On the Molecular Selection of Exopolysaccharide-Producing Lactic Acid Bacteria from Indigenous Fermented Plant-Based Foods and Further Fine Chemical Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fermented Foods and Drinks
2.2. Isolation and Characterization of Lactic Acid Bacteria from Plant-Based Fermented Foods
2.3. DNA Extraction
2.4. Molecular Identification of Bacteria by Sequencing of 16S rRNA
2.5. Screening of Lactic Acid Bacteria for Their Ability to Produce Exopolysaccharides
2.6. Selection of a Carbon Source for Exopolysaccharide Synthesis
2.6.1. Seed Cultures
2.6.2. Exopolysaccharide Synthesis
2.6.3. Determination of Biomass Optical Density
2.7. Isolation and Purification of Exopolysaccharides
2.7.1. Isolation of Exopolysaccharides
2.7.2. Qualitative Tests for Determination of Monosaccharide Unit in Polysaccharides
General Analytical Methods
Monosaccharide Composition Analysis
Molecular Weight Distribution Analysis
Fourier-Transform Infrared (FTIR) Spectroscopy
2.8. Statistics
3. Results
3.1. Isolation of Lactic Acid Bacteria from Plant-Based Foods
3.2. Molecular Identification of Lactic Acid Bacteria
3.3. Screening of Lactic Acid Bacteria for Their Ability to Produce Exopolysaccharides
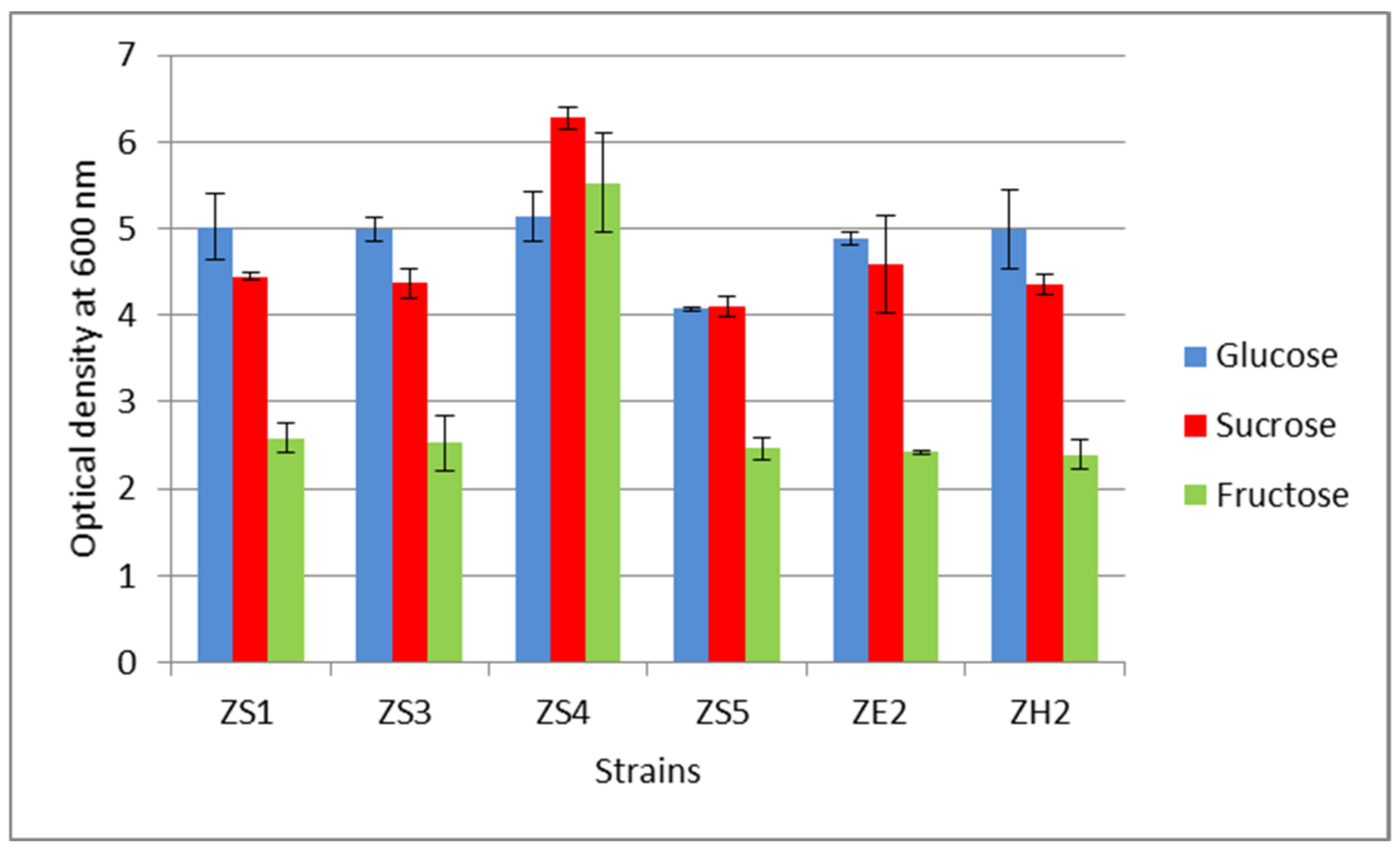
3.4. Selection of a Carbon Source for Exopolysaccharide Synthesis
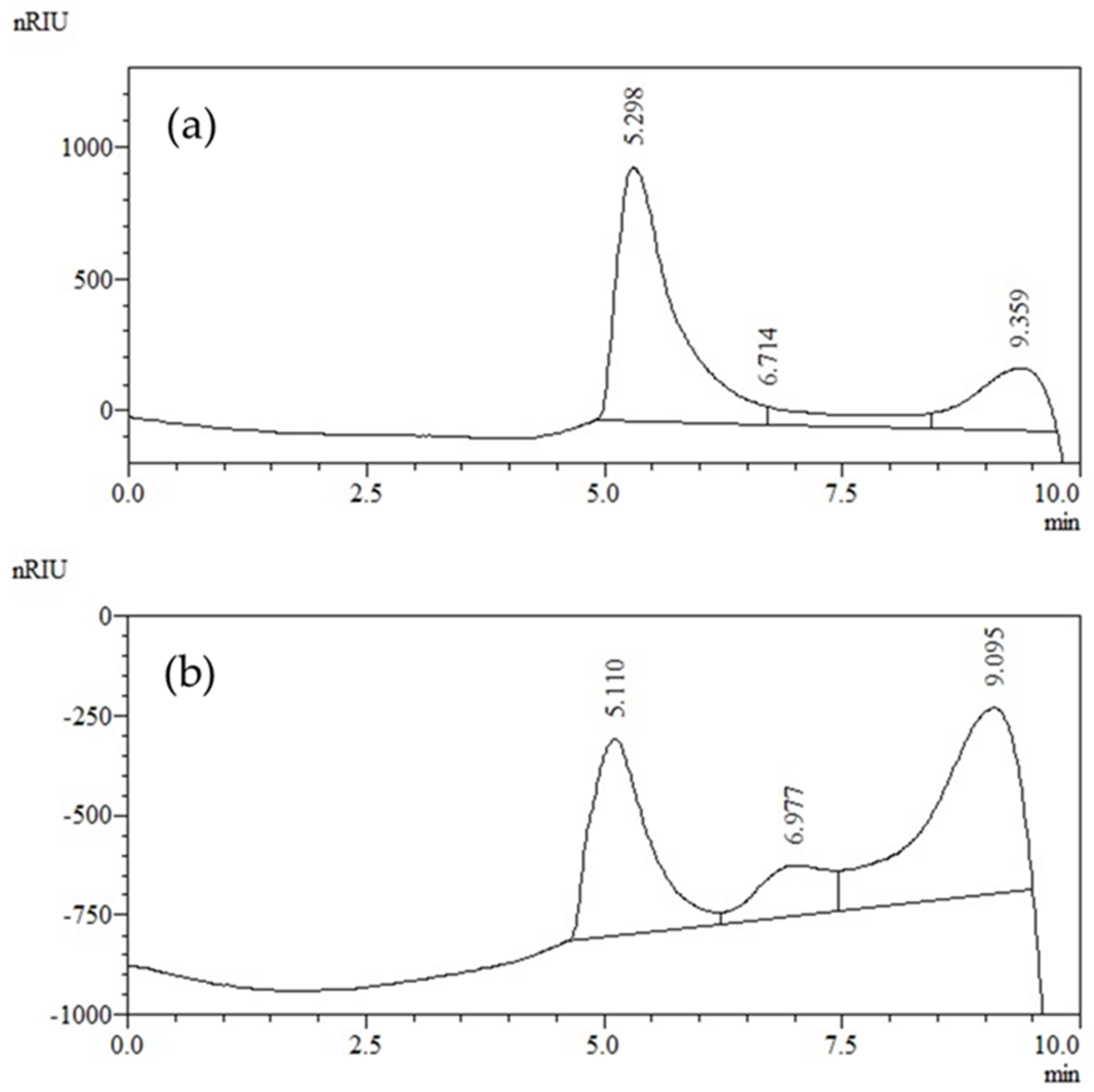
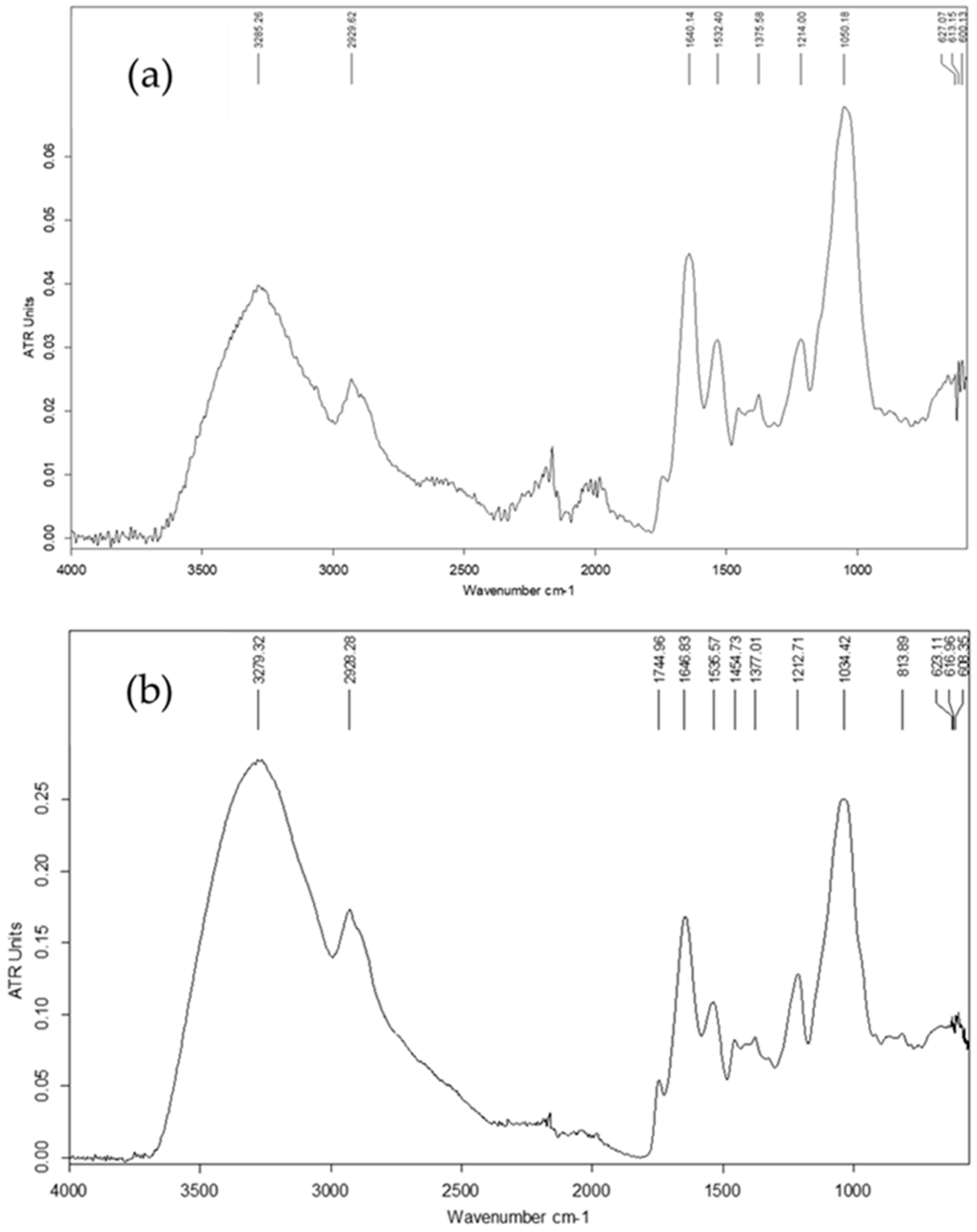
3.5. Yield and Chemical Characterization of Exopolysaccharides
Molecular Weight Distribution of Exopolysaccharides
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bautista-Gallego, J.; Medina, E.; Sánchezb, B.; Benítez-Cabello, A.; Arroyo-López, F.N. Role of lactic acid bacteria in fermented vegetables. Grasas Aceites 2020, 71, e358. [Google Scholar] [CrossRef]
- Bartkiene, B.; Starkute, V.; Jomantaite, I.; Zokaityte, E.; Mockus, E.; Tolpeznikaite, E.; Zokaityte, G.; Petrova, P.; Santini, A.; Rocha, J.M.; et al. Multifunctional nutraceuticals composition based on fermented Spirulina, apple cider vinegar, Jerusalem artichoke and bovine colostrum. Foods 2023, 12, 1690. [Google Scholar] [CrossRef]
- Mockus, E.; Zokaityte, E.; Starkute, V.; Klupsaite, D.; Ruibys, R.; Rocha, J.M.; Bartkevics, V.; Bartkiene, E. Influence of different lactic acid bacteria strains and milling process on the solid-state fermented green and red lentils (Lens culinaris L.) properties including gamma-aminobutyric acid formation. Front. Nutr. 2023, 10, 1118710. [Google Scholar] [CrossRef]
- Bartkiene, E.; Zokaityte, E.; Kentra, E.; Starkute, V.; Klupsaite, D.; Mockus, E.; Zokaityte, G.; Cernauskas, D.; Rocha, J.M.; Guiné, R.P.F. Characterisation of lacto-fermented edible cricket (Acheta domesticus) flour and their influence on the quality parameters and acrylamide formation in wheat biscuits. Fermentation 2023, 9, 153. [Google Scholar] [CrossRef]
- Özogul, F.; Rocha, J.M.; Bartkiene, E. Recent development of lactic acid bacteria and their metabolites on food quality, food safety and human health. Food Biosci. 2022, 50, 102183. [Google Scholar] [CrossRef]
- Yilmaz, B.; Bangara, S.P.; Echegaray, N.; Surib, S.; Tomasevic, I.; Lorenzo, J.M.; Melekoglu, E.; Rocha, J.M.; Özogul, F. The impacts of Lactiplantibacillus plantarum on the functional properties of fermented foods: A review of current knowledge. Microorganisms 2022, 10, 826. [Google Scholar] [CrossRef] [PubMed]
- Zokaityte, E.; Cernauskas, D.; Klupsaite, D.; Lele, V.; Starkute, V.; Zavistanaviciute, P.; Ruzauskas, M.; Gruzauskas, R.; Juodeikiene, G.; Rocha, J.M.; et al. Bioconversion of milk permeate with selected lactic acid bacteria strains and apple by-products into beverages with antimicrobial properties and enriched with galactooligosaccharides. Microorganisms 2020, 8, 1182. [Google Scholar] [CrossRef]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, D.M.; Weinert, E., Jr.; Chang, K.C.S.; McGinn, J.M.; Miller, S.A.; Keliihoomalu, C.; DuPonte, M.W. Natural Farming: Lactic Acid Bacteria. J. Sustain. Agric. 2013, 8, 3–4. [Google Scholar]
- Blandino, A.; Al-Aseeri, M.; Pandiella, S.; Cantero, D.; Webb, C. Cereal-based fermented foods and beverages. Food Res. Int. 2003, 36, 527–543. [Google Scholar] [CrossRef]
