Directed Modification of a GHF11 Thermostable Xylanase AusM for Enhancing Inhibitory Resistance towards SyXIP-I and Application of AusMPKK in Bread Making
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids and Primers
2.2. Chemicals
2.3. Homology Modeling and Computer-Aided Re-Design of AusM for Its Site-Directed Mutagenesis
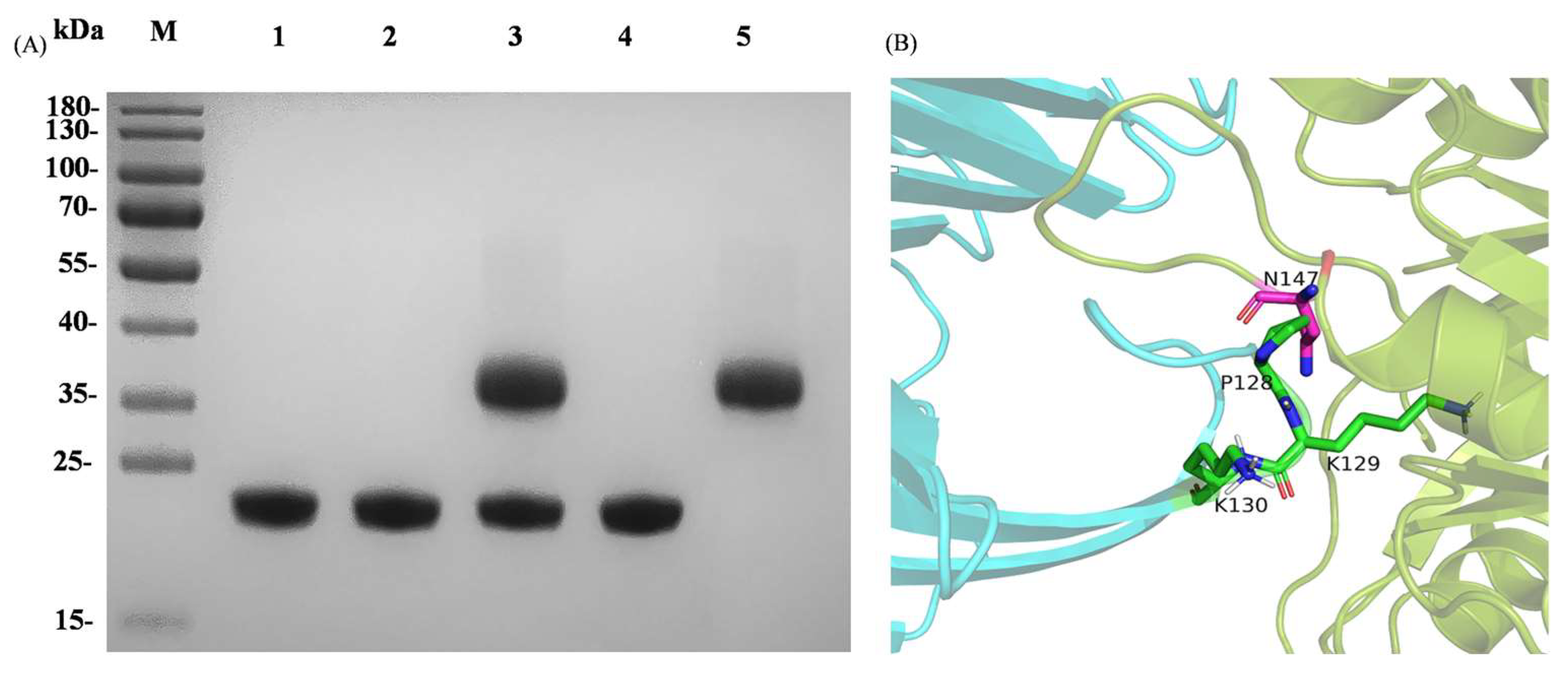
2.4. Cloning, Mutagenesis and Protein Expression
2.5. Purification of Xylanase Inhibitor SyXIP-I and Xylanases
2.6. Xylanase Activity and Inhibition Assays
2.7. Effect of Temperature on the Activity and Stability of Xylanses
2.8. Effect of EDTA and Metal Ions
2.9. Determination of Kinetic Parameters
2.10. Bread-Making Trials
2.11. Specific Volume and Texture of Bread Assays
2.12. Statistical Analysis
3. Results and Discussion
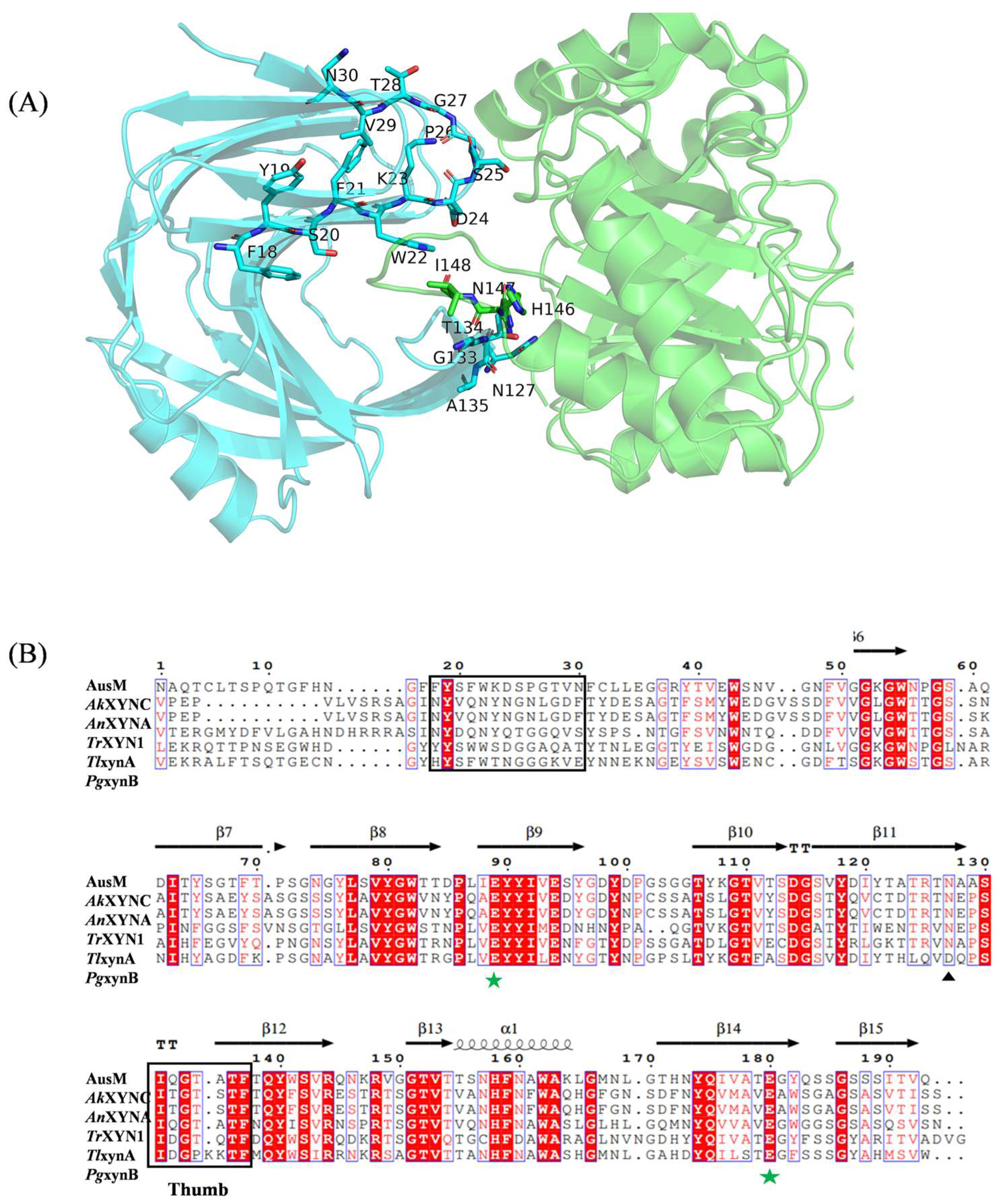
3.1. The Prediction of AusM-SyXIP Binding Sites
3.2. Inhibitor Sensitivity Assay
3.3. Interaction of AusM and Its Best Mutant and with Inhibitor SyXIP-I
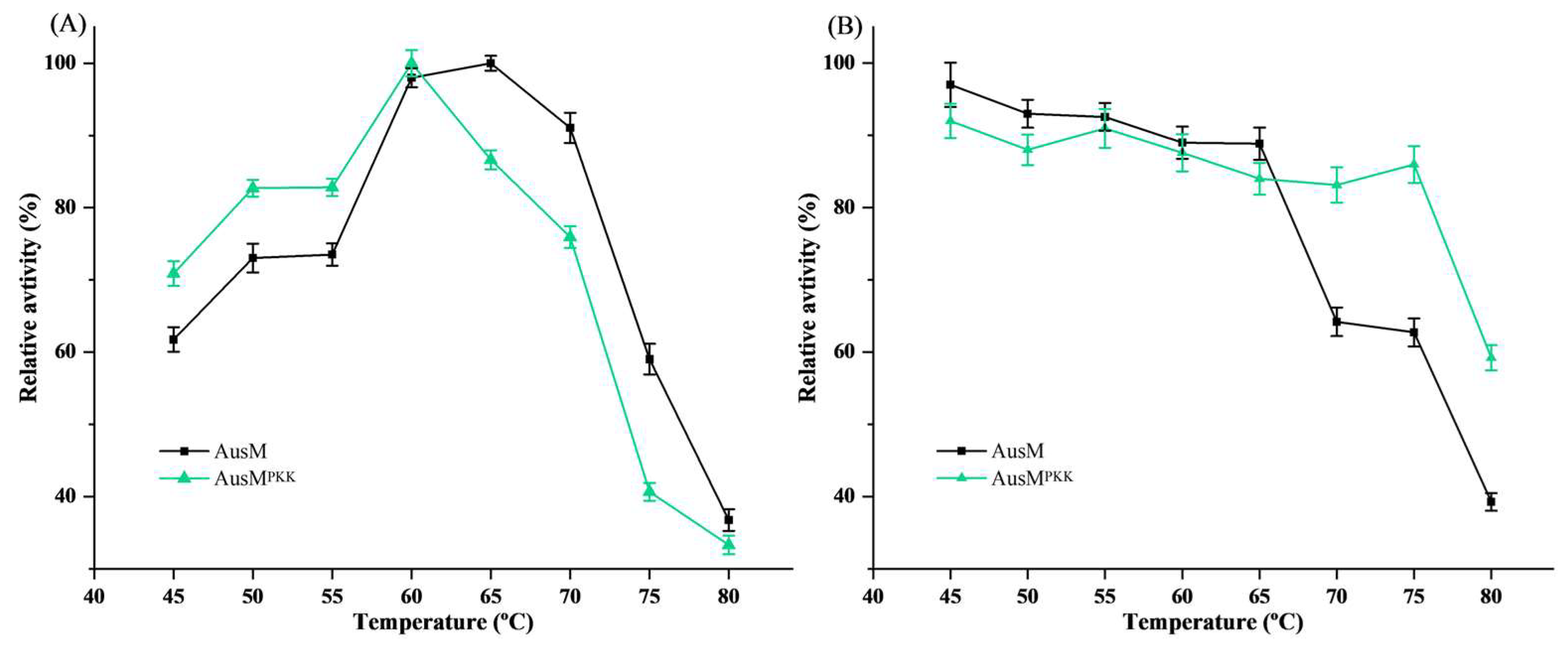
3.4. Effect of Temperature on the Activity and Stability of Xylanases
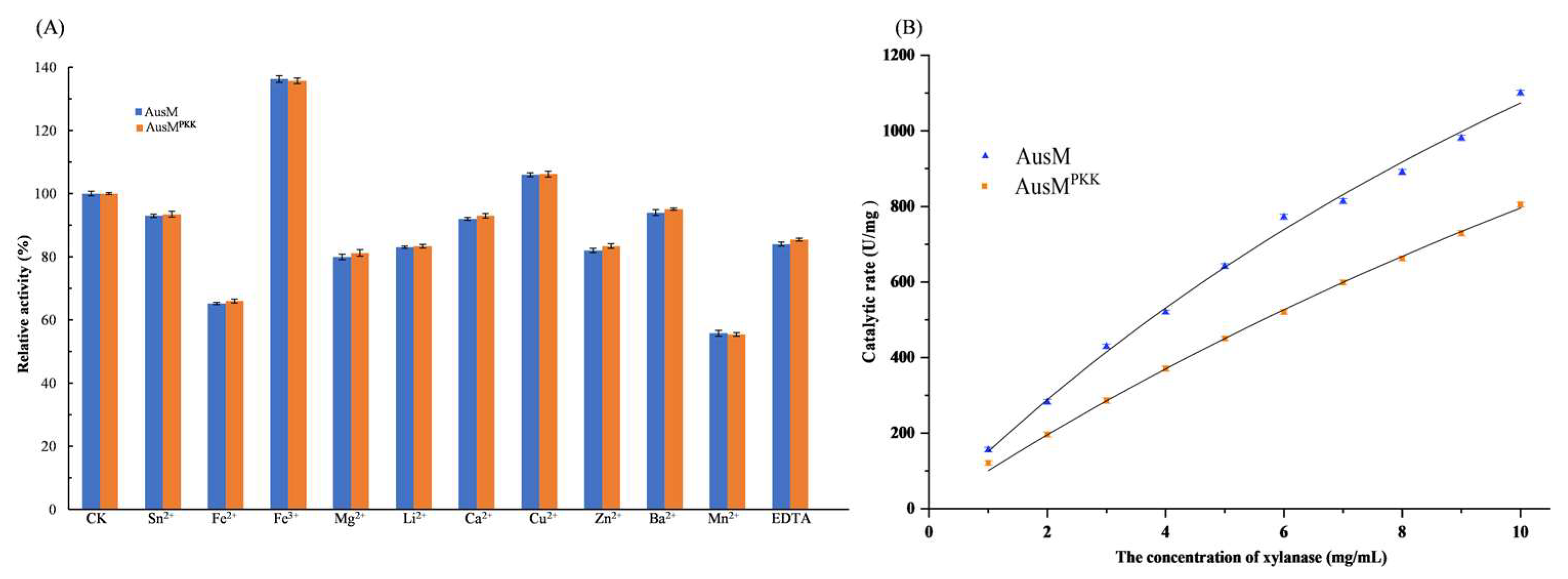
3.5. Enzymatic Properties of the Purified AusM and AusMPKK
3.6. Effect of Xylanase on Specific Bread Loaf Volume
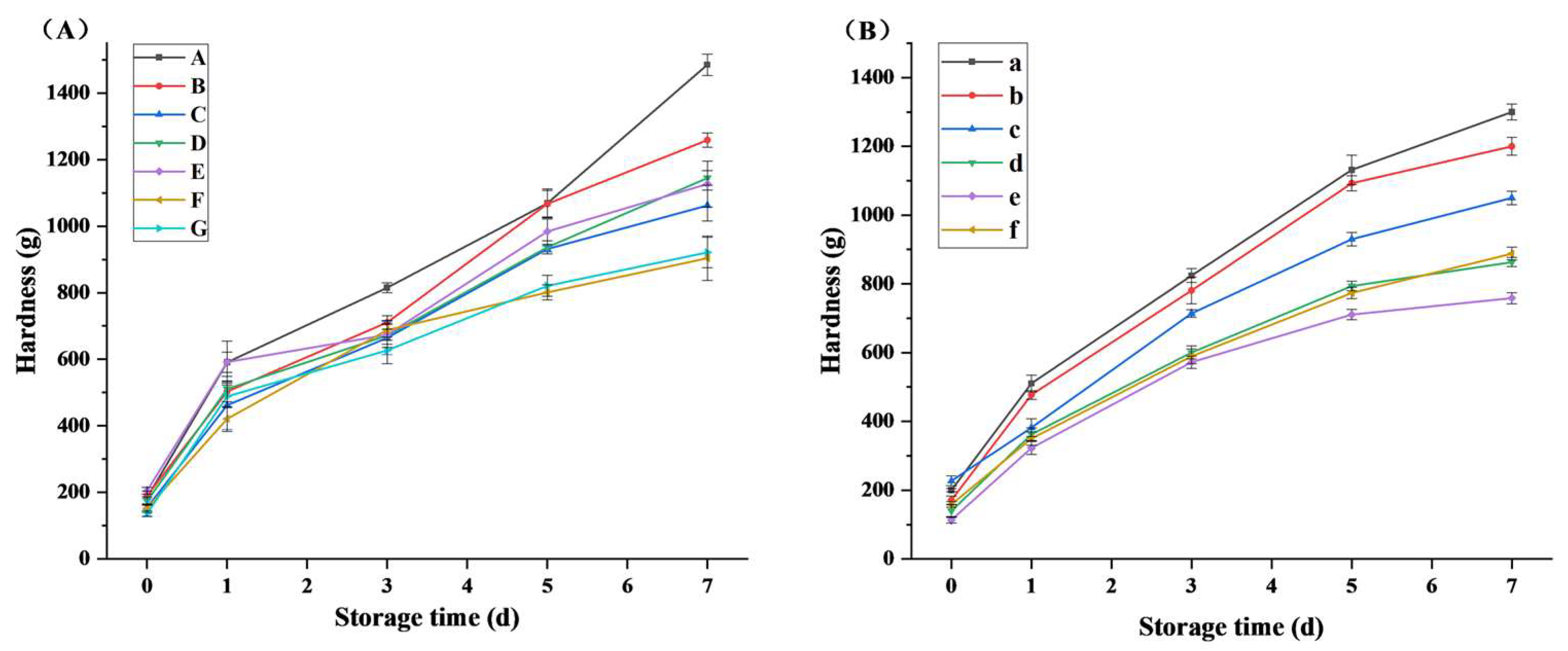
3.7. Effect of Xylanase on the Texture of Bread during Storage
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Leys, S.; Bondt, Y.D.; Bosmans, G.; Courtin, C.M. Assessing the impact of xylanase activity on the water distribution in wheat dough: A H1 NMR study. Food Chem. 2020, 325, 126828. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.M.; Cui, X.B.; Zhang, Z.H.; Zhou, T.; Gao, R.; Li, Y.X.; Ding, X.X. Effect of β-endoxylanase and α-arabinofuranosidase enzymatic hydrolysis on nutritional and technological properties of wheat brans. Food Chem. 2019, 302, 125332. [Google Scholar] [CrossRef]
