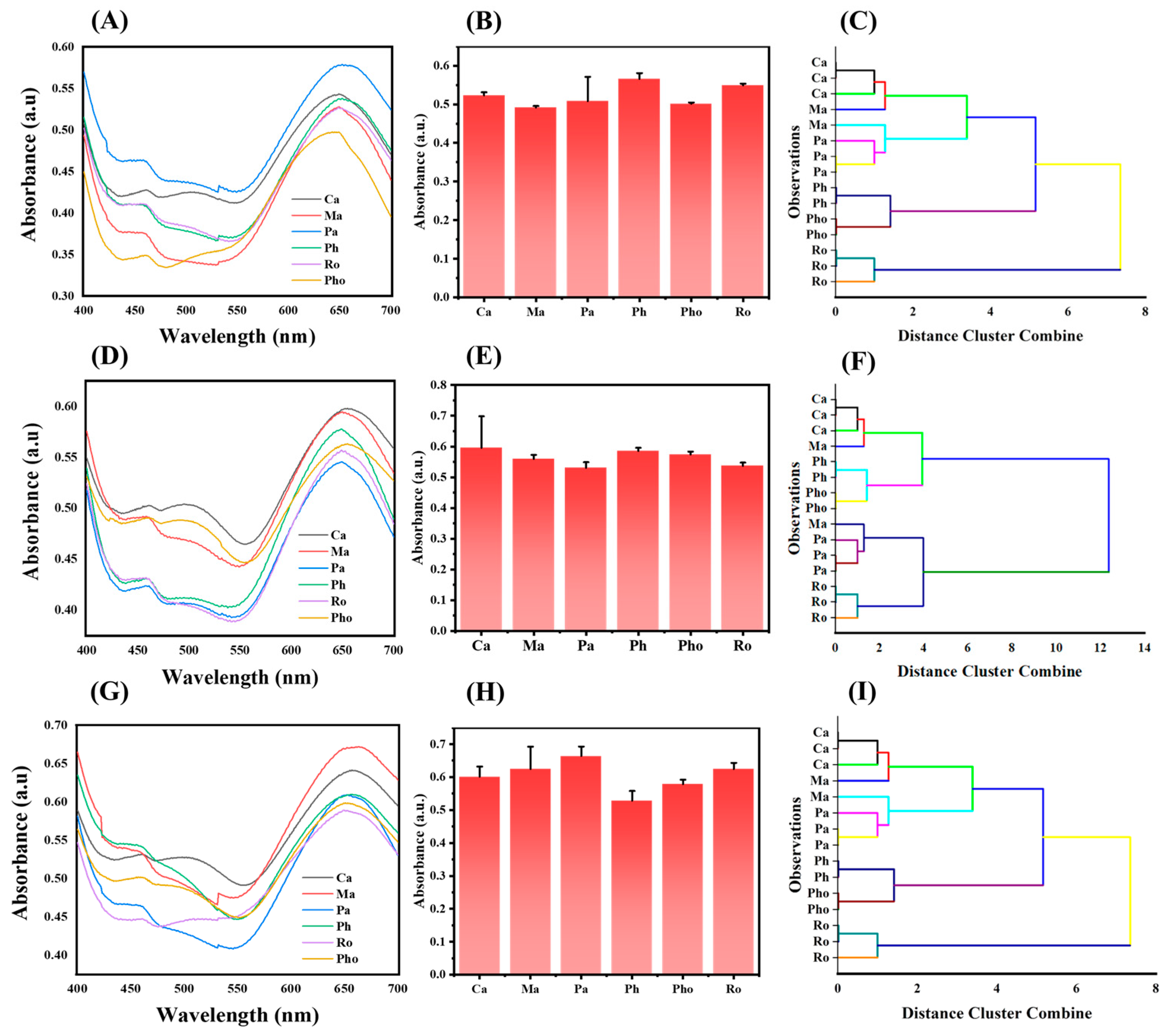
2.3. Discrimination of OPs
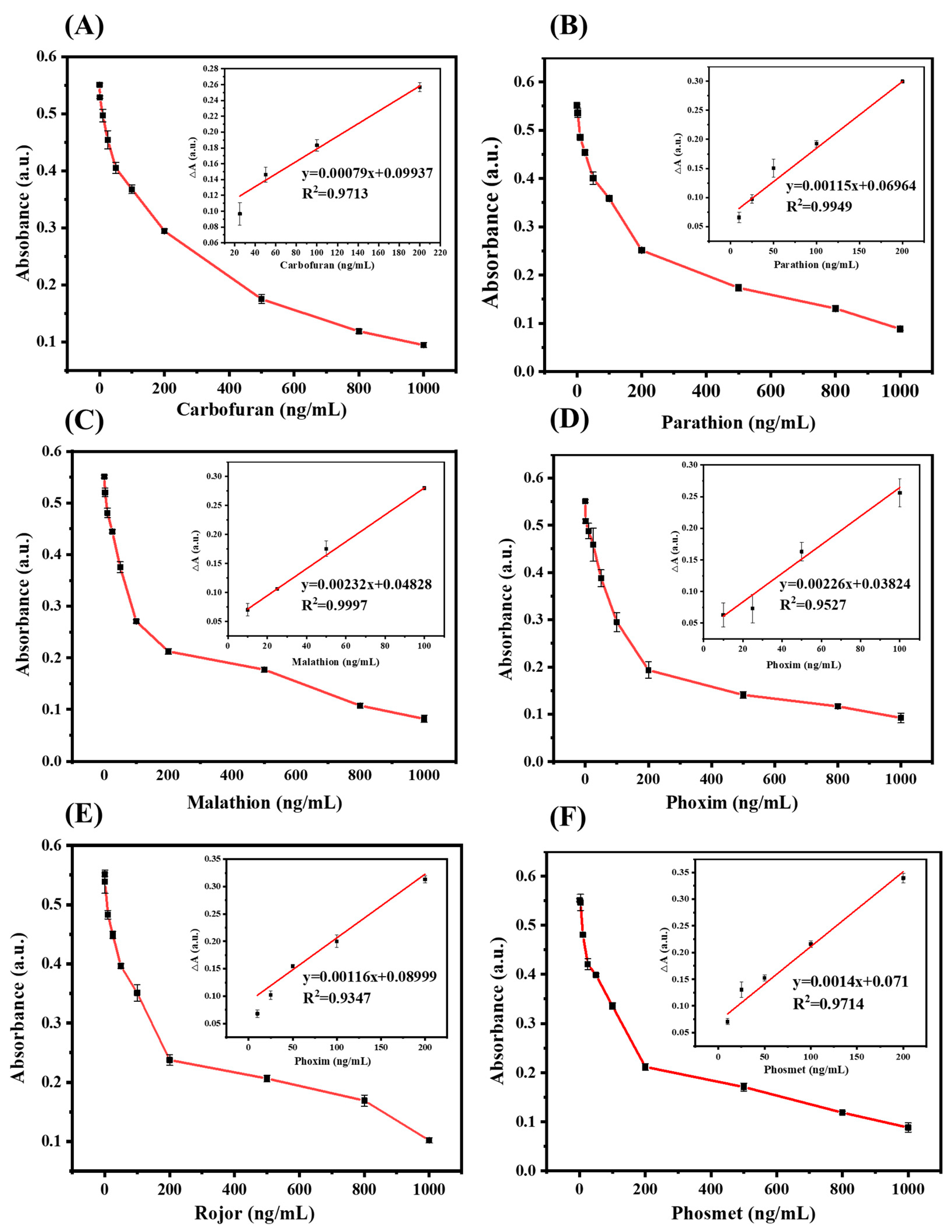
2.4. Quantitative Determination of OPs
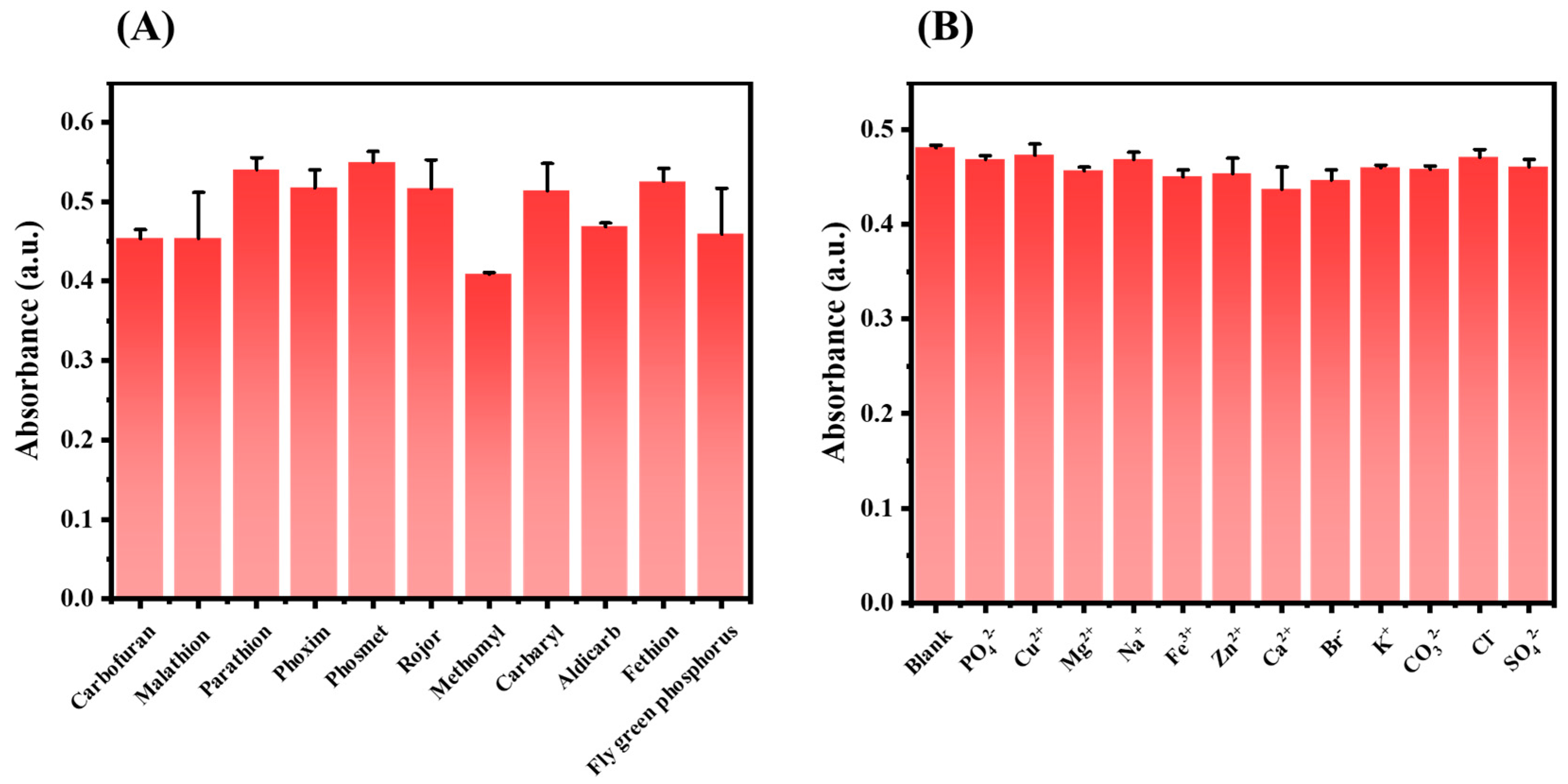
2.5. Specificity Analysis
2.6. Discrimination of the OPs in Real Samples
3. Results
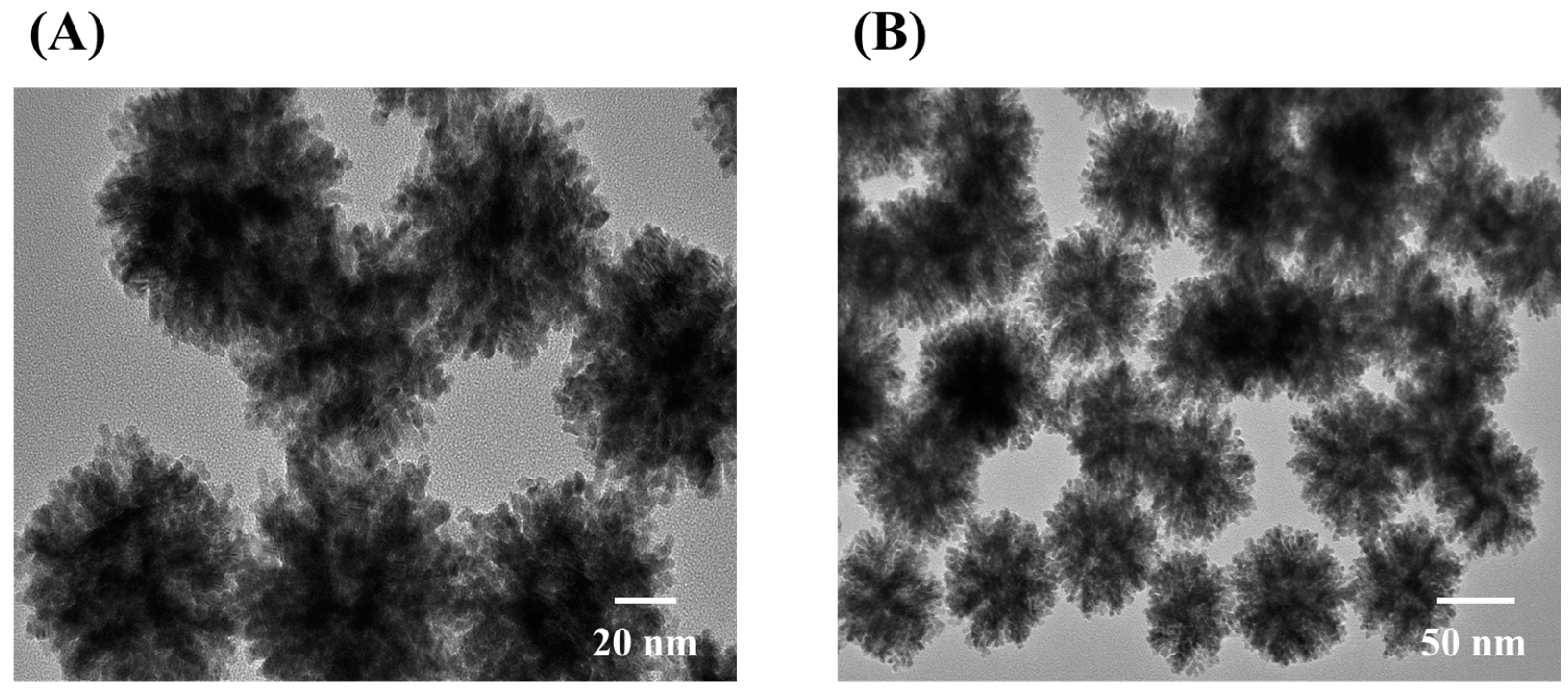
3.1. Characterization
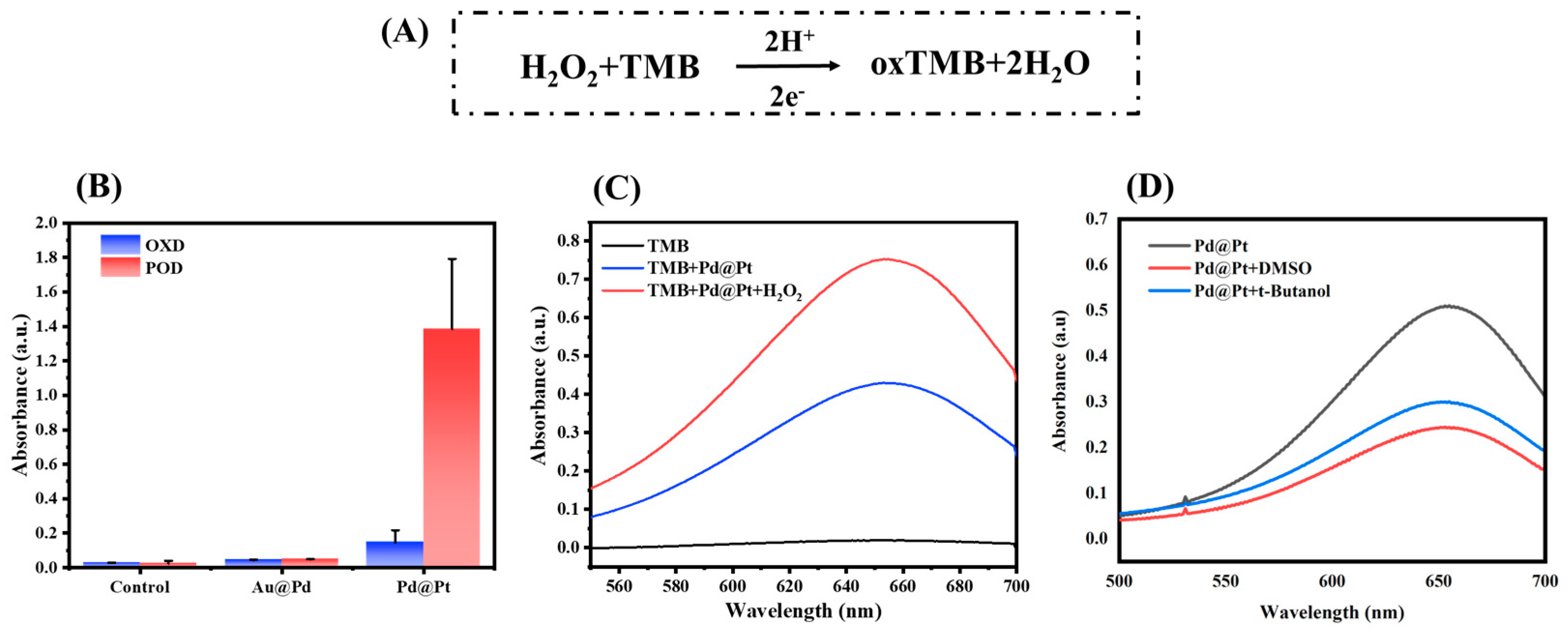
3.2. POD Activity Verification of Nanozymes
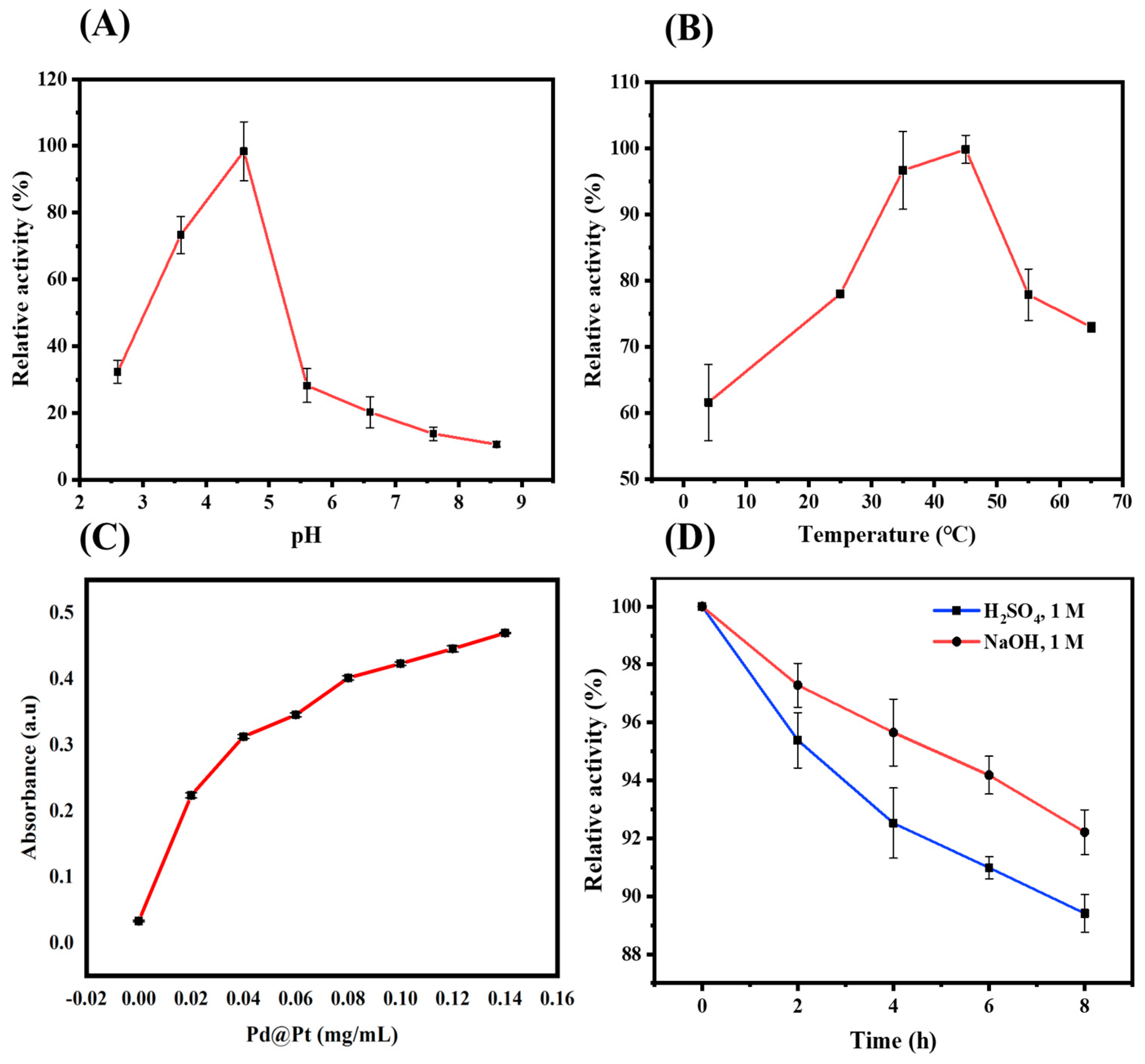
3.3. Condition Optimization of Pd@Pt
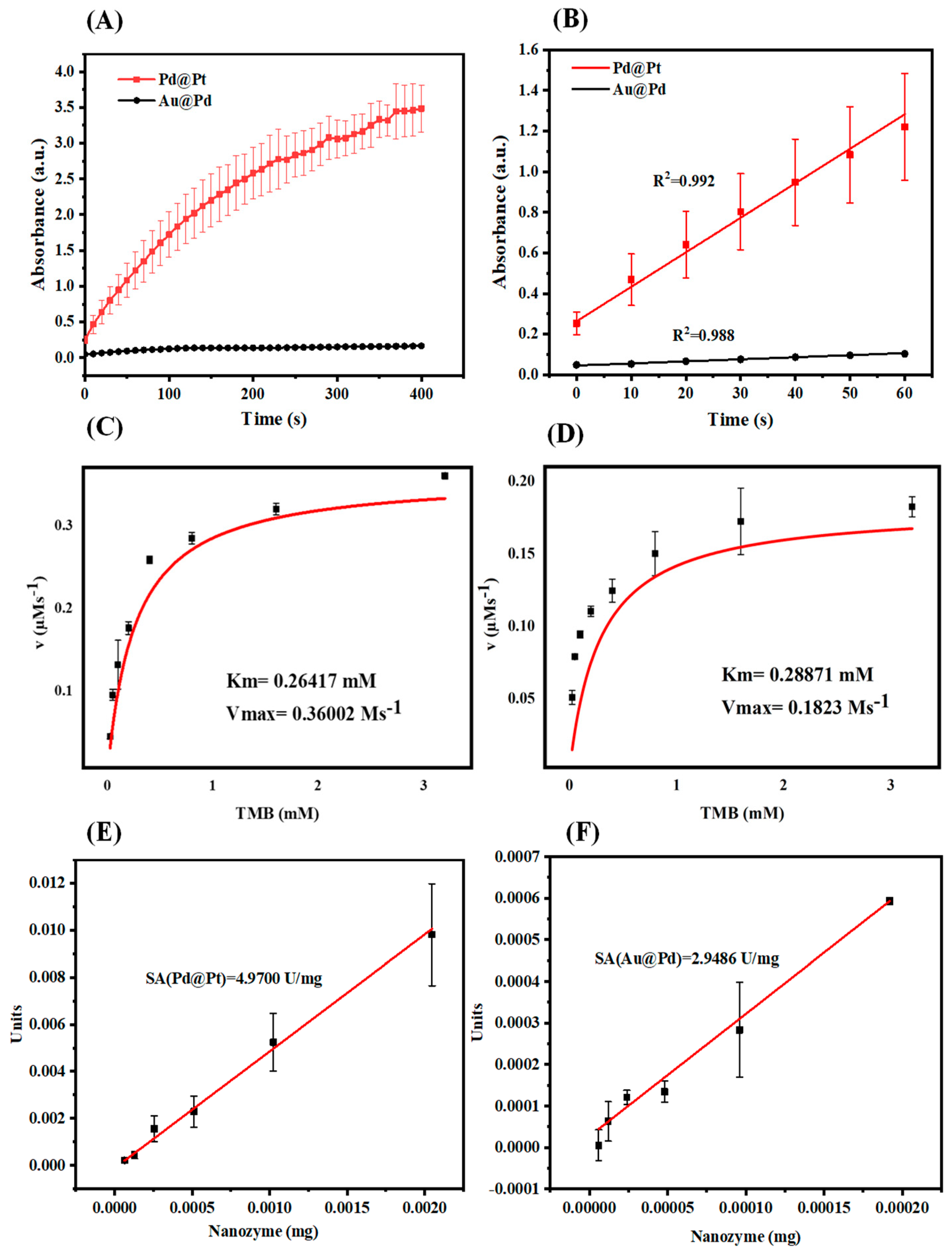
3.4. Catalytic Kinetics
3.5. Optimization
3.6. Sensitivity Analysis
3.7. Quantitative Determination of OPs
3.8. Specificity Analysis
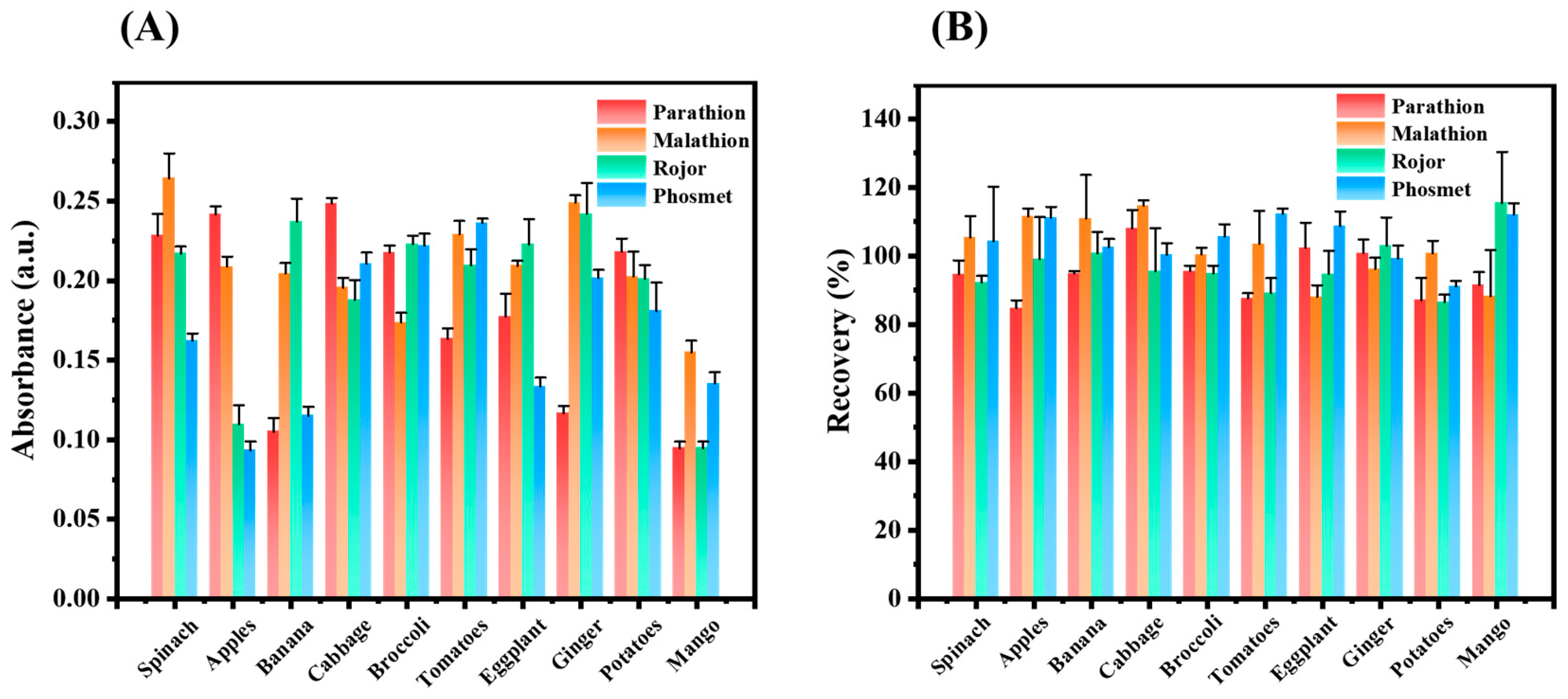
3.9. Discrimination of the OPs in Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muckoya, V.A.; Nomngongo, P.N.; Ngila, J.C. Determination of organophosphorus pesticides in wastewater samples using vor-tex-assisted dispersive liquid–liquid microextraction with liquid chromatography–mass spectrometry. Int. J. Environ. Sci. Technol. 2020, 17, 2325–2336. [Google Scholar] [CrossRef]
- Bala, R.; Dhingra, S.; Kumar, M.; Bansal, K.; Mittal, S.; Sharma, R.K.; Wangoo, N. Detection of organophosphorus pesticide—Malathion in environmental samples using peptide and aptamer based nanoprobes. Chem. Eng. J. 2017, 311, 111–116. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhou, Y.; Li, X.; Liu, S.; Tang, Z. Highly-sensitive organophosphorous pesticide biosensors based on nanostructured films of acetylcholinesterase and CdTe quantum dots. Biosens. Bioelectron. 2011, 26, 3081–3085. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, Y.; Zhao, Y.; Wang, J. Fluorescent and colorimetric dual-response sensor based on copper (II)-decorated graphitic carbon nitride nanosheets for detection of toxic organophosphorus. Food Chem. 2020, 345, 128560. [Google Scholar] [CrossRef]
- Singh, B.K. Organophosphorus-degrading bacteria: Ecology and industrial applications. Nat. Rev. Genet. 2009, 7, 156–164. [Google Scholar] [CrossRef]
