Recent and Advanced DNA-Based Technologies for the Authentication of Probiotic, Protected Designation of Origin (PDO) and Protected Geographical Indication (PGI) Fermented Foods and Beverages
Abstract
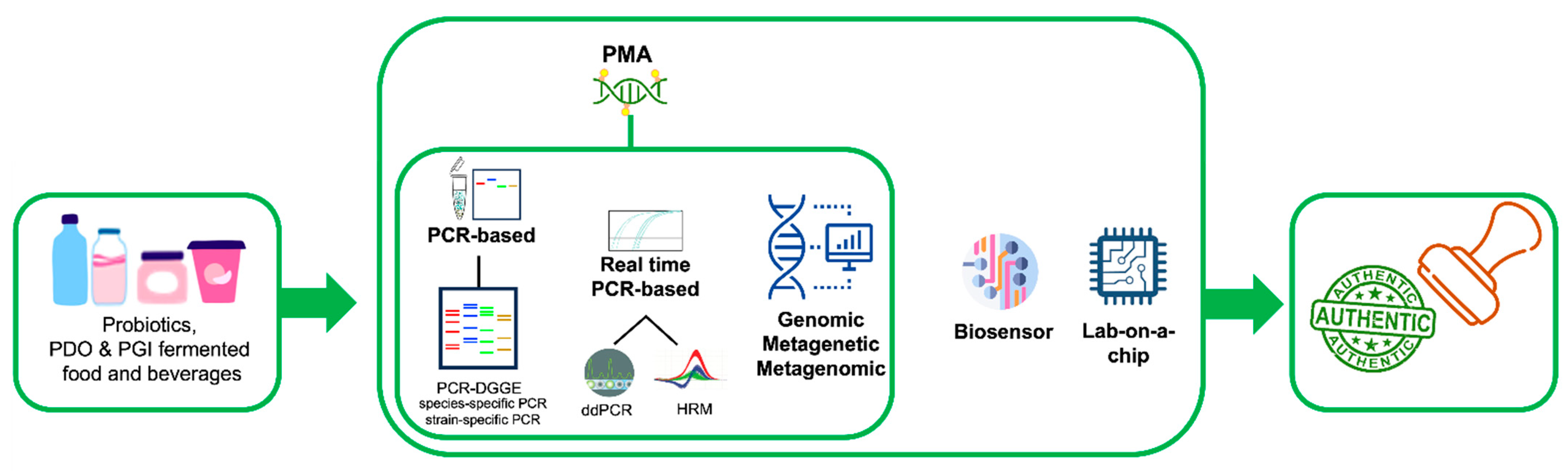
1. Introduction
2. Methods
2.1. Polymerase Chain Reaction (PCR)-Based Technologies
2.2. PCR-Based Typing Methods and Whole Genome Sequencing
2.3. Real-Time PCR-Based Methods
2.4. DNA-Based Biosensors and Lab-on-a-Chip Devices
2.5. PCR-DGGE
2.6. Metagenetics and Metagenomics
3. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Food and Drug Administration, FDA. Economically Motivated Adulteration (Food Fraud). 2023. Available online: https://www.fda.gov/food/compliance-enforcement-food/economically-motivated-adulteration-food-fraud (accessed on 1 June 2023).
- Marco, M.L.; Sanders, M.E.; Gänzle, M.; Arrieta, M.C.; Cotter, P.D.; De Vuyst, L.; Hill, C.; Holzapfel, W.; Lebeer, S.; Merenstein, D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 196–208. [Google Scholar] [CrossRef]
- European Union (EU). Geographical Indications and Quality Schemes Explained. 2023. Available online: https://agriculture.ec.europa.eu/farming/geographical-indications-and-quality-schemes/geographical-indications-and-quality-schemes-explained_en (accessed on 1 June 2023).
- European Union. Regulation (EU) No. 1151/2012 of the European Parliament and of the Council, of 21 November 2012, on Quality Schemes for Agricultural Products and Foodstuffs. 2023. Available online: https://eur-lex.europa.eu/legal-content/it/ALL/?uri=CELEX:32012R1151 (accessed on 1 June 2023).
- Dias, C.; Mendes, L. Protected Designation of Origin (PDO), Protected Geographical Indication (PGI) and Traditional Speciality Guaranteed (TSG): A bibiliometric analysis. Food Res. Int. 2018, 103, 492–508. [Google Scholar] [CrossRef] [PubMed]
- Sckokai, P.; Soregaroli, C.; Moro, D. Estimating market power by retailers in a dynamic framework: The Italian PDO cheese market. J. Agric. Econ. 2012, 64, 33–53. [Google Scholar] [CrossRef]
- Enjoy European Quality. Salumi. 2023. Available online: https://www.enjoyeuropeanquality.it/salumi/ (accessed on 1 June 2023).