- Leys, S.; Bondt, Y.D.; Schreurs, L.; Courtin, C.M. Sensitivity of the Bacillus subtilis Xyn A xylanase and its mutants to different xylanase inhibitors determines their activity profile and functionality during bread making. J. Agric. Food Chem. 2019, 67, 11198–11209. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, E.A.; dos Santos Góes, L.M.D.; Uetanabaro, A.P.T.; da Silva, E.G.P.; Rodrigues, L.B.; Pirovani, C.P.; da Costa, A.M. Thermoresistant xylanases from Trichoderma stromaticum: Application in bread making and manufacturing xylo-oligosaccharides. Food Chem. 2017, 221, 1499–1506. [Google Scholar] [CrossRef]
- Ktenioudaki, A.; Gallagher, E. Recent advances in the development of high-fibre baked products. Trends Food Sci. Tech. 2012, 28, 4–14. [Google Scholar] [CrossRef]
- Ordaz-Ortiz, J.J.; Saulnier, L. Structural variability of arabinoxylans from wheat flour. Comparison of water-extractable and xylanase-extractable arabinoxylans. J. Cereal Sci. 2005, 42, 119–125. [Google Scholar] [CrossRef]
- Izydorczyk, M.S.; Biliaderis, C.G. Cereal arabinoxylans: Advances in structure and physicochemical properties. Carbohyd. Polym. 1995, 28, 33–48. [Google Scholar] [CrossRef]
- Courtin, C.M.; Roelants, A.; Delcour, J.A. Fractionation-reconstitution experiments provide insight into the role of endoxylanases in bread-making. J. Agric. Food Chem. 1999, 47, 1870–1877. [Google Scholar] [CrossRef]
- Liu, W.J.; Brennan, M.A.; Serventi, L.; Brennan, C.S. Effect of cellulase, xylanase and α-amylase combinations on the rheological properties of Chinese steamed bread dough enriched in wheat bran. Food Chem. 2017, 234, 93–102. [Google Scholar] [CrossRef]
- Oliveira, D.S.; Telis-Romero, J.; Da-Silva, R.; Franco, C.M.L. Effect of a Thermoascus aurantiacus thermostable enzyme cocktail on wheat bread qualitiy. Food Chem. 2014, 143, 139–146. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef]