- Calderon, M.; Loiseau, G.; Guyot, J.P. Fermentation by Lactobacillus fermentum Ogi E1 of different combinations of carbohydrates occurring naturally in cereals: Consequences on growth energetics and alpha-amylase production. Int. J. Food Microbiol. 2003, 80, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Campbell-Platt, G. Fermented foods—A world perspective. Food Res. Int. 1994, 27, 253–257. [Google Scholar] [CrossRef]
- Caplice, E.; Fitzgerald, G.F. Food fermentations: Role of microorganisms in food production and preservation. Int. J. Food Microbiol. 1999, 50, 131–149. [Google Scholar] [CrossRef]
- Bartkiene, E.; Zarovaite, P.; Starkute, V.; Mockus, E.; Zokaityte, E.; Zokaityte, G.; Rocha, J.M.; Ruibys, R.; Klupsaite, D. Changes in Lacto-Fermented Agaricus bisporus (White and Brown Varieties) Mushroom Characteristics, Including Biogenic Amine and Volatile Compound Formation. Foods 2023, 12, 2441. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Rimsa, A.; Zokaityte, E.; Starkute, V.; Mockus, E.; Cernauskas, D.; Rocha, J.M.; Klupsaite, D. Changes in the Physicochemical Properties of Chia (Salvia hispanica L.) Seeds during Solid-State and Submerged Fermentation and Their Influence on Wheat Bread Quality and Sensory Profile. Foods 2023, 12, 2093. [Google Scholar] [CrossRef]
- Bartkiene, E.; Tolpeznikaite, E.; Klupsaite, D.; Starkute, V.; Bartkevics, V.; Skrastina, A.; Pavlenko, R.; Mockus, E.; Lele, V.; Batkeviciute, G.; et al. Bio-converted Spirulina for nutraceutical chewing candy formulations rich in L-glutamic and gamma-aminobutyric acids. Microorganisms 2023, 11, 441. [Google Scholar] [CrossRef] [PubMed]
- Tolpeznikaite, E.; Bartkevics, V.; Skrastina, A.; Pavlenko, R.; Ruzauskas, M.; Starkute, V.; Zokaityte, E.; Klupsaite, D.; Ruibys, R.; Rocha, J.M.; et al. Submerged and solid-state fermentation of Spirulina with lactobacilli strains: Antimicrobial properties and the formation of bioactive compounds of protein origin. Biology 2023, 12, 248. [Google Scholar] [CrossRef]
- Sharma, H.; Fidan, H.; Özogul, F.; Rocha, J.M. Recent development in the preservation effect of lactic acid bacteria and essential oils on chicken and seafood products. Front. Microbiol. 2022, 13, 092248. [Google Scholar] [CrossRef]
- Trakselyte-Rupsiene, K.; Juodeikiene, G.; Alzbergaite, G.; Zadeike, D.; Bartkiene, E.; Özogul, F.; Ruller, L.; Robert, J.; Rocha, J.M. Bio-refinery of plant drinks press cake permeate using ultrafiltration and lactobacillus fermentation into antimicrobials and its effect on the growth of wheatgrass in vivo. Food Biosci. 2022, 46, 101427. [Google Scholar] [CrossRef]
- Soro-Yao, A.A.; Brou, K.; Amani, G.; Thonart, P.; Djè, K.M. The use of lactic acid bacteria starter cultures during the processing of fermented cereal-based foods in West Africa: A review. Trop. Life Sci. Res. 2014, 25, 81–100. [Google Scholar]
- Jensen, M.P.; Ardö, Y.; Vogensen, F.K. Isolation of cultivable thermophilic lactic acid bacteria from cheeses made with mesophilic starter and molecular comparison with dairy-related Lactobacillus helveticus strains. Lett. Appl. Microbiol. 2009, 49, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; Bilancia, M.T.; Siragusa, S.; Gobbetti, M.; Caponio, F. Fermented goats’ milk produced with selected multiple starters as a potentially functional food. Food Microbiol. 2009, 26, 559–564. [Google Scholar] [CrossRef]
- Ağagündüz, D.; Yılmaz, B.; Şahin, T.O.; Güneşliol, B.E.; Ayten, S.; Russo, P.; Spano, G.; Rocha, J.M.; Bartkiene, E.; Özogul, F. Dairy Lactic Acid Bacteria and Their Potential Function in Dietetics: The Food–Gut-Health Axis. Foods 2021, 10, 3099. [Google Scholar] [CrossRef]
- Angelov, A.; Karadzhov, G.; Roshkova, Z. Strains selection of baker’s yeast with improved technological properties. Food Res. Int. 1996, 29, 235–239. [Google Scholar] [CrossRef]
- Bai, F.Y.; Han, D.Y.; Duan, S.F.; Wang, Q.M. The Ecology and Evolution of the Baker’s Yeast Saccharomyces cerevisiae. Genes 2022, 13, 230. [Google Scholar] [CrossRef]
- Lahue, C.; Madden, A.A.; Dunn, R.R.; Smukowski, H.C. History and Domestication of Saccharomyces cerevisiae in Bread Baking. Front. Genet. 2020, 11, 584718. [Google Scholar] [CrossRef] [PubMed]