- Gori, C.; Sottini, V.A. The role of the Consortia in the Italian wine production system and the impact of EU and national legislation. Wine Econ. Policy 2014, 3, 62–67. [Google Scholar] [CrossRef][Green Version]
- Danezis, G.P.; Tsagkaris, A.S.; Brusic, V.; Georgiou, C.A. Food authentication: State of the art and prospects. Curr. Opin. Food Sci. 2016, 10, 22–31. [Google Scholar] [CrossRef]
- Deng, L.; Liu, L.; Fu, T.; Li, C.; Jin, N.; Zhang, H.; Li, C.; Liu, Y.; Zhao, C. Genome sequence and evaluation of safety and probiotic potential of Lactiplantibacillus plantarum LPJZ-658. Microorganisms 2023, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- El Sheikha, A.F.; Hu, D.-M. Molecular techniques reveal more secrets of fermented foods. Crit. Rev. Food Sci. Nutr. 2018, 60, 11–32. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Fanelli, F.; de Souza, E.L. Editorial: Authenticity of Probiotic Foods and Dietary Supplements. Front. Microbiol. 2021, 12, 789049. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Fanelli, F.; Chieffi, D. Authenticity of probiotic foods and dietary supplements: A pivotal issue to address. Crit. Rev. Food Sci. Nutr. 2021, 62, 6854–6871, Erratum in Crit. Rev. Food Sci. Nutr. 2023, 63, 4210–4215. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Fanelli, F.; Chieffi, D. Authenticity of probiotic foods and supplements: Up-to-date situation and methods to assess it. In Probiotics for Human Nutrition in Health and Disease; Leite de Souza, E., de Brito Alves, J.L., Fusco, V., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 45–74. ISBN 9780323899086. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Saiki, R.K.; Scharf, S.J.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.B.; Faloona, F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987, 155, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Garibyan, L.; Avashia, N. Polymerase Chain Reaction. J. Investig. Dermatol. 2013, 133, 1–4. [Google Scholar] [CrossRef]
- Ansari, J.M.; Colasacco, C.; Emmanouil, E.; Kohlhepp, S.; Harriott, O. Strain-level diversity of commercial probiotic isolates of Bacillus, Lactobacillus, and Saccharomyces species illustrated by molecular identification and phenotypic profiling. PLoS ONE 2019, 14, e0213841. [Google Scholar] [CrossRef]
- Neviani, E.; Bottari, B.; Lazzi, C.; Gatti, M. New developments in the study of the microbiota of raw-milk, long-ripened cheeses by molecular methods: The case of Grana Padano and Parmigiano Reggiano. Front. Microbiol. 2013, 4, 36. [Google Scholar] [CrossRef]
- Lick, S.; Keller, M.; Bockelmann, W.; Heller, J. Rapid identification of Streptococcus thermophilus by primer-specific PCR amplification based on its lacZ gene. Syst. Appl. Microbiol. 1996, 19, 74–77. [Google Scholar] [CrossRef]
- Nikolaou, A.; Saxami, G.; Kourkoutas, Y.; Galanis, A. A new methodology for rapid detection of Lactobacillus delbrueckii subsp. bulgaricus based on multiplex PCR. J. Microbiol. Methods 2011, 84, 362–364. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Quero, G.M.; Chieffi, D.; Franz, C.M. Identification of Lactobacillus brevis using a species-specific AFLP-derived marker. Int. J. Food Microbiol. 2016, 232, 90–94. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Stea, G.; Morea, M.; Visconti, A. Novel PCR-based identification of Weissella confusa using an AFLP-derived marker. Int. J. Food Microbiol. 2011, 145, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.J.H.; Timmins, M.J. Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett. Appl. Microbiol. 1999, 29, 90–92. [Google Scholar] [CrossRef]
- Fortina, M.G.; Ricci, G.; Mora, D.; Parini, C.; Manachini, P.L. Specific identification of Lactobacillus helveticus by PCR with pepC, pepN and htrA targeted primers. FEMS Microbiol. Lett. 2001, 198, 85–89. [Google Scholar] [CrossRef][Green Version]
- Dickson, E.M.; Riggio, M.P.; Macpherson, L. A novel species-specific PCR assay for identifying Lactobacillus fermentum. J. Med Microbiol. 2005, 54, 299–303. [Google Scholar] [CrossRef][Green Version]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Yang, S.-M.; Lim, B.; Park, S.H.; Rackerby, B.; Kim, H.-Y. Design of PCR assays to specifically detect and identify 37 Lactobacillus species in a single 96 well plate. BMC Microbiol. 2020, 20, 96. [Google Scholar] [CrossRef]
- Guglielmotti, D.M.; Pujato, S.A.; Quiberoni, A.; Suárez, V.B. Hsp60 gene as a reliable target for taxonomical identification and discrimination of Leuconostoc species of dairy origin. Int. Dairy J. 2021, 126, 105227. [Google Scholar] [CrossRef]
- Xiang, X.; Lu, J.; Xu, X.; Hou, X.; Diao, E.; Qian, S.; Song, H.; Liang, L.; He, Y.; Shang, Y. Rapid identification of novel specific molecular targets for PCR detection of four Enterococcus species. LWT 2023, 173, 114356. [Google Scholar] [CrossRef]
- Council for Responsible Nutrition and International Probiotics Association. Best Practices Guidelines for Probiotics. 2017. Available online: https://internationalprobiotics.org/resources/guidelines/2017-best-practices-guidelines/ (accessed on 1 June 2023).
- FAO/WHO (Food and Agriculture Organization of the United Nations/World Health Organization). Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation. 2006. Available online: http://www.fao.org/3/a-a0512e.pdf (accessed on 1 June 2023).