- Driss, D.; Berrin, J.G.; Juge, N.; Bhiri, F.; Ghorbel, R.; Chaabouni, S.E. Functional characterization of Penicillium occitanis Pol6 and Penicillium funiculosum GH11 xylanases. Protein Expr. Purif. 2013, 90, 195–201. [Google Scholar] [CrossRef]
- Courtin, C.M.; Delcour, J.A. Relative activity of endoxylanases towards water-extractable and water-unextractable arabinoxylan. J. Cereal Sci. 2001, 33, 301–312. [Google Scholar] [CrossRef]
- Chadha, B.S.; Kaur, B.; Basotra, N.; Tsang, A.; Pandey, A. Thermostable xylanases from thermophilic fungi and bacteria: Current perspective. Bioresour. Technol. 2019, 277, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Rouau, X.; Daviet, S.; Tahir, T.; Cherel, B.; Saulnier, L. Effect of the proteinaceous wheat xylanase inhibitor XIP-I on the performances of an Aspergillus niger xylanase in breadmaking. J. Sci. Food Agric. 2006, 86, 1604–1609. [Google Scholar] [CrossRef]
- Gebruers, K.; Brijs, K.; Courtin, C.M.; Fierens, K.; Goesaert, H.; Rabijns, A.; Raedschelders, G.; Robben, J.; Sansen, S.; Sørensen, J.F.; et al. Properties of TAXI-type endoxylanase inhibitors. BBA Proteins Proteom. 2004, 1696, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Juge, N.; Payan, F.; Williamson, G. XIP-I, a xylanase inhibitor protein from wheat: A novel protein function. BBA Proteins Proteom. 2004, 1696, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Fierens, E.; Gebruers, K.; Voet, A.R.D.; De Maeyer, M.; Courtin, C.M.; Delcour, J.A. Biochemical and structural characterization of TLXI, the Triticum aestivum L. thaumatin-like xylanase inhibitor. J. Enzym. Inhib. Med. Chem. 2009, 24, 646–654. [Google Scholar] [CrossRef]
- Croes, E.; Gebruers, K.; Luyten, N.; Delcour, J.A.; Courtin, C.M. Immunoblot quantification of three classes of proteinaceous xylanase inhibitors in different wheat (Triticum aestivum) cultivars and milling fractions. J. Agric. Food Chem. 2009, 57, 1029–1035. [Google Scholar] [CrossRef]