- Klupsaite, K.; Starkute, V.; Zokaityte, E.; Cernauskas, D.; Mockus, E.; Kentra, E.; Sliazaite, R.; Abramaviciute, G.; Sakaite, P.; Komarova, V.; et al. The contribution of scalded and scalded-fermented rye wholemeal flour to quality parameters and acrylamide formation in semi wheat-rye bread. Foods 2023, 12, 937. [Google Scholar] [CrossRef]
- Klupsaite, D.; Kaminskaite, A.; Rimsa, A.; Gerybaite, A.; Stankaityte, A.; Sileikaite, A.; Svetlauskaite, E.; Cesonyte, E.; Urbone, G.; Pilipavicius, K.; et al. The contribution of new breed purple wheat (8526-2 and 8529-1) varieties wholemeal flour and sourdough to quality parameters and acrylamide formation in wheat bread. Fermentation 2022, 8, 724. [Google Scholar] [CrossRef]
- Lahtinen, S.; Ouwehand, A.C.; Salminen, S.; von Wright, A. Lactic Acid Bacteria: Microbiological and Functional Aspects, 4th ed.; CRC Press/Tayor & Francis Group LLC: Boca Raton, FL, USA, 2011. [Google Scholar]
- Picozzi, C.; Gallina, S.; Della Ferra, T.; Foschino, R. Comparison of cultural media for the enumeration of sourdough lactic acid bacteria. Ann. Microbiol. 2005, 55, 317–320. [Google Scholar]
- Rosemquist, H.; Hansen, A. The microbial stability of two bakery sourdoughs made from conventionally and organically grown rye. Food Microbiol. 2000, 17, 241–250. [Google Scholar] [CrossRef]
- Dardir, H.A. In vitro evaluation of probiotic activities of lactic acid bacteria strains isolated from novel probiotic dairy products. Glob. Vet. 2012, 8, 190–196. [Google Scholar]
- De Vuyst, L.; Leroy, F. Bacteriocins from lactic acid bacteria: Production, purification and food applications. J. Mol. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Klaenhammer, T.R.; Barrangou, R.; Buck, B.L.; Azcarate-Peril, M.A.; Altermann, E. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 2005, 29, 393–409. [Google Scholar] [CrossRef]
- Juodeikiene, G.; Trakselyte-Rupsiene, K.; Reikertaite, K.; Hajnal, E.J.; Bartkevics, V.; Pugajeva, I.; Gruzauskas, V.; Svazas, M.; Gruzauskas, R.; Santini, A.; et al. Influence of the biotreatment on Hordeum vulgare L. cereal wholemeal contamination and enzymatic activities. Foods 2023, 12, 1050. [Google Scholar] [CrossRef]
- Petkova, M.; Gotcheva, V.; Dimova, M.; Bartkiene, E.; Rocha, J.M.; Angelov, A. Screening of Lactiplantibacillus plantarum strains from sourdoughs for biosuppression of Pseudomonas syringae pv. syringae and Botrytis cinerea in table grapes. Microorganisms 2022, 10, 2094. [Google Scholar] [CrossRef] [PubMed]
- Tolpeznikaite, E.; Starkute, V.; Zokaityte, E.; Ruzauskas, M.; Pilkaityte, R.; Viskelis, P.; Urbonaviciene, D.; Ruibys, R.; Rocha, J.M.; Bartkiene, E. Effect of solid-state fermentation and ultrasonication processes on antimicrobial and antioxidant properties of algae extracts. Front. Nutr. 2022, 9, 990274. [Google Scholar] [CrossRef] [PubMed]
- Rathod, N.B.; Nirmal, N.P.; Pagarkar, A.; Özogul, F.; Rocha, J.M. Antimicrobial impacts of microbial metabolites on the preservation of fish and fishery products: A review with current knowledge. Microorganisms 2022, 10, 773. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Özogul, F.; Rocha, J.M. Bread sourdough lactic acid bacteria—Technological, antimicrobial, toxin-degrading, immune system- and faecal microbiota-modelling biological agents for the preparation of food, nutraceuticals and feed. Foods 2022, 11, 452. [Google Scholar] [CrossRef]
- Benkerroum, N.; Oubel, H.; Zahar, M.; Dlia, S.; Filali-Maltouf, A. Isolation of a bacteriocin-producing Lactococcus lactis subsp. lactis and application to control Listeria monocytogenes in Moroccan jben. J. Appl. Microbiol. 2000, 89, 960–968. [Google Scholar]
- Novotni, D.; Gänzle, M.; Rocha, J.M. Composition and activity of microbiota in sourdough and their effect on bread quality and safety. In Trends in Wheat and Bread Making; Galanakis, C.M., Ed.; Elsevier-Academic Press: Cambridge, MA, USA, 2021; pp. 129–172. [Google Scholar] [CrossRef]
- Ağagündüz, D.; Yılmaz, B.; Koçak, T.; Başar, H.B.; Rocha, J.M.; Özoğul, F. Novel Candidate Microorganisms for Fermentation Technology: From Potential Benefits to Safety Issues. Foods 2022, 11, 3074. [Google Scholar] [CrossRef]