- Fusco, V.; Quero, G.M. Culture-dependent and culture-independent nucleic-acid-based methods used in the microbial safety assessment of milk and dairy products. Compr. Rev. Food Sci. Food Saf. 2014, 13, 493–537. [Google Scholar] [CrossRef]
- Blandino, G.; Fazio, D.; Petronio, G.P.; Inturri, R.; Tempera, G.; Furneri, P.M. Labeling quality and molecular characterization studies of products containing Lactobacillus spp. strains. Int. J. Immunopathol. Pharmacol. 2015, 29, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Di Lena, M.; Quero, G.M.; Santovito, E.; Verran, J.; De Angelis, M.; Fusco, V. A selective medium for isolation and accurate enumeration of Lactobacillus casei-group lactobacilli in probiotic milks and dairy products. Int. Dairy J. 2015, 47, 27–36. [Google Scholar] [CrossRef]
- Masco, L.; Huys, G.; De Brandt, E.; Temmerman, R.; Swings, J. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 2005, 102, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Quero, G.M.; Poltronieri, P.; Morea, M.; Baruzzi, F. Autochthonous and probiotic lactic acid bacteria employed for production of “advanced traditional cheeses”. Foods 2019, 8, 412. [Google Scholar] [CrossRef]
- Quero, G.M.; Fusco, V.; Cocconcelli, P.S.; Owczarek, L.; Borcakli, M.; Fontana, C.; Skapska, S.; Jasinska, U.T.; Ozturk, T.; Morea, M. Microbiological, physico-chemical, nutritional and sensory characterization of traditional Matsoni: Selection and use of autochthonous multiple strain cultures to extend its shelf-life. Food Microbiol. 2014, 38, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Coppola, S.; Fusco, V.; Andolfi, R.; Aponte, M.; Blaiotta, G.; Ercolini, D.; Moschetti, G. Evaluating microbial diversity during the manufacture of “fior di latte di Agerola”, a traditional raw milk cheese of Naples area. J. Dairy Res. 2006, 73, 264–272. [Google Scholar] [CrossRef]
- Johnson, B.R.; Klaenhammer, T.R. Impact of genomics on the field of probiotic research: Historical perspectives to modern paradigms. Antonie van Leeuwenhoek 2014, 106, 141–156. [Google Scholar] [CrossRef]
- Peng, X.; Ed-Dra, A.; Yue, M. Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit. Rev. Food Sci. Nutr. 2022, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Manetsberger, J.; Caballero Gómez, N.; Benomar, N. In silico genomic analysis of the potential probiotic Lactiplantibacillus pentosus CF2-10N reveals promising beneficial effects with health promoting properties. Front. Microbiol. 2022, 13, 989824, Erratum in Front. Microbiol. 2023, 14, 1242095. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Pérez Montoro, B.; Casado Muñoz, M.D.C.; Knapp, C.W.; Gálvez, A.; Benomar, N. In silico genomic insights into aspects of food safety and defense mechanisms of a potentially probiotic Lactobacillus pentosus MP-10 isolated from brines of naturally fermented Aloreña green table olives. PLoS ONE 2017, 12, e0176801. [Google Scholar] [CrossRef] [PubMed]
- Alayande, K.A.; Aiyegoro, O.A.; Nengwekhulu, T.M.; Katata-Seru, L.; Ateba, C.N. Integrated genome-based probiotic relevance and safety evaluation of Lactobacillus reuteri PNW1. PLoS ONE 2020, 15, e0235873. [Google Scholar] [CrossRef]
- Arellano, K.; Vazquez, J.; Park, H.; Lim, J.; Ji, Y.; Kang, H.-J.; Cho, D.; Jeong, H.W.; Holzapfel, W.H. Safety evaluation and whole-genome annotation of Lactobacillus plantarum strains from different sources with special focus on isolates from green tea. Probiotics Antimicrob. Proteins 2020, 12, 1057–1070. [Google Scholar] [CrossRef]
- Aziz, T.; Naveed, M.; Jabeen, K.; Shabbir, M.A.; Sarwar, A.; Zhennai, Y.; Alharbi, M.; Alshammari, A.; Alasmari, A.F. Integrated genome based evaluation of safety and probiotic characteristics of Lactiplantibacillus plantarum YW11 isolated from Tibetan kefir. Front. Microbiol. 2023, 14, 1157615. [Google Scholar] [CrossRef]
- Baccouri, O.; Boukerb, A.M.; Farhat, L.B.; Zébré, A.; Zimmermann, K.; Domann, E.; Cambronel, M.; Barreau, M.; Maillot, O.; Rincé, I.; et al. Probiotic potential and safety evaluation of Enterococcus faecalis OB14 and OB15, isolated from Traditional tunisian testouri cheese and rigouta, using physiological and genomic analysis. Front. Microbiol. 2019, 10, 881. [Google Scholar] [CrossRef]
- Bae, W.-Y.; Lee, Y.J.; Jung, W.-H.; Shin, S.L.; Kim, T.-R.; Sohn, M. Draft genome sequence and probiotic functional property analysis of Lactobacillus gasseri LM1065 for food industry applications. Sci. Rep. 2023, 13, 12212. [Google Scholar] [CrossRef] [PubMed]
- Bang, W.Y.; Ban, O.-H.; Lee, B.S.; Oh, S.; Park, C.; Park, M.-K.; Jung, S.K.; Yang, J.; Jung, Y.H. Genomic-, phenotypic-, and toxicity-based safety assessment and probiotic potency of Bacillus coagulans IDCC 1201 isolated from green malt. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab026. [Google Scholar] [CrossRef] [PubMed]