- Berrin, J.G.; Juge, N. Factors affecting xylanase functionality in the degradation of arabinoxylans. Biotechnol. Lett. 2008, 30, 1139–1150. [Google Scholar] [CrossRef]
- Zhu, D.D.; Liu, X.Y.; Xie, X.; Yang, S.; Lin, H.; Chen, H.G. Characteristics of a XIP-resistant xylanase from Neocallimastix sp. GMLF 1 and its advantage in barley malt saccharification. Int. J. Food Sci. Tech. 2020, 55, 2152–2160. [Google Scholar] [CrossRef]
- Sancho, A.I.; Faulds, C.B.; Svensson, B.; Bartolomé, B.; Williamson, G.; Juge, N. Cross-inhibitory activity of cereal protein inhibitors against α-amylases and xylanases. BBA Proteins Proteom. 2003, 1650, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Iwashita, K.; Iwano, K. Cloning and sequencing of the xynC gene encoding acid xylanase of Aspergillus kawachii. Biosci. Biotech. Bioch. 1992, 56, 1338–1340. [Google Scholar] [CrossRef] [PubMed]
- Sansen, S.; De Ranter, C.J.; Gebruers, K.; Brijs, K.; Courtin, C.M.; Delcour, J.A.; Rabijns, A. Structural basis for inhibition of Aspergillus niger xylanase by Triticum aestivum xylanase inhibitor-I. J. Biol. Chem. 2004, 279, 36022–36028. [Google Scholar] [CrossRef] [PubMed]
- Törrönen, A.; Mach, R.L.; Messner, R.; Gonzalez, R.; Kalkkinen, N.; Harkki, A.; Kubicek, C.P. The two major xylanases from Trichoderma reesei: Characterization of both enzymes and genes. Nat. Biotechnol. 1992, 10, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Schlacher, A.; Holzmann, K.; Hayn, M.; Steiner, W.; Schwab, H. Cloning and characterization of the gene for the thermostable xylanase XynA from Thermomyces lanuginosus. J. Biotechnol. 1996, 49, 211–218. [Google Scholar] [CrossRef]