- Fraberger, V.; Özülkü, G.; Petrova, P.; Nada, K.; Petroc, K.; Domig, K.J.; Rocha, J.M. Sourdough as a source of technological, antimicrobial and probiotic microorganisms. In Sourdough Innovations: Novel Uses of Metabolites, Enzymes and Microbiota from Sourdough Processing; Garcia-Vaquero, M., Rocha, J.M., Eds.; CRC Press/Taylor and Francis Group: Boca Raton, FL, USA, 2023; p. 464. [Google Scholar] [CrossRef]
- Gibson, G.R. From probiotics to prebiotics and a healthy digestive system. J. Food Sci. 2004, 69, 141–143. [Google Scholar] [CrossRef]
- Horáčková, Š.; Žaludová, K.; Plocková, M. Stability of selected actobacilli in the conditions simulating those in the gastrointestinal tract. Czech J. Food Sci. 2011, 29, 30–35. [Google Scholar] [CrossRef]
- Huebner, J.; Wehling, R.L.; Hutkins, R.W. Functional activity of commercial prebiotics. Int. Dairy J. 2007, 17, 770–775. [Google Scholar] [CrossRef]
- Korcz, E.; Varga, L. Exopolysaccharides from lactic acid bacteria: Techno-functional application in the food industry. Trends Food Sci. Technol. 2021, 110, 375–384. [Google Scholar] [CrossRef]
- Berthold-Pluta, A.M.; Pluta, A.S.; Garbowska, M.; Stasiak-Różańska, L. Exopolysaccharide—Producing lactic acid bacteria—Health-promoting properties and application in the dairy industry. Adv. Microbiol. 2019, 58, 191–204. [Google Scholar] [CrossRef]
- Oleksy-Sobczak, M.; Klewicka, E.; Piekarska-Radzik, L. Exopolysaccharides production by Lactobacillus rhamnosus strains—Optimization of synthesis and extraction conditions. LWT—Food Sci. Technol. 2020, 122, 109055. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides produced by lactic acid bacteria: From biosynthesis to health-promoting properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Patel, A.; Prajapati, J. Food and health applications of exopolysaccharides produced by lactic acid bacteria. Adv. Dairy Res. 2013, 1, 107–114. [Google Scholar]
- Nguyen, P.; Nguyen, T.; Bui, D.C.; Hong, P.T.; Hoang, Q.K.; Nguyen, H.T. Exopolysaccharide production by lactic acid bacteria: The manipulation of environmental stresses for industrial applications. AIMS Microbiol. 2020, 6, 451–469. [Google Scholar] [CrossRef]
- Oerlemans, M.M.P.; Akkerman, R.; Ferrari, M.; Walvoort, M.T.C.; de Vos, P. Benefits of bacteria-derived exopolysaccharides on gastrointestinal microbiota, immunity and health. J. Funct. Foods 2021, 76, 104289. [Google Scholar] [CrossRef]
- Ziadi, M.; Bouzaiene, T.; M’Hir, S.; Zaafouri, K.; Mokhtar, F.; Hamdi, M.; Boisset-Helbert, C. Evaluation of the Efficiency of Ethanol Precipitation and Ultrafiltration on the Purification and Characteristics of Exopolysaccharides Produced by Three Lactic Acid Bacteria. BioMed Res. Int. 2018, 11, 896240. [Google Scholar] [CrossRef] [PubMed]
- Rühmann, B.; Schmid, J.; Sieber, V. Methods to identify the unexplored diversity of microbial exopolysaccharides. Front. Microbiol. 2015, 6, 565. [Google Scholar] [PubMed]
- Sanalibaba, P.; Çakmak, G.A. Exopolysaccharides Production by Lactic Acid Bacteria. Appl. Microbiol. 2016, 2, 000115. [Google Scholar] [CrossRef]
- Mıdık, F.; Tokatlı, M.; Elmacı, S.; Ozçelik, F. Influence of different culture conditions on exopolysaccharide production by indigenous lactic acid bacteria isolated from pickles. Arch. Microbiol. 2020, 202, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Macedo, M.G.; Lacroix, C.; Gardner, N.J.; Champagne, C.P. Effect of medium supplementation on exopolysaccharide production by Lactobacillus rhamnosus RW9595M in whey permeate. Int. Dairy J. 2002, 12, 419–426. [Google Scholar] [CrossRef]
- ISO 15214:1998; Microbiology of Food and Animal Feeding Stuff—Horizontal Method for the Identification—Colony Count Technique at 30 °C. International Organization for Standardization: Geneva, Switzerland, 1998.
- Minervini, F.; Di Cagno, R.; Lattanzi, A.A.; De Angelis, M.; Antonielli, L.; Cardinali, G.; Cappelle, S.; Gobbetti, M. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: Interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. 2012, 78, 251–1264. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Petkova, N.T.; Sherova, G.; Denev, P.P. Characterization of inulin from dahlia tubers isolated by microwave and ultrasound-assisted extractions. Int. Food Res. J. 2018, 25, 1876–1884. [Google Scholar]