- Pabbi, M.; Kaur, A.; Mittal, S.K.; Jindal, R. A surface expressed alkaline phosphatase biosensor modified with flower shaped ZnO for the detection of chlorpyrifos. Sensors Actuators B Chem. 2018, 258, 215–227. [Google Scholar] [CrossRef]
- Pundir, C.S.; Malik, A. Bio-sensing of organophosphorus pesticides: A review. Biosens. Bioelectron. 2019, 140, 111348. [Google Scholar] [CrossRef]
- Khatun, M.A.; Hoque, A.; Zhang, Y.; Lu, T.; Cui, L.; Zhou, N.-Y.; Feng, Y. Bacterial Consortium-Based Sensing System for Detecting Organophosphorus Pesticides. Anal. Chem. 2018, 90, 10577–10584. [Google Scholar] [CrossRef]
- Chang, J.; Li, H.; Hou, T.; Li, F. based fluorescent sensor for rapid naked-eye detection of acetylcholinesterase activity and or-ganophosphorus pesticides with high sensitivity and selectivity. Biosens. Bioelectron. 2016, 86, 971–977. [Google Scholar] [CrossRef]
- Wu, X.; Song, Y.; Yan, X.; Zhu, C.; Ma, Y.; Du, D.; Lin, Y. Carbon quantum dots as fluorescence resonance energy transfer sensors for organophosphate pesticides determination. Biosens. Bioelectron. 2017, 94, 292–297. [Google Scholar] [CrossRef]
- Long, Q.; Li, H.; Zhang, Y.; Yao, S. Upconversion nanoparticle-based fluorescence resonance energy transfer assay for organo-phosphorus pesticides. Biosens. Bioelectron. 2015, 68, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Jing, H.; Li, Y.; Yisibashaer, W.; Chen, J.; Li, B.N.; Zhang, Y. Smartphone based visual and quantitative assays on upconversional paper sensor. Biosens. Bioelectron. 2016, 75, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Bigdeli, A.; Ghasemi, F.; Golmohammadi, H.; Abbasi-Moayed, S.; Nejad, M.A.F.; Fahimi-Kashani, N.; Jafarinejad, S.; Shahrajabian, M.; Hormozi-Nezhad, M.R. Nanoparticle-based optical sensor arrays. Nanoscale 2017, 9, 16546–16563. [Google Scholar] [CrossRef]
- Askim, J.R.; Mahmoudi, M.; Suslick, K.S. Optical sensor arrays for chemical sensing: The optoelectronic nose. Chem. Soc. Rev. 2013, 42, 8649–8682. [Google Scholar] [CrossRef] [PubMed]