- Tison, M.C.; Andre-Leroux, G.; Lafond, M.; Georis, J.; Juge, N.; Berrin, J.G. Molecular determinants of substrate and inhibitor specificities of the Penicillium griseofulvum family 11 xylanases. BBA Proteins Proteom. 2009, 1794, 438–445. [Google Scholar] [CrossRef]
- Zhang, H.M.; Li, J.F.; Wang, J.Q.; Yang, Y.J.; Wu, M.C. Determinants for the improved thermostability of a mesophilic family 11 xylanase predicted by computational methods. Biotechnol. Biofuels 2014, 7, 3. [Google Scholar] [CrossRef]
- Fierens, K.; Geudens, N.; Brijs, K.; Courtin, C.M.; Gebruers, K.; Robben, J.; Campenhout, S.V.; Volckaert, G.; Delcour, J.A. High-level expression, purification, and characterization of recombinant wheat xylanase inhibitor TAXI-I secreted by the yeast Pichia pastoris. Protein Expr. Purif. 2004, 37, 39–46. [Google Scholar] [CrossRef]
- Zhang, B.L.; Yang, Z.X.; Huang, W.N.; Omedi, J.O.; Wang, F.; Zou, Q.B.; Zheng, J.X. Isoflavone aglycones enrichment in soybean sourdough bread fermented by lactic acid bacteria strains isolated from traditional Qu starters: Effects on in vitro gastrointestinal digestion, nutritional, and baking properties. Cereal Chem. 2019, 96, 129–141. [Google Scholar] [CrossRef]
- AACC. Approved Methods of the American Association of Cereal Chemists; American Association of Cereal Chemists (AACC): Egan, MN, USA, 2000. [Google Scholar]
- Payan, F.; Leone, P.; Porciero, S.; Furniss, C.; Tahir, T.; Williamson, G.; Durand, A.; Manzanares, P.; Gilbert, H.J.; Juge, N.; et al. The dual nature of the wheat xylanase protein inhibitor XIP-I: Structural basis for the inhibition of family 10 and family 11 xylanases. J. Biol. Chem. 2004, 279, 36029–36037. [Google Scholar] [CrossRef] [PubMed]
- Belien, T.; Campenhout, S.V.; Acker, M.V.; Robben, J.; Courtin, C.M.; Delcour, J.A.; Volckaert, G. Mutational analysis of endoxylanases XylA and XylB from the phytopathogen Fusarium graminearum reveals comprehensive insights into their inhibitor insensitivity. Appl. Environ. Microb. 