- Boucard, A.-S.; Florent, I.; Polack, B.; Langella, P.; Bermúdez-Humarán, L.G. Genome sequence and assessment of safety and potential probiotic traits of Lactobacillus johnsonii CNCM I-4884. Microorganisms 2022, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, F.; Montemurro, M.; Verni, M.; Garbetta, A.; Bavaro, A.R.; Chieffi, D.; Cho, G.-S.; Franz, C.M.A.P.; Rizzello, C.G.; Fusco, V. Probiotic potential and safety assessment of type strains of Weissella and Periweissella species. Microbiol. Spectr. 2023, 11, e0304722. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Falasconi, I.; Molinari, P.; Treu, L.; Basile, A.; Vezzi, A.; Campanaro, S.; Morelli, L. Genomic comparison of Lactobacillus helveticus strains highlights probiotic potential. Front. Microbiol. 2019, 10, 1380. [Google Scholar] [CrossRef]
- Heo, S.; Kim, J.-H.; Kwak, M.-S.; Jeong, D.-W.; Sung, M.-H. Functional genomic insights into probiotic Bacillus siamensis strain B28 from traditional Korean fermented kimchi. Foods 2021, 10, 1906. [Google Scholar] [CrossRef] [PubMed]
- Hussein, W.E.; Abdelhamid, A.G.; Rocha-Mendoza, D.; García-Cano, I.; Yousef, A.E. Assessment of safety and probiotic traits of Enterococcus durans OSY-EGY, isolated from Egyptian artisanal cheese, using comparative genomics and Phenotypic analyses. Front. Microbiol. 2020, 11, 608314. [Google Scholar] [CrossRef]
- Jiang, Y.-H.; Yang, R.-S.; Lin, Y.-C.; Xin, W.-G.; Zhou, H.-Y.; Wang, F.; Zhang, Q.-L.; Lin, L.-B. Assessment of the safety and probiotic characteristics of Lactobacillus salivarius CGMCC20700 based on whole-genome sequencing and phenotypic analysis. Front. Microbiol. 2023, 14, 1120263. [Google Scholar] [CrossRef]
- Lee, Y.-R.; Bang, W.Y.; Baek, K.-R.; Kim, G.-H.; Kang, M.-J.; Yang, J.; Seo, S.-O. Safety evaluation by phenotypic and genomic characterization of four Lactobacilli strains with probiotic properties. Microorganisms 2022, 10, 2218. [Google Scholar] [CrossRef]
- Li, B.; Zhan, M.; Evivie, S.E.; Jin, D.; Zhao, L.; Chowdhury, S.; Sarker, S.K.; Huo, G.; Liu, F. Evaluating the safety of potential probiotic Enterococcus durans KLDS6.0930 using whole genome sequencing and oral toxicity study. Front. Microbiol. 2018, 9, 1943. [Google Scholar] [CrossRef]
- Mileriene, J.; Aksomaitiene, J.; Kondrotiene, K.; Asledottir, T.; Vegarud, G.E.; Serniene, L.; Malakauskas, M. Whole-genome sequence of Lactococcus lactis subsp. lactis LL16 confirms safety, probiotic potential, and reveals functional traits. Microorganisms 2023, 11, 1034. [Google Scholar] [CrossRef]
- Oh, Y.J.; Kim, S.-A.; Yang, S.H.; Kim, D.H.; Cheng, Y.-Y.; Kang, J.I.; Lee, S.Y.; Han, N.S. Integrated genome-based assessment of safety and probiotic characteristics of Lactiplantibacillus plantarum PMO 08 isolated from kimchi. PLoS ONE 2022, 17, e0273986. [Google Scholar] [CrossRef] [PubMed]
- Raethong, N.; Santivarangkna, C.; Visessanguan, W.; Santiyanont, P.; Mhuantong, W.; Chokesajjawatee, N. Whole-genome sequence analysis for evaluating the safety and probiotic potential of Lactiplantibacillus pentosus 9D3, a gamma-aminobutyric acid (GABA)-producing strain isolated from Thai pickled weed. Front. Microbiol. 2022, 13, 969548. [Google Scholar] [CrossRef]
- Salvetti, E.; Orrù, L.; Capozzi, V.; Martina, A.; Lamontanara, A.; Keller, D.; Cash, H.; Felis, G.E.; Cattivelli, L.; Torriani, S.; et al. Integrate genome-based assessment of safety for probiotic strains: Bacillus coagulans GBI-30, 6086 as a case study. Appl. Microbiol. Biotechnol. 2016, 100, 4595–4605. [Google Scholar] [CrossRef] [PubMed]
- Saroj, D.B.; Gupta, A.K. Genome based safety assessment for Bacillus coagulans strain LBSC (DSM 17654) for probiotic application. Int. J. Food Microbiol. 2020, 318, 108523. [Google Scholar] [CrossRef] [PubMed]
- Senan, S.; Prajapati, J.B.; Joshi, C.G. Feasibility of genome-wide screening for biosafety assessment of probiotics: A case study of Lactobacillus helveticus MTCC 5463. Probiotics Antimicrob. Proteins 2015, 7, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Stergiou, O.S.; Tegopoulos, K.; Kiousi, D.E.; Tsifintaris, M.; Papageorgiou, A.C.; Tassou, C.C.; Chorianopoulos, N.; Kolovos, P.; Galanis, A. Whole-genome sequencing, phylogenetic and genomic analysis of Lactiplantibacillus pentosus L33, a potential probiotic strain isolated from fermented sausages. Front. Microbiol. 2021, 12, 746659. [Google Scholar] [CrossRef] [PubMed]
