1H NMR Metabolomics on Pigs’ Liver Exposed to Antibiotics Administration: An Explorative Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Experimental Design
2.3. Sample Preparation
2.4. 1H NMR Spectroscopy Analysis
2.5. Data Processing and Multivariate and Univariate Data Analysis
3. Results
3.1. Assignment of 1H NMR Spectra Signals of Hepatic Polar and Non-Polar Extracts
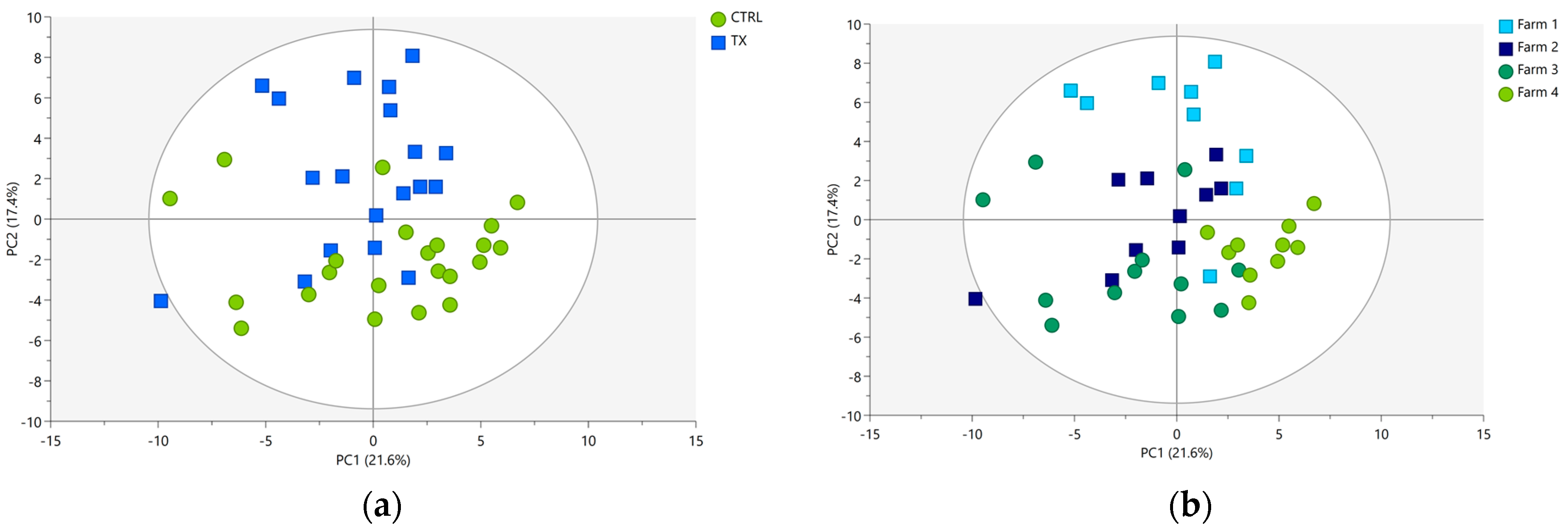
3.2. Data Exploration
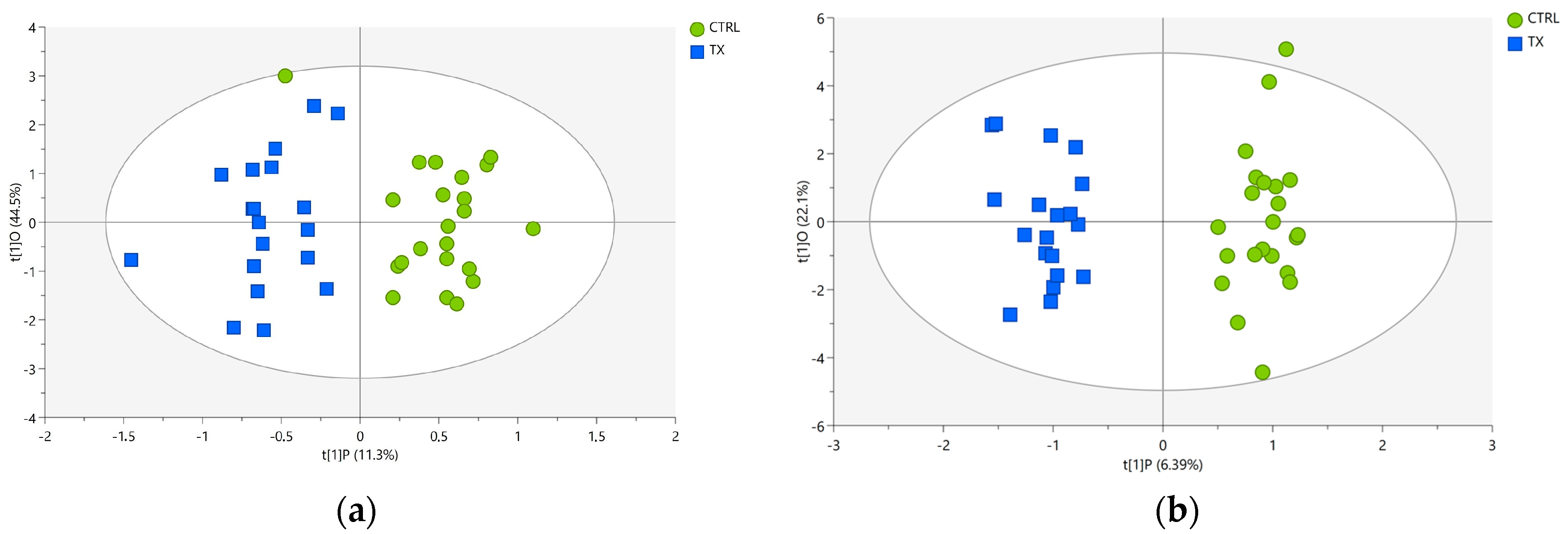
3.3. Discriminant Models and Feature Selection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grunert, K.G.; Sonntag, W.I.; Glanz-Chanos, V.; Forum, S. Consumer interest in environmental impact, safety, health and animal welfare aspects of modern pig production: Results of a cross-national choice experiment. Meat Sci. 2018, 137, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Whelan, R.; Tönges, S.; Böhl, F.; Lyko, F. Epigenetic biomarkers for animal welfare monitoring. Front. Vet. Sci. 2023, 9, 1107843. [Google Scholar] [CrossRef] [PubMed]
- Capuano, E.; Boerrigter-Eenling, R.; van der Veer, G.; van Ruth, S.M. Analytical authentication of organic products: An overview of markers. J. Sci. Food Agric. 2013, 93, 12–28. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Action Plan on Antimicrobial Resistance. WHO, 2015. Available online: https://apps.who.int/iris/handle/10665/193736 (accessed on 14 June 2023).
- Regulation (EC), n. 470/2009 of the European Parliament and of the Council of 6 May 2009 Laying Down Community Procedures for the Establishment of Residue Limits of Pharmacologically Active Substances in Foodstuffs of Animal Origin, Repealing Council Regulation (EEC) No 2377/90 and Amending Directive 2001/82/EC of the European Parliament and of the Council and Regulation (EC) No 726/2004 of the European Parliament and of the Council. Off. J. Eur. Union 2009, L152, 11–22.
- Commission Regulation (EU), n. 37/2010 of 22 December 2009 on pharmacologically active substances and their classification regarding maximum residue limits in foodstuffs of animal origin. Off. J. Eur. Union 2009, L15, 1–72.
- Regulation (EU) 2019/6 of the European Parliament and of the Council of 11 December 2018 on veterinary medicinal products and repealing Directive 2001/82/EC. Off. J. Eur. Union 2018, L4, 43–167.
- Regulation (EU) 2019/4 of the European Parliament and of the Council of 11 December 2018 on the manufacture, placing on the market and use of medicated feed, amending Regulation (EC) No 183/2005 of the European Parliament and of the Council and repealing Council Directive 90/167/EEC. Off. J. Eur. Union 2018, L4, 1–23.