2007, 73, 4602–4608. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.M.A.; Cuong, N.T.; Nguyen, T.T.; Nguyen, N.P.D.; Tuyen, D.T. Purification, identification, and characterization of a glycoside hydrolase family 11-xylanase with high activity from Aspergillus niger VTCC 017. Mol. Biotechnol. 2022, 64, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Sengupta, R.; Sahoo, R.; Ray, S.S.; Bhattacharjee, A.; Ghosh, S. A novel cellulase free alkaliphilic xylanase from alkali tolerant Penicillium citrinum: Production, purification and characterization. Lett. Appl. Microbiol. 2007, 44, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.A.O.; Terrasan, C.R.F.; Carmona, E.C. Purification and characterization of xylanases from Trichoderma inhamatum. Electron. J. Biotechnol. 2015, 18, 307–313. [Google Scholar] [CrossRef]
- Liao, H.P.; Sun, S.W.; Wang, P.; Bi, W.L.; Tan, S.Y.; Wei, Z.; Mei, X.L.; Liu, D.Y.; Raza, W.; Shen, Q.R.; et al. A new acidophilic endo-β-1,4-xylanase from Penicillium oxalicum: Cloning, purification, and insights into the influence of metal ions on xylanase activity. J. Ind. Microbiol. Biotechnol. 2014, 41, 1071–1083. [Google Scholar] [CrossRef]
- de Amo, G.S.; Bezerra-Bussoli, C.; da Silva, R.R.; Kishi, L.T.; Ferreira, H.; Mariutti, R.B.; Arni, R.K.; Gomes, E.; Bonilla-Rodriguez, G.O. Heterologous expression, purification and biochemical characterization of a new xylanase from Myceliophthora heterothallica F.2.1.4. Int. J. Biol. Macromol. 2019, 131, 798–805. [Google Scholar] [CrossRef]
- Intasit, R.; Cheirsilp, B.; Suyotha, W.; Boonsawang, P. Purification and characterization of a highly-stable fungal xylanase from Aspergillus tubingensis cultivated on palm wastes through combined solid-state and submerged fermentation. Prep. Biochem. Biotechnol. 2022, 52, 311–317. [Google Scholar] [CrossRef]