- Surachat, K.; Kantachote, D.; Deachamag, P.; Wonglapsuwan, M. Genomic insight into Pediococcus acidilactici HN9, a potential probiotic strain isolated from the traditional Thai-style fermented beef Nhang. Microorganisms 2020, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Toropov, V.; Demyanova, E.; Shalaeva, O.; Sitkin, S.; Vakhitov, T. Whole-genome sequencing of Lactobacillus helveticus D75 and D76 confirms safety and probiotic potential. Microorganisms 2020, 8, 329. [Google Scholar] [CrossRef]
- Umanets, A.; Surono, I.S.; Venema, K. I am better than I look: Genome based safety assessment of the probiotic Lactiplantibacillus plantarum IS-10506. BMC Genom. 2023, 24, 518. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Y.; Liu, Y.; Zhong, J.; Zhang, D. Assessing the safety and probiotic characteristics of Lacticaseibacillus rhamnosus X253 via complete genome and phenotype analysis. Microorganisms 2023, 11, 140. [Google Scholar] [CrossRef]
- Castro-López, C.; García, H.S.; Martínez-Ávila, G.C.G.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Genomics-based approaches to identify and predict the health-promoting and safety activities of promising probiotic strains—A probiogenomics review. Trends Food Sci. Technol. 2020, 108, 148–163. [Google Scholar] [CrossRef]
- Fusco, V.; Riccardi, M.; Quero, G.M. Thin agar layer-versus most probable number-PCR to enumerate viable and stressed Escherichia coli O157:H7 and application in a traditional raw milk pasta filata cheese. Int. J. Food Microbiol. 2012, 159, 1–8. [Google Scholar] [CrossRef]
- Zand, E.; Froehling, A.; Schoenher, C.; Zunabovic-Pichler, M.; Schlueter, O.; Jaeger, H. Potential of flow cytometric approaches for rapid microbial detection and characterization in the food industry—A review. Foods 2021, 10, 3112. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Giffard, P.M. Microbiological applications of high-resolution melting analysis. J. Clin. Microbiol. 2012, 50, 3418–3421. [Google Scholar] [CrossRef]
- Wilhelm, J.; Pingoud, A. Real-time polymerase chain reaction. ChemBioChem 2003, 4, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T. High-resolution DNA melting analysis: Advancements and limitations. Hum. Mutat. 2009, 30, 857–859. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Chen, S.; Zheng, Y.; Zheng, X.; Lin, J.-M. Droplet-based digital PCR (ddPCR) and its applications. TrAC Trends Anal. Chem. 2023, 158, 116897. [Google Scholar] [CrossRef]
- Achilleos, C.; Berthier, F. Quantitative PCR for the specific quantification of Lactococcus lactis and Lactobacillus paracasei and its interest for Lactococcus lactis in cheese samples. Food Microbiol. 2013, 36, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Herbel, S.; Lauzat, B.; von Nickisch-Rosenegk, M.; Kuhn, M.; Murugaiyan, J.; Wieler, L.; Guenther, S. Species-specific quantification of probiotic lactobacilli in yoghurt by quantitative real-time PCR. J. Appl. Microbiol. 2013, 115, 1402–1410. [Google Scholar] [CrossRef]
- Iacumin, L.; Ginaldi, F.; Manzano, M.; Anastasi, V.; Reale, A.; Zotta, T.; Rossi, F.; Coppola, R.; Comi, G. High resolution melting analysis (HRM) as a new tool for the identification of species belonging to the Lactobacillus casei group and comparison with species-specific PCRs and multiplex PCR. Food Microbiol. 2015, 46, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kim, D.; Yang, S.-M.; Kim, H.-Y. Validation of probiotic species or subspecies identity in commercial probiotic products using high-resolution PCR method based on large-scale genomic analysis. Food Res. Int. 2022, 154, 111011. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Yang, S.-M.; Choi, C.H.; Shin, M.-K.; Kim, H.-Y. Droplet digital PCR method for the absolute quantitative detection and monitoring of Lacticaseibacillus casei. Food Microbiol. 2023, 113, 104265. [Google Scholar] [CrossRef]
- Kurbakov, K.A.; Konorov, E.A.; Minaev, M.Y.; Kuznetsova, O.A. Multiplex Real-Time PCR with HRM for Detection of Lactobacillus sakei and Lactobacillus curvatus in Food Samples. Food Technol. Biotechnol. 2019, 57, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Le Dréan, G.; Mounier, J.; Vasseur, V.; Arzur, D.; Habrylo, O.; Barbier, G. Quantification of Penicillium camemberti and P. roqueforti mycelium by real-time PCR to assess their growth dynamics during ripening cheese. Int. J. Food Microbiol. 2010, 138, 100–107. [Google Scholar] [CrossRef]
- Masco, L.; Vanhoutte, T.; Temmerman, R.; Swings, J.; Huys, G. Evaluation of real-time PCR targeting the 16S rRNA and recA genes for the enumeration of bifidobacteria in probiotic products. Int. J. Food Microbiol. 2007, 113, 351–357. [Google Scholar] [CrossRef]