- Commission Delegated Regulation (EU) 2022/1644 of 7 July 2022 supplementing Regulation (EU) 2017/625 of the European Parliament and of the Council with specific requirements for the performance of official controls on the use of pharmacologically active substances authorised as veterinary medicinal products or as feed additives and of prohibited or unauthorised pharmacologically active substances and residues thereof. Off. J. Eur. Union 2022, L248, 3–17.
- Commission Implementing Regulation (EU) 2022/1646 of 23 September 2022 on uniform practical arrangements for the performance of official controls as regards the use of pharmacologically active substances authorised as veterinary medicinal products or as feed additives and of prohibited or unauthorised pharmacologically active substances and residues thereof, on specific content of multi-annual national control plans and specific arrangements for their preparation. Off. J. Eur. Union 2022, L248, 32–45.
- Commission Implementing Regulation (EU) 2021/808 of March 2021 on the performance of the analytical methods for residues of pharmacologically active substances used in food-producing animals and on the interpretation of results as well as on the methods to be used for sampling and repealing Decisions 2002/657/EC and 98/179/EC. Off. J. Eur. Union 2021, L180, 84–109.
- Chen, J.; Ying, G.G.; Deng, W.J. Antibiotic residues in food: Extraction, analysis, and human health concerns. J. Agric. Food Chem. 2019, 67, 7569–7586. [Google Scholar] [CrossRef] [PubMed]
- Pena, A.; Lino, C.M.; Alonso, R.; Barceló, D. Determination of tetracycline antibiotic residues in edible swine tissues by liquid chromatography with spectrofluorometric detection and confirmation by mass spectrometry. J. Agric. Food Chem. 2007, 27, 4973–4979. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, L.M.; Nobile, M.; Panseri, S.; Arioli, F. Antibiotic use in heavy pigs: Comparison between urine and muscle samples from food chain animals analysed by HPLC-MS/MS. Food Chem. 2017, 235, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Masiá, A.; Suarez-Varela, M.M.; Llopis-Gonzalez, A.; Picó, Y. Determination of pesticides and veterinary drug residues in food by liquid chromatography-mass spectrometry: A review. Anal. Chim. Acta. 2016, 936, 40–61. [Google Scholar] [CrossRef] [PubMed]
- Cruz Barros, S.; Sanches Silva, A.; Torres, D. Multiresidues multiclass analytical methods for determination of antibiotics in animal arigin food: A critical analysis. Antibiotics 2023, 12, 202. [Google Scholar] [CrossRef]
- Li, X.; Chi, Q.; Xia, S.; Pan, Y.; Chen, Y.; Wang, K. Untargeted multi-residue method for the simultaneous determination of 141 veterinary drugs and their metabolites in pork by high-performance liquid chromatography time-of-flight mass spectrometry. J. Chromatogr. A 2020, 1634, 461671. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Report for 2021 on the Results from the Monitoring of Veterinary Medicinal Product Residues and Other Substances in Live Animals and Animal Products. EFSA Support. Publ. 2023, EN-7886, 111. [Google Scholar] [CrossRef]
- Jo, H.E.; Kwon, M.S.; Whon, T.W.; Kim, D.W.; Yun, M.; Lee, J.; Shin, M.Y.; Kim, S.H.; Choi, H.J. Alteration of gut microbiota after antibiotic exposure in finishing swine. Front. Microbiol. 2021, 12, 596002. [Google Scholar] [CrossRef]
- Zeineldin, M.; Aldridge, B.; Lowe, J. Antimicrobial Effects on Swine Gastrointestinal Microbiota and Their Accompanying Antibiotic Resistome. Front. Microbiol. 2019, 10, 1035. [Google Scholar] [CrossRef]
- Yang, J.H.; Bhargava, P.; McCloskey, D.; Mao, N.; Palsson, B.O.; Collins, J.J. Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell. Host Microbe. 2017, 22, 757–765.e3. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Yu, K.; Yu, M.; Zhang, C.; Su, Y.; Zhu, W. Alteration of metabolomic markers of amino-acid metabolism in piglets with in-feed antibiotics. Amino Acids 2017, 49, 771–781. [Google Scholar] [CrossRef]