- Diehl, K.L.; Anslyn, E.V. Array sensing using optical methods for detection of chemical and biological hazards. Chem. Soc. Rev. 2013, 42, 8596–8611. [Google Scholar] [CrossRef]
- Anzenbacher, P., Jr.; Lubal, P.; Buček, P.; Palacios, M.A.; Kozelkova, M.E. A practical approach to optical cross-reactive sensor arrays. Chem. Soc. Rev. 2010, 39, 3954–3979. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Huang, W.; Shen, F.; Li, T.; Li, S.; Xu, W.; Lv, C.; Luo, Q.; Liu, J. Graphene oxide-based colorimetric detection of organophosphorus pesticides via a multi-enzyme cascade reaction. Nanoscale 2020, 12, 5829–5833. [Google Scholar] [CrossRef]
- Yang, W.; Yang, X.; Zhu, L.; Chu, H.; Li, X.; Xu, W. Nanozymes: Activity origin, catalytic mechanism, and biological application. Coord. Chem. Rev. 2021, 448, 214170. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J. Nanozyme’s catching up: Activity, specificity, reaction conditions and reaction types. Mater. Horizons 2021, 8, 336–350. [Google Scholar] [CrossRef]
- Sun, A.; Mu, L.; Hu, X. Graphene Oxide Quantum Dots as Novel Nanozymes for Alcohol Intoxication. ACS Appl. Mater. Interfaces 2017, 9, 12241–12252. [Google Scholar] [CrossRef]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S.; et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tian, Y.; Huang, P.; Wu, F.-Y. Using target-specific aptamers to enhance the peroxidase-like activity of gold nanoclusters for colorimetric detection of tetracycline antibiotics. Talanta 2020, 208, 120342. [Google Scholar] [CrossRef]
- Zhao, X.E.; Zuo, Y.N.; Qu, X.; Sun, J.; Liu, L.; Zhu, S. Colorimetric determination of the activities of tyrosinase and catalase via substrate-triggered decomposition of MnO2 nanosheets. Microchim. Acta 2019, 186, 848. [Google Scholar] [CrossRef]
- Xi, J.; Wei, G.; An, L.; Xu, Z.; Xu, Z.; Fan, L.; Gao, L. Copper/Carbon Hybrid Nanozyme: Tuning Catalytic Activity by the Copper State for Antibacterial Therapy. Nano Lett. 2019, 19, 7645–7654. [Google Scholar] [CrossRef]