- GB 5009.5-2010; Determination of Protein in Foods. National Food Safety Standard of the People’s Republic of China. China National Center for Food Safety Risk Assessment: Beijing, China, 2010.
- Sosulski, F.W.; Imafidon, G.I. Amino acid composition and nitrogen-to-protein conversion factors for animal and plant foods. J. Agric. Food Chem. 1990, 38, 1351–1356. [Google Scholar] [CrossRef]
- Karkhanis, Y.D.; Zeltner, J.Y.; Jackson, J.J.; Carlo, D.J. A new and improved microassay to determine 2-keto-3-deoxyoctonate in lipopolysaccharide of gram-negative bacteria. Anal. Biochem. 1978, 85, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Ognyanov, M.; Georgiev, Y.; Petkova, N.; Ivanov, I.; Vasileva, I.; Kratchanova, M. Isolation and characterization of pectic polysaccharide fraction from in vitro suspension culture of Fumaria officinalis L. Int. J. Polym. Sci. 2018, 2018, 5705036. [Google Scholar] [CrossRef]
- Davidson, A.E. Analysis of sugars found in mucopolysaccharides. Methods Enzymol. 1966, 8, 52–60. [Google Scholar]
- McComb, E.A.; McCready, R.M. Determination of acetyl in pectin and in acetylated carbohydrate polymers. Anal. Chem. 1957, 29, 819–821. [Google Scholar] [CrossRef]
- Gotcheva, V.; Pandiella, S.S.; Angelov, A.; Roshkova, Z.; Webb, C. Monitoring the fermentation of the traditional Bulgarian beverage boza. Int. J. Food Sci. Technol. 2001, 36, 129–135. [Google Scholar] [CrossRef]
- Angelov, A.; Blagoeva, G.; Angelov, A.D.; Stefanova, P.; Bokossa, I.Y.; Tchekessi, C.K.C.; Marco, M.L.; Gotcheva, V. Molecular identification of yeasts and lactic acid bacteria involved in the production of Beninese fermented food Degue. Open Biotechnol. J. 2017, 11, 3–13. [Google Scholar] [CrossRef]
- Plengvidhya, V.; Breidt, F., Jr.; Lu, Z.; Fleming, H.P. DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl. Environ. Microbiol. 2007, 73, 7697–7702. [Google Scholar] [CrossRef]
- Pederson, C.S.; Albury, M.N. The Sauerkraut Fermentation; Technical Bulletin 824; New York State Agricultural Experiment Station: Geneva, NY, USA, 1969. [Google Scholar]
- Cui, Y.; Wang, M.; Zheng, Y.; Miao, K.; Qu, X. The Carbohydrate Metabolism of Lactiplantibacillus plantarum. Int. J. Mol. Sci. 2021, 22, 13452. [Google Scholar] [CrossRef]
- Satora, P.; Skotniczny, M.; Strnad, S. Chemical composition and sensory quality of sauerkraut produced from different cabbage varieties. LWT—Food Sci. Technol. 2021, 136, 110325. [Google Scholar] [CrossRef]
- Hamidi, M.; Okoro, O.V.; Milan, P.B.; Khalili, M.R.; Samadian, H.; Nie, L. Fungal exopolysaccharides: Properties, sources, modifications, and biomedical applications. Carbohydr. Polym. 2022, 284, 119152. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Copikova, J.; Matejka, P.; Machovic, V. Fourier transform Raman and infrared spectroscopy of pectins. Carbohydr. Polym. 2003, 54, 97–106. [Google Scholar] [CrossRef]
- Torres, S.; Verón, H.; Contreras, L.; Isla, M. An overview of plant-autochthonous microorganisms and fermented vegetable foods. Food Sci. Hum. Wellness 2020, 9, 112–123. [Google Scholar] [CrossRef]
- Ibarburu, I.; Puertas, A.I.; Berregi, I.; Rodríguez-Carvajal, M.A.; Prieto, A.; Dueñas, M.T. Production and partial characterization of exopolysaccharides produced by two Lactobacillus suebicus strains isolated from cider. Int. J. Food Microbiol. 2015, 214, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Bengoa, A.A.; Goretti, M.; Iraporda, C.; Dueñas, M.T.; Abraham, A.G.; Garrote, G.L. Impact of growth temperature on exopolysaccharide production and probiotic properties of Lactobacillus paracasei strains isolated from kefir grains. Food Microbiol. 2018, 69, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.I.; Martínez, B.; Guillén, R.; Jiménez-Díaz, R.; Rodríguez, A. Culture conditions determine the balance between two different exopolysaccharides produced by Lactobacillus pentosus LPS26. Appl. Environ. Microbiol. 2006, 72, 7495–7502. [Google Scholar] [CrossRef]