- Pontonio, E.; Di Cagno, R.; Mahony, J.; Lanera, A.; De Angelis, M.; van Sinderen, D.; Gobbetti, M. Sourdough authentication: Quantitative PCR to detect the lactic acid bacterial microbiota in breads. Sci. Rep. 2017, 7, 624. [Google Scholar] [CrossRef]
- Shehata, H.R.; Newmaster, S.G. A Validated real-time PCR method for the specific identification of probiotic strain Lactobacillus rhamnosus GG (ATCC 53103). J. AOAC Int. 2020, 103, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Shehata, H.R.; Newmaster, S.G. Combined targeted and non-targeted PCR based methods reveal high levels of compliance in probiotic products sold as dietary supplements in United States and Canada. Front. Microbiol. 2020, 11, 1095. [Google Scholar] [CrossRef]
- Shehata, H.R.; Newmaster, S.G. Enumeration of probiotic strain Lacticaseibacillus rhamnosus GG (ATCC 53103) using viability real-time PCR. Probiotics Antimicrob. Proteins 2021, 13, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Shehata, H.R.; Ragupathy, S.; Allen, S.; Leyer, G.; Newmaster, S.G. Real-time PCR assays for the specific identification of probiotic strains Lactobacillus gasseri BNR17 and Lactobacillus reuteri LRC (NCIMB 30242). Probiotics Antimicrob. Proteins 2020, 13, 837–846. [Google Scholar] [CrossRef]
- Sheu, S.-J.; Hwang, W.-Z.; Chiang, Y.-C.; Lin, W.-H.; Chen, H.-C.; Tsen, H.-Y. Use of tuf gene-based primers for the PCR detection of probiotic Bifidobacterium species and enumeration of bifidobacteria in fermented milk by cultural and quantitative real-time PCR methods. J. Food Sci. 2010, 75, M521–M527. [Google Scholar] [CrossRef] [PubMed]
- Udomsil, N.; Chen, S.; Rodtong, S.; Yongsawatdigul, J. Quantification of viable bacterial starter cultures of Virgibacillus sp. and Tetragenococcus halophilus in fish sauce fermentation by real-time quantitative PCR. Food Microbiol. 2016, 57, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Vaccalluzzo, A.; Pino, A.; Bosco, G.; Caggia, C.; Randazzo, C.L. Quantitative PCR Assay as a tool for the detection of lactobacilli in sicilian table olives produced at an industrial scale. Fermentation 2023, 9, 355. [Google Scholar] [CrossRef]
- Bilen, M.; Dufour, J.-C.; Lagier, J.-C.; Cadoret, F.; Daoud, Z.; Dubourg, G.; Raoult, D. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome 2018, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Barzideh, Z.; Siddiqi, M.; Mohamed, H.M.; LaPointe, G. Dynamics of starter and non-starter lactic acid bacteria populations in long-ripened cheddar cheese using propidium monoazide (PMA) treatment. Microorganisms 2022, 10, 1669. [Google Scholar] [CrossRef] [PubMed]
- Desfossés-Foucault, E.; Dussault-Lepage, V.; Le Boucher, C.; Savard, P.; LaPointe, G.; Roy, D. Assessment of probiotic viability during cheddar cheese manufacture and ripening using propidium monoazide-PCR quantification. Front. Microbiol. 2012, 3, 350. [Google Scholar] [CrossRef]
- Scariot, M.C.; Venturelli, G.L.; Prudêncio, E.S.; Arisi, A.C.M. Quantification of Lactobacillus paracasei viable cells in probiotic yoghurt by propidium monoazide combined with quantitative PCR. Int. J. Food Microbiol. 2018, 264, 1–7. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, Z.; Bao, Q.; Zhang, H. Application of propidium monoazide quantitative real-time PCR to quantify the viability of Lactobacillus delbrueckii ssp. bulgaricus. J. Dairy Sci. 2016, 99, 9570–9580. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, M.L.M.; Padilha, M.; Vieira, A.D.S.; Franco, B.D.; Martinez, R.C.R.; Saad, S.M.I. Advantageous direct quantification of viable closely related probiotics in petit-suisse cheeses under in vitro gastrointestinal conditions by propidium monoazide—qPCR. PLoS ONE 2013, 8, e82102. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on biosensors and recent development of nanostructured materials-enabled biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Ma, J.; Li, D.; Wang, R. DNA-based biosensors for the biochemical analysis: A review. Biosensors 2022, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.Y. A review on microscale polymerase chain reaction based methods in molecular diagnosis, and future prospects for the fabrication of fully integrated portable biomedical devices. Microchim. Acta 2018, 185, 285. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal amplification of nucleic acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, M.C.; Spoto, G. Integration of isothermal amplification methods in microfluidic devices: Recent advances. Biosens. Bioelectron. 2017, 90, 174–186. [Google Scholar] [CrossRef]
- Vidic, J.; Vizzini, P.; Manzano, M.; Kavanaugh, D.; Ramarao, N.; Zivkovic, M.; Radonic, V.; Knezevic, N.; Giouroudi, I.; Gadjanski, I. Point-of-need DNA testing for detection of foodborne pathogenic bacteria. Sensors 2019, 19, 1100. [Google Scholar] [CrossRef]