- Uribe, L.; Diezemann, G.; Gauss, J.; Morth, J.P.; Cascella, M. Structural origin of metal specificity in isatin hydrolase from Labrenzia aggregata investigated by computer simulations. Chem. A Eur. J. 2018, 24, 5074–5077. [Google Scholar] [CrossRef]
- Tao, X.; Wang, X.; Liu, B.; Liu, J. Conjugation of antibodies and aptamers on nanozymes for developing biosensors. Biosens. Bioelectron. 2020, 168, 112537. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Chen, M.; Zhao, Y.; Xu, J.; Hou, X. Triple enzyme mimetic activity of Fe3O4@C@MnO2 composites derived from metal–organic frameworks and their application to colorimetric biosensing of dopamine. Microchim. Acta 2022, 189, 12. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ma, Z.-R.; Xiao, Y.-Y.; Yin, Y.-C.; Huang, W.-M.; Huang, Z.-C.; Zheng, Y.-Z.; Mu, F.-Y.; Huang, R.; Shi, G.-Y.; et al. Electronic metal–support interaction modulates single-atom platinum catalysis for hydrogen evolution reaction. Nat. Commun. 2021, 12, 3021. [Google Scholar] [CrossRef]
- Yılmaz, G.E.; Saylan, Y.; Göktürk, I.; Yılmaz, F.; Denizli, A. Selective Amplification of Plasmonic Sensor Signal for Cortisol Detection Using Gold Nanoparticles. Biosensors 2022, 12, 482. [Google Scholar] [CrossRef]
- Göktürk, I.; Bakhshpour, M.; Çimen, D.; Yılmaz, F.; Bereli, N.; Denizli, A. SPR Signal Enhancement with Silver Nanoparti-cle-Assisted Plasmonic Sensor for Selective Adenosine Detection. IEEE Sens. J. 2022, 22, 14862–14869. [Google Scholar] [CrossRef]
- Yuan, C.-X.; Fan, Y.-R.; Zhang, T.; Guo, H.-X.; Zhang, J.-X.; Wang, Y.-L.; Shan, D.-L.; Lu, X.-Q. A New Electrochemical Sensor of Nitro Aromatic Compound Based on Three-Dimensional Porous Pt–Pd Nanoparticles Supported by Graphene–Multiwalled Carbon Nanotube Composite. Biosens. Bioelectron. 2014, 58, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.F.; Park, J.Y. Amperometric Glucose Biosensor Based on Pt-Pd Nanoparticles Supported by Reduced Graphene Oxide and Integrated with Glucose Oxidase. Electroanalysis 2014, 26, 940–951. [Google Scholar] [CrossRef]