- Salazar, N.; Prieto, A.; Leal, J.A.; Mayo, B.; Bada-Gancedo, J.C.; de los Reyes-Gavilán, C.G.; Ruas-Madiedo, P. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J. Dairy Sci. 2009, 92, 4158–4168. [Google Scholar] [CrossRef]
- Paulo, E.; Vasconcelos, M.; Oliveira, I.; Affe, H.; Nascimento, R.; Melo, I.; Roque, M.; Assis, S. An alternative method for screening lactic acid bacteria for the production of exopolysaccharides with rapid confirmation. Ciência Tecnol. Aliment. 2012, 32, 710–714. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, K.; Yin, S.; Liu, S.; Zhu, Y.; Yang, Y.; Wang, C. Purification and characterization of an exopolysaccharide produced by Lactobacillus plantarum HY isolated from home-made Sichuan Pickle. Int. J. Biol. Macromol. 2019, 134, 516–526. [Google Scholar] [CrossRef]
- Liua, Z.; Penga, Z.; Huanga, T.; Xiaoa, Y.; Lia, J.; Xiea, M.; Xionga, T. Comparison of bacterial diversity in traditionally homemade paocai and Chinese spicy cabbage. Food Microbiol. 2019, 83, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Qian, Y.; Tao, Y.; She, X.; Li, Y.; Chen, X.; Guo, S.; Xiang, W.; Liu, L.; Du, H.; et al. Characterization of the microbial communities and their correlations with chemical profiles in assorted vegetable Sichuan pickles. Food Control 2020, 113, 107174. [Google Scholar] [CrossRef]
- Oleksy-Sobczak, M.; Klewicka, E. Optimization of Media Composition to Maximize the Yield of Exopolysaccharides Production by Lactobacillus rhamnosus Strains. Probiotics Antimicrob. Proteins 2020, 12, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ji, J.; Rui, X.; Yu, J.; Tang, W.; Chen, X.; Jiang, M.; Dong, M. Production of exopolysaccharides by Lactobacillus helveticus MB2-1 and its functional characteristics in vitro. LWT—Food Sci. Technol. 2014, 59, 732–739. [Google Scholar] [CrossRef]
- Endo, A.; Maeno, S.; Tanizawa, Y.; Kneifel, W.; Arita, M.; Dicks, L.; Salminen, S. Fructophilic lactic acid bacteria, a unique group of fructose-fermenting microbes. Appl. Environ. Microbiol. 2018, 84, e01290-18. [Google Scholar] [CrossRef] [PubMed]
- Kowsalya, M.; Velmurugan, T.; Mythili, R.; Kim, W.; Sudha, K.G.; Ali, S.; Kalpana, B.; Ramalingam, S.; Rajeshkumar, M.P. Extraction and characterization of exopolysaccharides from Lactiplantibacillus plantarum strain PRK7 and PRK 11, and evaluation of their antioxidant, emulsion, and antibiofilm activities. Int. J. Biol. Macromol. 2023, 242, 124842. [Google Scholar] [CrossRef] [PubMed]
- Saif, F.A.A.A.; Sakr, E.A.E. Characterization and bioactivities of exopolysaccharide produced from probiotic Lactobacillus plantarum 47FE and Lactobacillus pentosus 68FE. Bioact. Carbohydr. Diet. Fibre 2020, 24, 100231. [Google Scholar] [CrossRef]
- Wang, X.; Shao, C.; Liu, L.; Guo, X.; Xu, Y.; Lü, X. Optimization, partial characterization and antioxidant activity of an exopolysaccharide from Lactobacillus plantarum KX041. Int. J. Biol. Macromol. 2017, 103, 1173–1184. [Google Scholar] [CrossRef]

| No. | Isolate Index | Cell Shape | Gram | Catalase | Food Source |
|---|---|---|---|---|---|
| 1 | BB1 | Cocci | Gram (+) | Catalase (−) | Rye Boza |
| 2 | BB3 | Cocci | Gram (+) | Catalase (−) | |
| 3 | T1A | Cocci | Gram (+) | Catalase (−) | |
| 4 | T3 | Rods | Gram (+) | Catalase (−) | |
| 5 | ED1 | Cocci | Gram (+) | Catalase (−) | Wheat Boza |
| 6 | ED2 | Cocci | Gram (+) | Catalase (−) | |
| 7 | ED3 | Rods | Gram (+) | Catalase (−) | |
| 8 | ED4 | Cocci | Gram (+) | Catalase (−) | |
| 9 | ED6 | Cocci | Gram (+) | Catalase (−) | |
| 10 | LP | Cocci | Gram (+) | Catalase (−) | Wheat Boza Bena |
| 11 | LIN1 | Cocci | Gram (+) | Catalase (−) | |
| 12 | LIN2 | Cocci | Gram (+) | Catalase (−) | |
| 13 | BZ3 | Cocci | Gram (+) | Catalase (−) | Rye-Wheat Boza |
| 14 | BZ8 | Cocci | Gram (+) | Catalase (−) | |
| 15 | BZ5 | Rods | Gram (+) | Catalase (−) | |
| 16 | A1 | Rods | Gram (+) | Catalase (−) | |
| 17 | BMT | Cocci | Gram (+) | Catalase (−) | |
| 18 | CBZ | Cocci | Gram (+) | Catalase (−) | |
| 19 | B17 | Cocci | Gram (+) | Catalase (−) | Beninese White Sorghum Degue |
| 20 | B31 | Rods | Gram (+) | Catalase (−) | |
| 21 | B16 | Cocci | Gram (+) | Catalase (−) | |
| 22 | B21 | Cocci | Gram (+) | Catalase (−) | |