- Kasper, C.; Ribeiro, D.; de Almeida, A.M.; Larzul, C.; Liaubet, L.; Murani, E. Omics application in animal science-A special emphasis on stress response and damaging behaviour in pigs. Genes 2020, 11, 920. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, H.R.; Kim, K.H.; Ji, S.Y.; Kim, M.; Lee, Y.; Lee, S.D.; Jeong, J.Y. Effects of acute heat stress on salivary metabolites in growing pigs: An analysis using nuclear magnetic resonance-based metabolomics profiling. J. Anim. Sci. Technol. 2021, 63, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wang, C.; Hao, Y.; Gu, X.; Wang, H. Chronic Heat Stress Induces Acute Phase Responses and Serum Metabolome Changes in Finishing Pigs. Animals 2019, 9, 395. [Google Scholar] [CrossRef] [PubMed]
- Fabrile, M.P.; Ghidini, S.; Conter, M.; Varrà, M.O.; Ianieri, A.; Zanardi, E. Filling gaps in animal welfare assessment through metabolomics. Front. Vet. Sci. 2023, 10, 1129741. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019, 58, 968–994. [Google Scholar] [CrossRef] [PubMed]
- Castle, A.L.; Fiehn, O.; Kaddurah-daouk, R.; Lindon, J.C. Metabolomics standards workshop and the development of international standards for reporting metabolomics experimental results. Brief Bioinform. 2006, 7, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Scali, F.; Santucci, G.; Maisano, A.M.; Giudici, F.; Guadagno, F.; Tonni, M.; Amicabile, A.; Formenti, N.; Giacomini, E.; Lazzaro, M.; et al. The Use of Antimicrobials in Italian Heavy Pig Fattening Farms. Antibiotics 2020, 9, 892. [Google Scholar] [CrossRef]
- AACTING. Guidelines for Collection, Analysis and Reporting of Farm-Level Antimicrobial Use, in the Scope of Antimicrobial Stewardship—Version 1.2. 2019. Available online: https://aacting.org/swfiles/files/AACTING_Guidelines_V1.2_2019.07.02_54.pdf (accessed on 11 July 2023).
- ClassyFarm—Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia-Romagna “BRUNO UBERTINI”. Available online: https://www.classyfarm.it/ (accessed on 10 November 2022).
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Wagner, L.; Trattner, S.; Pickova, J.; Gómez-Requeni, P.; Moazzami, A.A. 1H NMR-based metabolomics studies on the effect of sesamin in Atlantic salmon (Salmo salar). Food Chem. 2014, 147, 98–105. [Google Scholar] [CrossRef]
- Merrifield, C.A.; Lewis, M.; Claus, S.P.; Beckonert, O.P.; Dumas, M.E.; Duncker, S.; Kochhar, S.; Rezzi, S.; Lindon, J.C.; Bailey, M.; et al. A metabolic system-wide characterisation of the pig: A model for human physiology. Mol. BioSys. 2011, 7, 2577–2588. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Vosegaard, T.; Guo, Z. Applications of nuclear magnetic resonance in lipid analyses: An emerging powerful tool for lipidomics studies. Prog. Lipid Res. 2017, 68, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Dargue, R.; Zia, R.; Lau, C.; Nicholls, A.W.; Dare, T.O.; Lee, K.; Jalan, R.; Coen, M.; Wilson, I.D. Metabolism and effects on endogenous metabolism of paracetamol (Acetaminophen) in a porcine model of liver failure. Toxicol. Sci. 2020, 175, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, C.I.; Mappley, L.J.; La Ragione, R.M.; Woodward, M.J.; Claus, S.P. NMR-based metabolic characterization of chicken tissues and biofluids: A model for avian research. Metabolomics 2016, 12, 157. [Google Scholar] [CrossRef]
- Jiang, C.Y.; Yang, K.M.; Yang, L.; Miao, Z.X.; Wang, Y.H.; Zhu, H.B. A (1)H NMR-based metabonomic investigation of time-related metabolic trajectories of the plasma, urine and liver extracts of hyperlipidemic hamsters. PLoS ONE 2013, 8, e66786. [Google Scholar] [CrossRef] [PubMed]
- Fathi, F.; Brun, A.; Rott, K.H.; Cobra, P.F.; Tonelli, M.; Eghbalnia, H.R.; Caviedes-Vidal, E.; Karasov, W.H.; Markley, J.L. NMR-based identification of metabolites in polar and non-polar extracts of avian liver. Metabolites 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- BMRB—Biological Magnetic Resonance Bank. Available online: https://bmrb.io/ (accessed on 17 November 2022).