- Cheng, N.; Song, Y.; Zeinhom, M.M.A.; Chang, Y.-C.; Sheng, L.; Li, H.; Du, D.; Li, L.; Zhu, M.-J.; Luo, Y.; et al. Nanozyme-mediated dual immunoassay integrated with smartphone for use in simultaneous detection of pathogens. ACS Appl. Mater. Interfaces 2017, 9, 40671–40680. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Cheng, N.; Zhang, Q.; Yang, Z.; Liu, B.; Wang, X.; Huang, K.; Luo, Y. Pd@Pt nanoparticles: Trienzyme catalytic mechanisms, surface-interface effect with DNA and application in biosensing. Sens. Actuators B Chem. 2022, 364, 131907. [Google Scholar] [CrossRef]
- Ataee-Esfahani, H.; Imura, M.; Yamauchi, Y. All-metal mesoporous nanocolloids: Solution-phase synthesis of core-shell pd@pt nanoparticles with a designed concave surface. Angew. Chem. 2013, 125, 13856–13860. [Google Scholar] [CrossRef]
- Shim, K.; Lee, W.-C.; Heo, Y.-U.; Shahabuddin, M.; Park, M.-S.; Hossain, S.A.; Kim, J.H. Rationally designed bimetallic Au@Pt nanoparticles for glucose oxidation. Sci. Rep. 2019, 9, 894. [Google Scholar] [CrossRef]
- Zhu, W.; Wu, Z.; Foo, G.S.; Gao, X.; Zhou, M.; Liu, B.; Veith, G.M.; Wu, P.; Browning, K.L.; Lee, H.N.; et al. Taming interfacial electronic properties of platinum nanoparticles on vacancy-abundant boron nitride nanosheets for enhanced catalysis. Nat. Commun. 2017, 8, 15291. [Google Scholar] [CrossRef]
- Cheng, N.; Song, Y.; Shi, Q.; Du, D.; Liu, D.; Luo, Y.; Xu, W.; Lin, Y. Au@Pd Nanopopcorn and aptamer nanoflower assisted lateral flow strip for thermal detection of exosomes. Anal. Chem. 2019, 91, 13986–13993. [Google Scholar] [CrossRef] [PubMed]
- Bruno, L.; Scuderi, M.; Priolo, F.; Falciola, L.; Mirabella, S. Enlightening the bimetallic effect of Au@Pd nanoparticles on Ni oxide nanostruc-tures with enhanced catalytic activity. Sci. Rep. 2023, 13, 3203. [Google Scholar] [CrossRef]