- Silva, P.O.D.; Guimaraes, N.C.D.A.; Serpa, J.D.M.; Masui, D.C.; Marchetti, C.R.; Verbisck, N.V.; Zanoelo, F.F.; Ruller, R.; Giannesi, G.C. Application of an endo-xylanase from Aspergillus japonicus in the fruit juice clarification and fruit peel waste hydrolysis. Biocatal. Agric. Biotech. 2019, 21, 101312. [Google Scholar] [CrossRef]
- Pescador-Piedra, J.C.; Garrido-Castro, A.; Chanona-Perez, J.; Farrera-Rebollo, R.; Gutierrez-Lopez, G.; Calderon-Dominguez, G. Effect of the addition of mixtures of glucose oxidase, peroxidase and xylanase on rheological and breadmaking properties of wheat flour. Int. J. Food Prop. 2009, 12, 748–765. [Google Scholar] [CrossRef]
- Li, Y.Y.; Li, C.; Huang, H.; Rao, S.Q.; Zhang, Q.; Zhou, J.W.; Li, J.H.; Du, G.C.; Liu, L. Significantly enhanced thermostability of Aspergillus niger xylanase by modifying its highly flexible regions. J. Agr. Food Chem. 2022, 70, 4620–4630. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, J.C.; Teng, C.; Li, X.T. Cloning, overexpression and characterization of a xylanase gene from a novel Streptomyces rameus L2001 in Pichia pastoris. J. Mol. Catal. B Enzym. 2016, 131, 85–93. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, D.; Varghese, L.M.; Mahajan, R. Fast flow rate processes for purification of alkaline xylanase isoforms from Bacillus pumilus AJK and their biochemical characterization for industrial application purposes. Biotechnol. Progr. 2020, 36, e2898. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Q.; Weng, X.Y.; Sun, J.Y. Expression of recombinant Aspergillus niger xylanase A in Pichia pastoris and its action on xylan. Protein Expr. Purif. 2006, 48, 292–299. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Primer Sequence (5′→3′) |
|---|---|
| AusM-F | CTTTAAGAAGGAGATATACCATGAACGCTCAAACTTGT |
| AusM-R | GTGGTGGTGGTGGTGGTGCTGAACAGTGATGGACGAAGA |
| r-pET-28a-F | CATGGTATATCTCCTTCTTAAAG |
| r-pET-28a-R | CACCACCACCACCACCAC |