- Ercolini, D. PCR-DGGE fingerprinting: Novel strategies for detection of microbes in food. J. Microbiol. Methods 2004, 56, 297–314. [Google Scholar] [CrossRef]
- Chen, T.; Wu, Q.; Zhou, H.; Deng, K.; Wang, X.; Meng, F.; Yang, S.; Wang, X.; Shah, N.P.; Wei, H. Assessment of commercial probiotic products in China for labelling accuracy and probiotic characterisation of selected isolates. Int. J. Dairy Technol. 2016, 70, 119–126. [Google Scholar] [CrossRef]
- Elliot, E.; Teversham, K. An evaluation of nine probiotics available in South Africa, August 2003. S. Afr. Med. J. 2004, 94, 121–124. [Google Scholar]
- Fasoli, S.; Marzotto, M.; Rizzotti, L.; Rossi, F.; Dellaglio, F.; Torriani, S. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int. J. Food Microbiol. 2002, 82, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Patrone, V.; Molinari, P.; Morelli, L. Microbiological and molecular characterization of commercially available probiotics containing Bacillus clausii from India and Pakistan. Int. J. Food Microbiol. 2016, 237, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, J.; Britz, T.; Torriani, S.; Witthuhn, R. Identification of probiotic microorganisms in South African products using PCR-based DGGE analysis. Int. J. Food Microbiol. 2005, 98, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Aponte, M.; Fusco, V.; Andolfi, R.; Coppola, S. Lactic acid bacteria occurring during manufacture and ripening of Provolone del Monaco cheese: Detection by different analytical approaches. Int. Dairy J. 2008, 18, 403–413. [Google Scholar] [CrossRef]
- Flórez, A.B.; Mayo, B. Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE. Int. J. Food Microbiol. 2006, 110, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Arcuri, E.F.; El Sheikha, A.F.; Rychlik, T.; Piro-Métayer, I.; Montet, D. Determination of cheese origin by using 16S rDNA fingerprinting of bacteria communities by PCR–DGGE: Preliminary application to traditional Minas cheese. Food Control 2013, 30, 1–6. [Google Scholar] [CrossRef]
- Dufossé, L.; Donadio, C.; Valla, A.; Meile, J.-C.; Montet, D. Determination of speciality food salt origin by using 16S rDNA fingerprinting of bacterial communities by PCR–DGGE: An application on marine salts produced in solar salterns from the French Atlantic Ocean. Food Control 2013, 32, 644–649. [Google Scholar] [CrossRef]
- El Sheikha, A.F.; Bouvet, J.-M.; Montet, D. Biological bar code for determining the geographical origin of fruits using 28S rDNA fingerprinting of fungal communities by PCR-DGGE: An application to Shea tree fruits. Qual. Assur. Saf. Crop. Foods 2011, 3, 40–47. [Google Scholar] [CrossRef]
- El Sheikha, A.F.; Condur, A.; Métayer, I.; Le Nguyen, D.D.; Loiseau, G.; Montet, D. Determination of fruit origin by using 26S rDNA fingerprinting of yeast communities by PCR-DGGE: Preliminary application to Physalis fruits from Egypt. Yeast 2009, 26, 567–573. [Google Scholar] [CrossRef]
- El Sheikha, A.F.; Durand, N.; Sarter, S.; Okullo, J.B.; Montet, D. Study of the microbial discrimination of fruits by PCR-DGGE: Application to the determination of the geographical origin of Physalis fruits from Colombia, Egypt, Uganda and Madagascar. Food Control 2012, 24, 57–63. [Google Scholar] [CrossRef]
- Ercolini, D.; Frisso, G.; Mauriello, G.; Salvatore, F.; Coppola, S. Microbial diversity in Natural Whey Cultures used for the production of Caciocavallo Silano PDO cheese. Int. J. Food Microbiol. 2008, 124, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Mauriello, G.; Blaiotta, G.; Moschetti, G.; Coppola, S. PCR-DGGE fingerprints of microbial succession during a manufacture of traditional water buffalo mozzarella cheese. J. Appl. Microbiol. 2004, 96, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Osimani, A.; Milanović, V.; Aquilanti, L.; De Filippis, F.; Stellato, G.; Di Mauro, S.; Turchetti, B.; Buzzini, P.; Ercolini, D.; et al. Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 2015, 49, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Hamdouche, Y.; Guehi, T.; Durand, N.; Kedjebo, K.B.D.; Montet, D.; Meile, J.C. Dynamics of microbial ecology during cocoa fermentation and drying: Towards the identification of molecular markers. Food Control 2015, 48, 117–122. [Google Scholar] [CrossRef]
- Mauriello, G.; Moio, L.; Genovese, A.; Ercolini, D. Relationships between flavoring capabilities, bacterial composition, and geographical origin of natural whey cultures used for traditional water-buffalo mozzarella cheese manufacture. J. Dairy Sci. 2003, 86, 486–497. [Google Scholar] [CrossRef]