- HMDB—Human Metabolome Database. Available online: https://hmdb.ca/ (accessed on 5 December 2022).
- Andersen, C.M.; Bro, R. Variable selection in regression—A tutorial. J. Chemom. 2010, 24, 728–737. [Google Scholar] [CrossRef]
- Vinaixa, M.; Samino, S.; Saez, I.; Duran, J.; Guinovart, J.J.; Yanes, O. A Guideline to Univariate Statistical Analysis for LC/MS-Based Untargeted Metabolomics-Derived Data. Metabolites 2012, 2, 775–795. [Google Scholar] [CrossRef]
- Boccard, J.; Rutledge, D.N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal. Chim. Acta 2013, 769, 30–39. [Google Scholar] [CrossRef]
- Vaja, R.; Rana, M. Drugs and the liver. Anaesth. Intensive Care Med. 2020, 21, 517–523. [Google Scholar] [CrossRef]
- Odoi, A.; Samuels, R.; Carter, C.N.; Smith, J. Antibiotic prescription practices and opinions regarding antimicrobial resistance among veterinarians in Kentucky, USA. PLoS ONE 2021, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Mellor, D.J. Moving beyond the “Five Freedoms” by Updating the “Five Provisions” and Introducing Aligned “Animal Welfare Aims”. Animals 2016, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Guevara, R.D.; Pastor, J.J.; Manteca, X.; Tedo, G.; Llonch, P. Systematic review of animal-based indicators to measure thermal, social, and immune-related stress in pigs. PLoS ONE 2022, 17, e0266524. [Google Scholar] [CrossRef] [PubMed]
- Batchu, P.; Naldurtiker, A.; Kouakou, B.; Terrill, T.H.; McCommon, G.W.; Kannan, G. Metabolomic exploration of the effects of habituation to livestock trailer and extended transportation in goats. Front. Mol. Biosci. 2022, 9, 1027069. [Google Scholar] [CrossRef] [PubMed]
- Gjorgjieva, M.; Mithieux, G.; Rajas, F. Hepatic stress associated with pathologies characterized by disturbed glucose production. Cell. Stress 2019, 3, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.X.; Wang, Z.; Yu, T. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 2022, 7, 305. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Xiao, H.; Ren, W.; Yin, J.; Hu, J.; Duan, J.; Liu, G.; Tan, B.; Xiong, X.; Oso, A.O.; et al. An NMR-based metabolomic approach to investigate the effects of supplementation with glutamic acid in piglets challenged with deoxynivalenol. PLoS ONE 2014, 9, e113687. [Google Scholar] [CrossRef] [PubMed]
- Mazzetti, A.P.; Fiorile, M.C.; Primavera, A.; Lo Bello, M. Glutathione transferases and neurodegenerative diseases. Neurochem. Int. 2015, 82, 10–18. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- KEGG—Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.genome.jp/kegg/ (accessed on 7 January 2023).