| F18H-F | TCACAACGGTTTCCACTACTCTTTCTGGAAG |
| Y19A-F | AACGGTTTCTTCGCGTCTTTCTGGAAG |
| S20A-F | GGTTTCTTCTACGCGTTCTGGAAGGAC |
| F21A-F | TTCTTCTACTCTGCGTGGAAGGACAGT |
| W22A-F | TTCTACTCTTTCGCGAAGGACAGTCCA |
| K23T-F | TTCTACTCTTTCTGGACCGACAGTCCAGGT |
| D24A-F | TCTTTCTGGAAGGCGAGTCCAGGTACT |
| S25A-F | TTCTGGAAGGACGCGCCAGGTACTGTT |
| P26A-F G27A-F T28K-F V29A-F N30E-F N127A-F | CTGGAAGGACAGTGCGGGTACTGTTAATTTT AAGGACAGTCCAGCGACTGTTAATTTT GACAGTCCAGGTAAGGTTAATTTTTGT AGTCCAGGTACTGCGAATTTTTGTCTG CCAGGTACTGTTGAGTTTTGTCTGTTG CGGCTACCCGTACCAATGCGGCTTCCATTCA |
| PKK-F | GCTGCTTCCATTCAGGGACCAAAGAAGACCTTCACTCAGTAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Huang, J.; Liu, Y.; Chen, X.; Gao, T.; Li, N.; Huang, W.; Wu, M. Directed Modification of a GHF11 Thermostable Xylanase AusM for Enhancing Inhibitory Resistance towards SyXIP-I and Application of AusMPKK in Bread Making. Foods 2023, 12, 3574. https://doi.org/10.3390/foods12193574
Zhang D, Huang J, Liu Y, Chen X, Gao T, Li N, Huang W, Wu M. Directed Modification of a GHF11 Thermostable Xylanase AusM for Enhancing Inhibitory Resistance towards SyXIP-I and Application of AusMPKK in Bread Making. Foods. 2023; 12(19):3574. https://doi.org/10.3390/foods12193574
Chicago/Turabian StyleZhang, Dong, Jing Huang, Youyi Liu, Xingyi Chen, Tiecheng Gao, Ning Li, Weining Huang, and Minchen Wu. 2023. "Directed Modification of a GHF11 Thermostable Xylanase AusM for Enhancing Inhibitory Resistance towards SyXIP-I and Application of AusMPKK in Bread Making" Foods 12, no. 19: 3574. https://doi.org/10.3390/foods12193574
APA StyleZhang, D., Huang, J., Liu, Y., Chen, X., Gao, T., Li, N., Huang, W., & Wu, M. (2023). Directed Modification of a GHF11 Thermostable Xylanase AusM for Enhancing Inhibitory Resistance towards SyXIP-I and Application of AusMPKK in Bread Making. Foods, 12(19), 3574. https://doi.org/10.3390/foods12193574