| 23 | B262 | Cocci | Gram (+) | Catalase (−) | |
| 24 | ZM1 | Rods | Gram (+) | Catalase (−) | Sauerkraut |
| 25 | ZM2 | Rods | Gram (+) | Catalase (−) | |
| 26 | ZM3 | Rods | Gram (+) | Catalase (−) | |
| 27 | ZM4 | Rods | Gram (+) | Catalase (−) | |
| 28 | ZM5 | Rods | Gram (+) | Catalase (−) | |
| 29 | ZM6 | Rods | Gram (+) | Catalase (−) | |
| 30 | ZM7 | Rods | Gram (+) | Catalase (−) | |
| 31 | ZM8 | Rods | Gram (+) | Catalase (−) | |
| 32 | ZJ1 | Cocci | Gram (+) | Catalase (−) | |
| 33 | ZJ2 | Cocci | Gram (+) | Catalase (−) | |
| 34 | ZJ3 | Cocci | Gram (+) | Catalase (−) | |
| 35 | ZJ4 | Cocci | Gram (+) | Catalase (−) | |
| 36 | ZJ5 | Cocci | Gram (+) | Catalase (−) | |
| 37 | ZS1 | Rods | Gram (+) | Catalase (−) | |
| 38 | ZS3 | Rods | Gram (+) | Catalase (−) | |
| 39 | ZS4 | Rods | Gram (+) | Catalase (−) | |
| 40 | ZS5 | Rods | Gram (+) | Catalase (−) | |
| 41 | ZA3 | Rods | Gram (+) | Catalase (−) | |
| 42 | ZH2 | Rods | Gram (+) | Catalase (−) | |
| 43 | ZE2 | Rods | Gram (+) | Catalase (−) |
| No. | Strain Index | Species | Similarity, % | Food Source |
|---|---|---|---|---|
| 1 | BB1 | Pediococcus acidilactici | 99.35 | Rye boza |
| 2 | BB3 | Pediococcus acidilactici | 99.47 | Rye boza |
| 3 | T1A | Pediococcus acidilactici | 98.48 | Rye boza |
| 4 | T3 | Limosilactobacillus fermentum | 99.49 | Rye boza |
| 5 | ED1 | Pediococcus acidilactici | 99.32 | Wheat Boza |
| 6 | ED2 | Pediococcus pentosaceus | 99.61 | Wheat Boza |
| 7 | ED3 | Limosilactobacillus fermentum | 99.83 | Wheat Boza |
| 8 | ED4 | Pediococcus acidilactici | 99.66 | Wheat Boza |
| 9 | ED6 | Pediococcus pentosaceus | 100.00 | Wheat Boza |
| 10 | LP | Enterococcus faecium | 100.00 | Wheat Boza Bena |
| 11 | LIN1 | Pediococcus pentosaceus | 99.22 | Wheat Boza Bena |
| 12 | LIN2 | Pediococcus acidilactici | 99.48 | Wheat Boza Bena |
| 13 | BZ3 | Pediococcus acidilactici | 99.40 | Rye-Wheat Boza |
| 14 | BZ8 | Enterococcus faecium | 99.49 | Rye-Wheat Boza |
| 15 | BZ5 | Lactiplantibacillus plantarum | 99.40 | Rye-Wheat Boza |
| 16 | A1 | Levilactobacillus brevis | 100.00 | Rye-Wheat Boza |
| 17 | BMT | Pediococcus acidilactici | 99.50 | Rye-Wheat Boza |
| 18 | CBZ | Pediococcus acidilactici | 98.83 | Rye-Wheat Boza |
| 19 | B17 | Pediococcus acidilactici | 97.9 | Beninese White Sorghum Degue |
| 20 | B31 | Limosilactobacillus fermentum | 99.78 | Beninese White Sorghum Degue |
| 21 | B16 | Pediococcus acidilactici | 96.85 | Beninese White Sorghum Degue |
| 22 | B21 | Enterococcus faecium | 99.56 | Beninese White Sorghum Degue |
| 23 | B262 | Pediococcus acidilactici | 97.79 | Beninese White Sorghum Degue |
| 24 | ZM1 | Lactiplantibacillus plantarum | 99.91 | Sauerkraut |
| 25 | ZM2 | Lactiplantibacillus plantarum | 99.57 | Sauerkraut |
| 26 | ZM3 | Lactiplantibacillus plantarum | 99.83 | Sauerkraut |
| 27 | ZM4 | Lactiplantibacillus plantarum | 99.56 | Sauerkraut |
| 28 | ZM5 | Lactiplantibacillus plantarum | 100.00 | Sauerkraut |
| 29 | ZM6 | Lactiplantibacillus plantarum | 99.83 | Sauerkraut |
| 30 | ZM7 | Lactiplantibacillus plantarum | 99.40 | Sauerkraut |
| 31 | ZM8 | Lactiplantibacillus plantarum | 99.4 | Sauerkraut |
| 32 | ZJ1 | Pediococcus pentosaceus | 99.66 | Sauerkraut |
| 33 | ZJ2 | Pediococcus pentosaceus | 99.58 | Sauerkraut |
| 34 | ZJ3 | Pediococcus pentosaceus | 99.66 | Sauerkraut |
| 35 | ZJ4 | Pediococcus pentosaceus | 99.28 | Sauerkraut |
| 36 | ZJ5 | Pediococcus pentosaceus | 99.74 | Sauerkraut |
| 37 | ZS1 | Lactiplantibacillus plantarum | 98.40 | Sauerkraut |
| 38 | ZS3 | Lactiplantibacillus plantarum | 99.66 | Sauerkraut |
| 39 | ZS4 | Lactiplantibacillus plantarum | 99.08 | Sauerkraut |
| 40 | ZS5 | Lactiplantibacillus plantarum | 99.66 | Sauerkraut |
| 41 | ZA3 | Lactiplantibacillus plantarum | 100.00 | Sauerkraut |
| 42 | ZH2 | Lactiplantibacillus plantarum | 99.24 | Sauerkraut |
| 43 | ZE2 | Lactiplantibacillus plantarum | 99.77 | Sauerkraut |
| № | Strain | Species | EPS Production | No. | Strain | Species | EPS Production |
|---|---|---|---|---|---|---|---|
| 1 | BB1 | Pediococcus acidilactici | ND | 23 | B262 | Pediococcus acidilactici | ND |
| 2 | BB3 | Pediococcus acidilactici | ND | 24 | ZM1 | Lactiplantibacillus plantarum | ND |