- Bleeker, E.A.; de Jong, W.H.; Geertsma, R.E.; Groenewold, M.; Heugens, E.H.; Koers-Jacquemijns, M.; van de Meent, D.; Popma, J.R.; Rietveld, A.G.; Wijnhoven, S.W.P. e al. Considerations on the EU definition of a nanomaterial: Science to support policy making. Regul. Toxicol. Pharmacol. 2013, 65, 119–125. [Google Scholar] [CrossRef]
- Sabale, S.; Kandesar, P.; Jadhav, V.; Komorek, R.; Motkuri, R.K.; Yu, X.-Y. Recent developments in the synthesis, properties, and biomedical applications of core/shell superparamagnetic iron oxide nanoparticles with gold. Biomater. Sci. 2017, 5, 2212–2225. [Google Scholar] [CrossRef]
- Maddela, S.; Makula, A.; Jayarambabu, N. Fe3O4 nanoparticles mediated synthesis of novel isatin-dihydropyrimidinone hybrid molecules as antioxidant and cytotoxic agents. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem. Anti-Cancer Agents) 2017, 17, 456–463. [Google Scholar] [CrossRef]
- Cecarini, V.; Gee, J.; Fioretti, E.; Amici, M.; Angeletti, M.; Eleuteri, A.M.; Keller, J.N. Protein oxidation and cellular homeostasis: Emphasis on metabolism. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yuan, M.; Qiao, L.; Guo, R. An efficient colorimetric biosensor for glucose based on peroxidase-like protein-Fe3O4 and glucose oxidase nanocomposites. Biosens. Bioelectron. 2014, 52, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Suslick, B.A.; Feng, L.; Suslick, K.S. Discrimination of complex mixtures by a colorimetric sensor array: Coffee aromas. Anal. Chem. 2010, 82, 2067–2073. [Google Scholar] [CrossRef]
- Zhang, C.; Suslick, K.S. Colorimetric sensor array for soft drink analysis. J. Agric. Food Chem. 2006, 55, 237–242. [Google Scholar] [CrossRef]
- Zhang, C.; Bailey, D.P.; Suslick, K.S. Colorimetric sensor arrays for the analysis of beers: A feasibility study. J. Agric. Food Chem. 2006, 54, 4925–4931. [Google Scholar] [CrossRef]
- Li, Z.; Fang, M.; LaGasse, M.K.; Askim, J.R.; Suslick, K.S. Colorimetric recognition of aldehydes and ketones. Angew. Chem. 2017, 129, 9992–9995. [Google Scholar] [CrossRef]
- Li, Z.; Suslick, K.S. A Hand-held optoelectronic nose for the identification of liquors. ACS Sensors 2018, 3, 121–127. [Google Scholar] [CrossRef]