- Rychlik, T.; Szwengiel, A.; Bednarek, M.; Arcuri, E.; Montet, D.; Mayo, B.; Nowak, J.; Czarnecki, Z. Application of the PCR-DGGE technique to the fungal community of traditional Wielkopolska fried ripened curd cheese to determine its PGI authenticity. Food Control 2017, 73, 1074–1081. [Google Scholar] [CrossRef]
- Ercolini, D.; Moschetti, G.; Blaiotta, G.; Coppola, S. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo Mozzarella cheese production: Bias of culture-dependent and culture-independent analyses. Syst. Appl. Microbiol. 2001, 24, 610–617. [Google Scholar] [CrossRef]
- Yap, M.; Ercolini, D.; Álvarez-Ordóñez, A.; O’Toole, P.W.; O’Sullivan, O.; Cotter, P.D. Next-generation food research: Use of meta-omic approaches for characterizing microbial communities along the food chain. Annu. Rev. Food Sci. Technol. 2022, 13, 361–384. [Google Scholar] [CrossRef]
- Celano, G.; Costantino, G.; Calasso, M.; Randazzo, C.; Minervini, F. Distinctive traits of four apulian traditional Agri-food product (TAP) cheeses manufactured at the same dairy plant. Foods 2022, 11, 425. [Google Scholar] [CrossRef]
- Ullah, M.; Raza, A.; Ye, L.; Yu, Z. Viability and composition validation of commercial probiotic products by selective culturing combined with next-generation sequencing. Microorganisms 2019, 7, 188. [Google Scholar] [CrossRef]
- Shehata, H.R.; Newmaster, S.G. The power of DNA based methods in probiotic authentication. Front. Microbiol. 2023, 14, 1158440. [Google Scholar] [CrossRef] [PubMed]
- Tamang, J.P.; Das, S.; Kharnaior, P.; Pariyar, P.; Thapa, N.; Jo, S.-W.; Yim, E.-J.; Shin, D.-H. Shotgun metagenomics of Cheonggukjang, a fermented soybean food of Korea: Community structure, predictive functionalities and amino acids profile. Food Res. Int. 2021, 151, 110904. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Al-Zahrani, I.A.; Bibi, F.; El Ghany, M.A.; Azhar, E.I. New insights of bacterial communities in fermented vegetables from shotgun metagenomics and identification of antibiotic resistance genes and probiotic bacteria. Food Res. Int. 2022, 157, 111190. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef] [PubMed]
| Species | Target Gene | Product Encoded by the Gene | References |
|---|---|---|---|
| Streptococcus thermophylus | lacZ | Β-galactosidase enzyme | [21] |
| Lactobacillus delbrueckii subsp. bulgaricus | tuf 16S rRNA | elongation factor Tu 16S rRNA | [22] |
| Levilactobacillus brevis | Gene encoding the aldo/keto reductase of the diketogulonate-reductase family of L. brevis | aldo/keto reductase of the diketogulonate-reductase family of L. brevis | [23] |
| Weissella confusa | lepA gene | GTPbinding protein LepA (Elongation Factor 4) | [24] |
| Lacticaseibacillus casei | 16S rRNA | Ribosomal RNA | [25] |
| Lacticaseibacillus paracasei | |||
| Lacticaseibacillus rhamnosus | |||
| Lactobacillus helveticus | pepC pepN htrA | aminopeptidases C aminopeptidase N, trypsin-like serine protease | [26] |
| Limosilactobacillus fermentum | 16S rRNA | Ribosomal RNA | [27] |
| Lactiplantibacillus plantarum Lactiplantibacillus pentosus Lactiplantibacillus paraplantarum | recA | RecA | [28] |
| 37 Lactobacillus species | 16S-23S rRNA gene | [29] | |
| Leuconostoc (Ln.) mesenteroides, Ln. pseudomesenteroides, Ln. lactis and Ln. citreum | Hsp60 | [30] | |
| Enterococcus (E.) faecalis, E. faecium, E. hirae, and E. casseliflavus | 58 specific molecular targets obtained by pan-genome analysis | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fusco, V.; Fanelli, F.; Chieffi, D. Recent and Advanced DNA-Based Technologies for the Authentication of Probiotic, Protected Designation of Origin (PDO) and Protected Geographical Indication (PGI) Fermented Foods and Beverages. Foods 2023, 12, 3782. https://doi.org/10.3390/foods12203782
Fusco V, Fanelli F, Chieffi D. Recent and Advanced DNA-Based Technologies for the Authentication of Probiotic, Protected Designation of Origin (PDO) and Protected Geographical Indication (PGI) Fermented Foods and Beverages. Foods. 2023; 12(20):3782. https://doi.org/10.3390/foods12203782
Chicago/Turabian StyleFusco, Vincenzina, Francesca Fanelli, and Daniele Chieffi. 2023. "Recent and Advanced DNA-Based Technologies for the Authentication of Probiotic, Protected Designation of Origin (PDO) and Protected Geographical Indication (PGI) Fermented Foods and Beverages" Foods 12, no. 20: 3782. https://doi.org/10.3390/foods12203782
APA StyleFusco, V., Fanelli, F., & Chieffi, D. (2023). Recent and Advanced DNA-Based Technologies for the Authentication of Probiotic, Protected Designation of Origin (PDO) and Protected Geographical Indication (PGI) Fermented Foods and Beverages. Foods, 12(20), 3782. https://doi.org/10.3390/foods12203782