| 3 | T1A | Pediococcus acidilactici | ND | 25 | ZM2 | Lactiplantibacillus plantarum | ND |
| 4 | T3 | Limosilactobacillus fermentum | ND | 26 | ZM3 | Lactiplantibacillus plantarum | ND |
| 5 | ED1 | Pediococcus acidilactici | ND | 27 | ZM4 | Lactiplantibacillus plantarum | ND |
| 6 | ED2 | Pediococcus pentosaceus | ND | 28 | ZM5 | Lactiplantibacillus plantarum | ND |
| 7 | ED3 | Limosilactobacillus fermentum | ND | 29 | ZM6 | Lactiplantibacillus plantarum | ND |
| 8 | ED4 | Pediococcus acidilactici | ND | 30 | ZM7 | Lactiplantibacillus plantarum | ND |
| 9 | ED6 | Pediococcus pentosaceus | ND | 31 | ZM8 | Lactiplantibacillus plantarum | ND |
| 10 | LP | Enterococcus faecium | ND | 32 | ZJ1 | Pediococcus pentosaceus | ND |
| 11 | LIN1 | Pediococcus pentosaceus | ND | 33 | ZJ2 | Pediococcus pentosaceus | ND |
| 12 | LIN2 | Pediococcus acidilactici | ND | 34 | ZJ3 | Pediococcus pentosaceus | ND |
| 13 | BZ3 | Pediococcus acidilactici | ND | 35 | ZJ4 | Pediococcus pentosaceus | ND |
| 14 | BZ8 | Enterococcus faecium | ND | 36 | ZJ5 | Pediococcus pentosaceus | ND |
| 15 | BZ5 | Lactiplantibacillus plantarum | ND | 37 | ZS1 | Lactiplantibacillus plantarum | ++ |
| 16 | A1 | Levilactobacillus brevis | ND | 38 | ZS3 | Lactiplantibacillus plantarum | + |
| 17 | BMT | Pediococcus acidilactici | ND | 39 | ZS4 | Lactiplantibacillus plantarum | +++ |
| 18 | CBZ | Pediococcus acidilactici | ND | 40 | ZS5 | Lactiplantibacillus plantarum | ++ |
| 19 | B17 | Pediococcus acidilactici | ND | 41 | ZA3 | Lactiplantibacillus plantarum | ND |
| 20 | B31 | Limosilactobacillus fermentum | ND | 42 | ZH2 | Lactiplantibacillus plantarum | ++ |
| 21 | B16 | Pediococcus acidilactici | ND | 43 | ZE2 | Lactiplantibacillus plantarum | +++ |
| 22 | B21 | Enterococcus faecium | ND |
| Strain | EPS Concentration, mg/L | ||
|---|---|---|---|
| Glucose | Sucrose | Fructose | |
| ZS1 | 145.81 ± 16.92 | 128.91 ± 55.85 | 152.34 ± 15.63 |
| ZS3 | 178.08 ± 2.86 | 193.06 ± 19.30 | 168.81 ± 32.23 |
| ZS4 | 168.35 ± 18.74 | 182.74 ± 2.60 | 173.24 ± 41.69 |
| ZS5 | 107.9 ± 41.74 | 181.93± 14.13 | 141.78 ± 49.07 |
| ZE2 | 159.23 ± 6.33 | 211.53 ± 16.50 | 147.58 ± 25.57 |
| ZH2 | 154.6 ± 17.89 | 171.95 ± 13.51 | 161.18 ± 6.14 |
| ЕPS-1 | EPS-2 | |
|---|---|---|
| Yield (g EPS/L) 1 | 0.73 | 0.36 |
| Total carbohydrates 2, w/w% | 46 ± 0.6 | 60 ± 1.0 |
| Rhamnose | 1.8 ± 0.3 (8.8) 3 | 2.1 ± 0.1 (8.4) |
| Mannose | 1.1 ± 0.1 (5.6) | 1.0 ± 0.05 (4.6) |
| Glucose | 18.4 ± 0.1 (80) | 22.3 ± 0.4 (83) |
| Xylose | 1.2 ± 0.2 (5.6) | 1.0 ± 0.1 (4.2) |
| Uronic acids | <0.5 (0) | <0.5 (0) |
| Hexosamines | 12.6 ± 0.1 | 6.7 ± 0.2 |
| Rare sugars test | + | + |
| Acetyl content, % (w/w) | 0.7 ± 0.05 | 0.8 ± 0.0 |
| Protein, % (w/w) (N × 5.7) | 41 ± 0.5 | 41 ± 0.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelov, A.; Georgieva, A.; Petkova, M.; Bartkiene, E.; Rocha, J.M.; Ognyanov, M.; Gotcheva, V. On the Molecular Selection of Exopolysaccharide-Producing Lactic Acid Bacteria from Indigenous Fermented Plant-Based Foods and Further Fine Chemical Characterization. Foods 2023, 12, 3346. https://doi.org/10.3390/foods12183346
Angelov A, Georgieva A, Petkova M, Bartkiene E, Rocha JM, Ognyanov M, Gotcheva V. On the Molecular Selection of Exopolysaccharide-Producing Lactic Acid Bacteria from Indigenous Fermented Plant-Based Foods and Further Fine Chemical Characterization. Foods. 2023; 12(18):3346. https://doi.org/10.3390/foods12183346
Chicago/Turabian StyleAngelov, Angel, Aneliya Georgieva, Mariana Petkova, Elena Bartkiene, João Miguel Rocha, Manol Ognyanov, and Velitchka Gotcheva. 2023. "On the Molecular Selection of Exopolysaccharide-Producing Lactic Acid Bacteria from Indigenous Fermented Plant-Based Foods and Further Fine Chemical Characterization" Foods 12, no. 18: 3346. https://doi.org/10.3390/foods12183346
APA StyleAngelov, A., Georgieva, A., Petkova, M., Bartkiene, E., Rocha, J. M., Ognyanov, M., & Gotcheva, V. (2023). On the Molecular Selection of Exopolysaccharide-Producing Lactic Acid Bacteria from Indigenous Fermented Plant-Based Foods and Further Fine Chemical Characterization. Foods, 12(18), 3346. https://doi.org/10.3390/foods12183346