- Jiang, B.; Duan, D.; Gao, L.; Zhou, M.; Fan, K.; Tang, Y.; Xi, J.; Bi, Y.; Tong, Z.; Gao, G.F.; et al. Standardized assays for determining the catalytic activity and kinetics of peroxidase-like nanozymes. Nat. Protoc. 2018, 13, 1506–1520. [Google Scholar] [CrossRef]
- Salelles, L.; Floury, J.; Le Feunteun, S. Pepsin activity as a function of pH and digestion time on caseins and egg white proteins under static in vitro conditions. Food Funct. 2021, 12, 12468–12478. [Google Scholar] [CrossRef]
- Li, D.; Liu, B.; Huang, P.-J.J.; Zhang, Z.; Liu, J. Highly active fluorogenic oxidase-mimicking NiO nanozymes. Chem. Commun. 2018, 54, 12519–12522. [Google Scholar] [CrossRef]
- Gupta, P.K.; Son, S.E.; Seong, G.H. L-Cysteine-Meditated Self-Assembled PtRu Derived Bimetallic Metal–Carbon Hybrid: An Excellent Peroxidase Mimics for Colorimetric and Fluorometric Detection of Hydrogen Peroxide and Cholesterol. Adv. Mater. Interfaces 2021, 8, 2101115. [Google Scholar] [CrossRef]
- GB 2763-2021; State Administration for Market Regulation of China. National Food Safety Standard Maximum Residue Limits for Pesticides in Foods. Office of Agricultural Affairs: Beijing, China, 2021.
- Soares, S.; Rosado, T.; Barroso, M.; Vieira, D.N.; Gallardo, E. Organophosphorus pesticide determination in biological specimens: Bioanalytical and toxicological aspects. Int. J. Leg. Med. 2019, 133, 1763–1784. [Google Scholar] [CrossRef]
- Weerathunge, P.; Behera, B.K.; Zihara, S.; Singh, M.; Prasad, S.N.; Hashmi, S.; Mariathomas, P.R.D.; Bansal, V.; Ramanathan, R. Dynamic interactions between peroxidase-mimic silver NanoZymes and chlorpyrifos-specific aptamers enable highly-specific pesticide sensing in river water. Anal. Chim. Acta 2019, 1083, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Zhang, J.; Cheng, Y.; Zhang, C.; Fei, X.; Xian, Y. Platinum Nanoparticle Encapsulated Metal–Organic Frameworks for Colorimetric Measurement and Facile Removal of Mercury(II). ACS Appl. Mater. Interfaces 2017, 9, 40716–40725. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, X.; Yang, Y.; Tan, Y.; He, Y.; Liu, M.; Liu, X.; Yuan, Q. Peroxidase-Mimicking Nanozyme with Enhanced Activity and High Stability Based on Metal-Support Interactions. Chemistry 2017, 24, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zheng, S.; Zhang, J.; Li, W.; Fu, Y. Peroxidase Mimicking Activity of Palladium Nanocluster Altered by Heparin. Catal. Lett. 2021, 151, 2537–2546. [Google Scholar] [CrossRef]
- Feng, J.; Huang, P.; Wu, F.-Y. Gold–platinum bimetallic nanoclusters with enhanced peroxidase-like activity and their integrated agarose hydrogel-based sensing platform for the colorimetric analysis of glucose levels in serum. Analyst 2017, 142, 4106–4115. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majid, Z.; Zhang, Q.; Yang, Z.; Che, H.; Cheng, N. A Multi-Enzyme Cascade Response for the Colorimetric Recognition of Organophosphorus Pesticides Utilizing Core-Shell Pd@Pt Nanoparticles with High Peroxidase-like Activity. Foods 2023, 12, 3319. https://doi.org/10.3390/foods12173319
Majid Z, Zhang Q, Yang Z, Che H, Cheng N. A Multi-Enzyme Cascade Response for the Colorimetric Recognition of Organophosphorus Pesticides Utilizing Core-Shell Pd@Pt Nanoparticles with High Peroxidase-like Activity. Foods. 2023; 12(17):3319. https://doi.org/10.3390/foods12173319
Chicago/Turabian StyleMajid, Zainabu, Qi Zhang, Zhansen Yang, Huilian Che, and Nan Cheng. 2023. "A Multi-Enzyme Cascade Response for the Colorimetric Recognition of Organophosphorus Pesticides Utilizing Core-Shell Pd@Pt Nanoparticles with High Peroxidase-like Activity" Foods 12, no. 17: 3319. https://doi.org/10.3390/foods12173319
APA StyleMajid, Z., Zhang, Q., Yang, Z., Che, H., & Cheng, N. (2023). A Multi-Enzyme Cascade Response for the Colorimetric Recognition of Organophosphorus Pesticides Utilizing Core-Shell Pd@Pt Nanoparticles with High Peroxidase-like Activity. Foods, 12(17), 3319. https://doi.org/10.3390/foods12173319