- Rubio Gomez, M.A.; Ibba, M. Aminoacyl-tRNA synthetases. RNA 2020, 8, 910–936. [Google Scholar] [CrossRef]
- Hong, S.; Moreno-Navarrete, J.M.; Wei, X.; Kikukawa, Y.; Tzameli, I.; Prasad, D.; Lee, Y.; Asara, J.M.; Fernandez-Real, J.M.; Maratos-Flier, E.; et al. Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nat. Med. 2015, 21, 887–894. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Liu, C.; Qin, R.; Gong, D.; Wang, R.; Zhang, D.; Che, L.; Chen, D.; Xin, G.; et al. Multi-omics profiling highlights lipid metabolism alterations in pigs fed low-dose antibiotics. BMC Genet. 2020, 21, 112. [Google Scholar] [CrossRef]
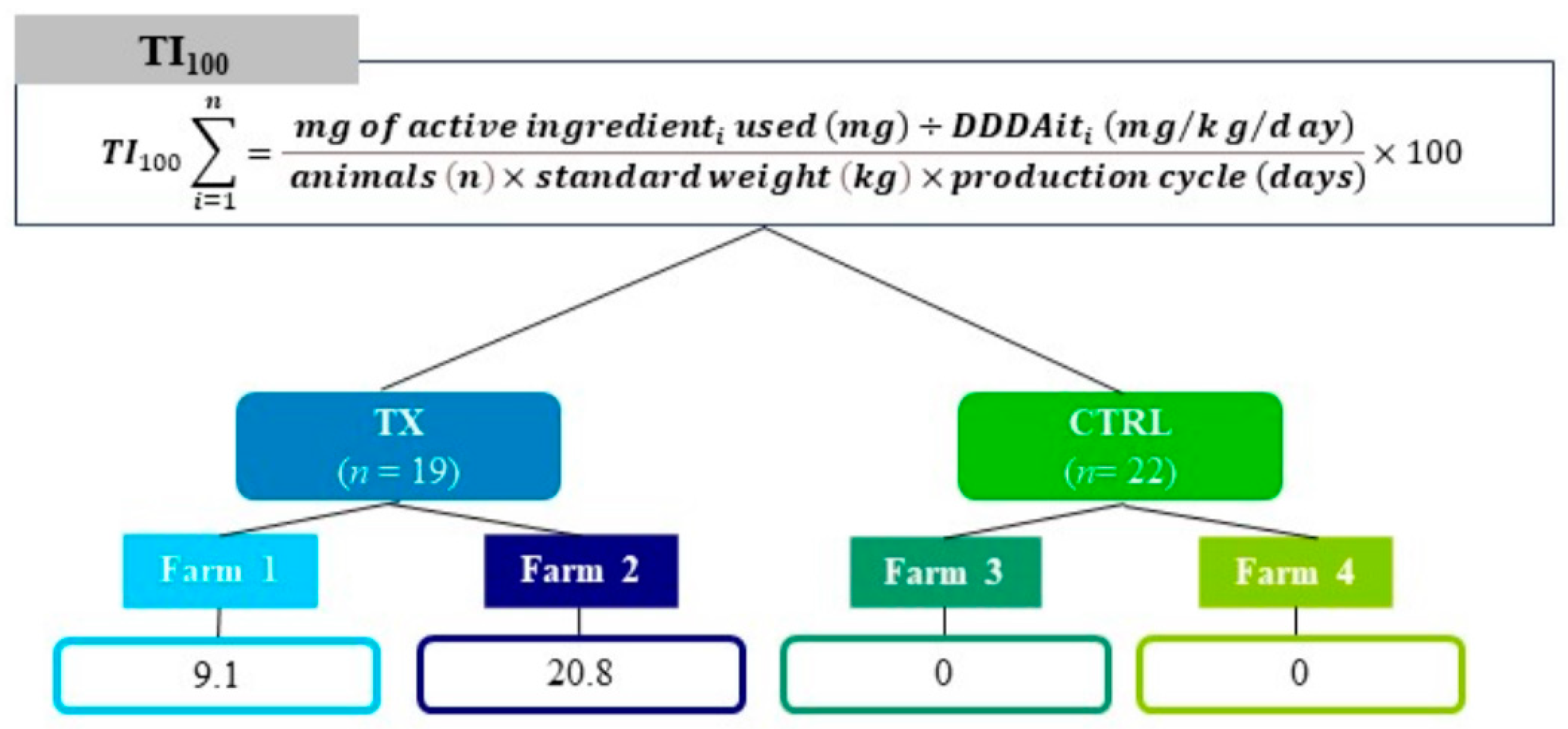
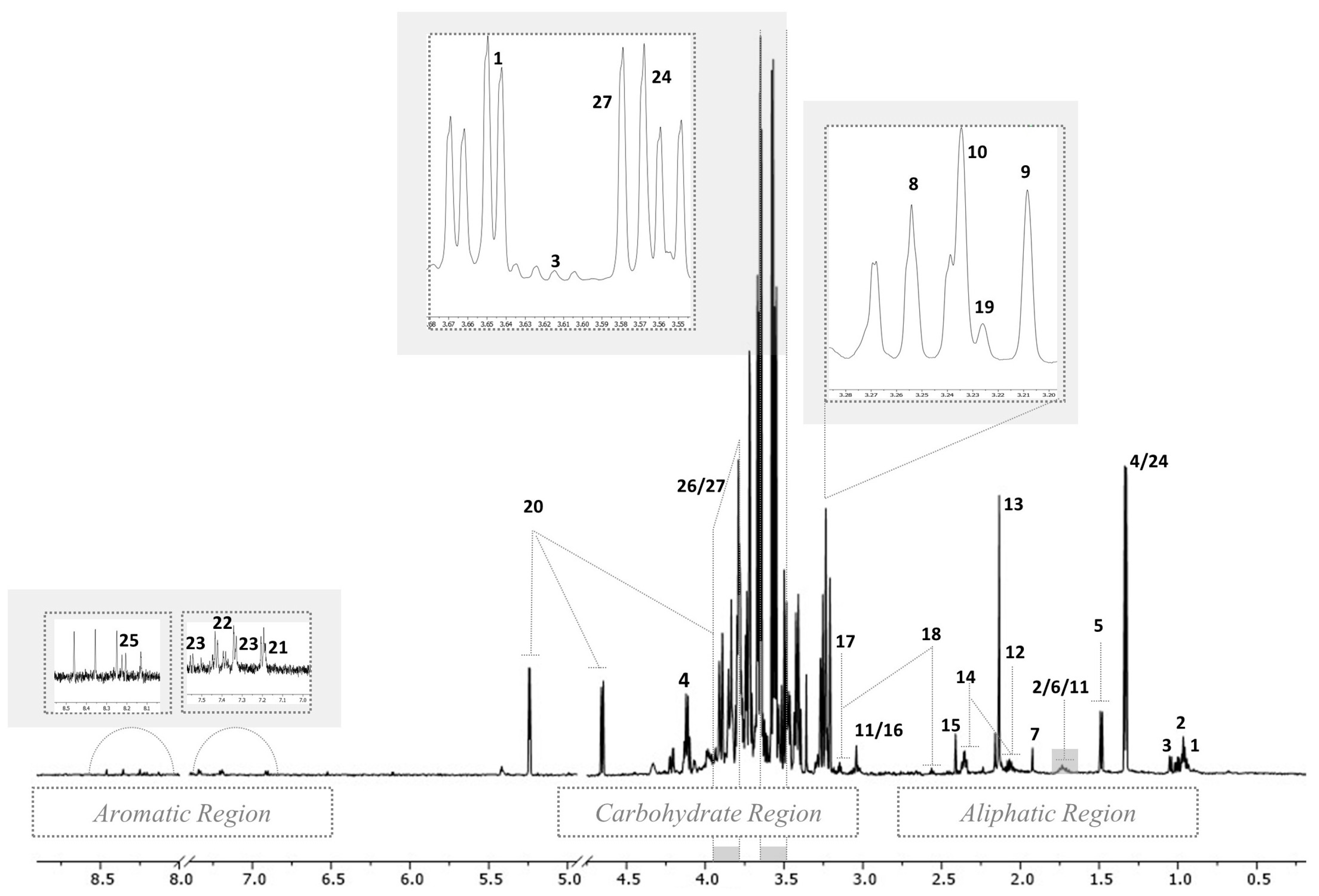
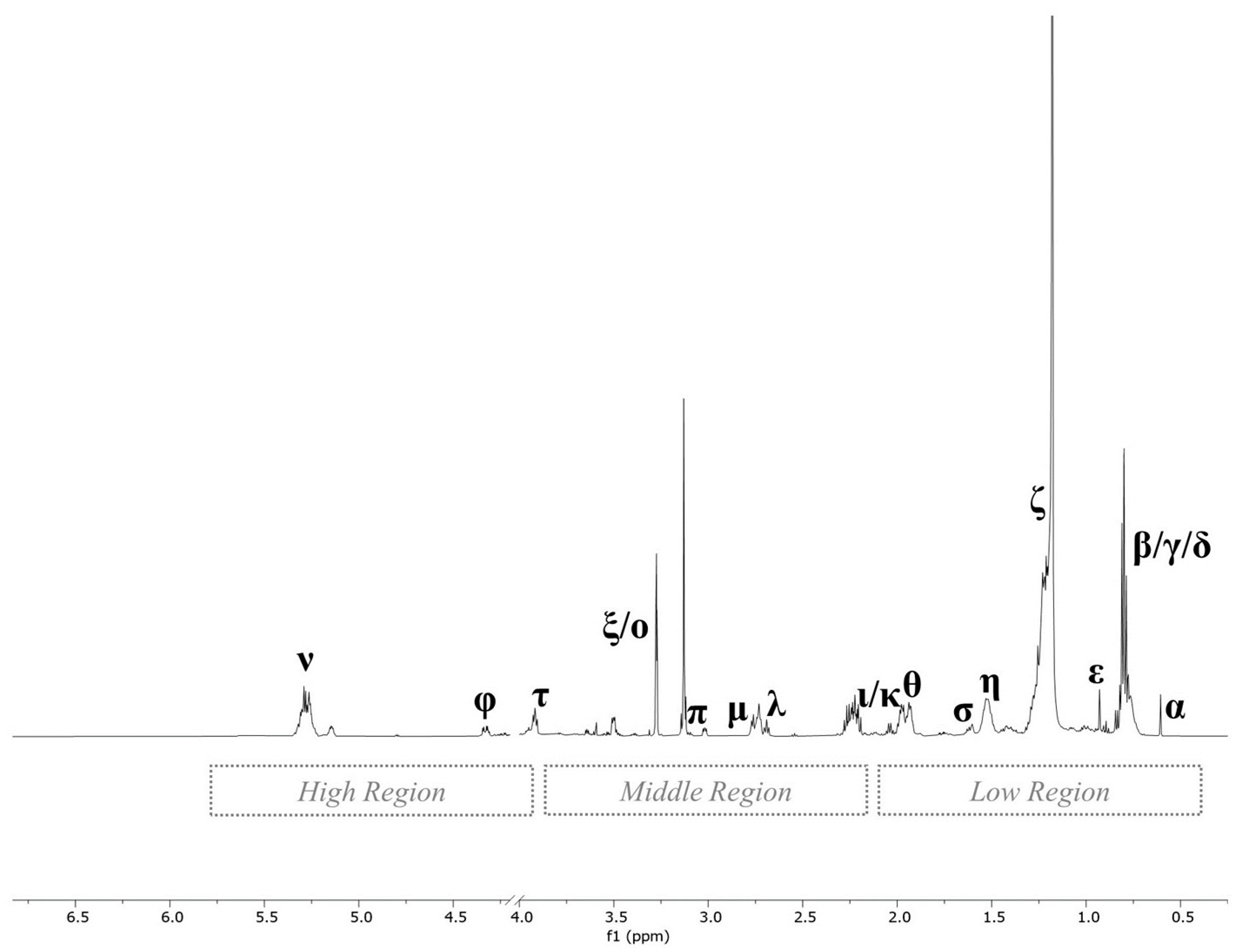
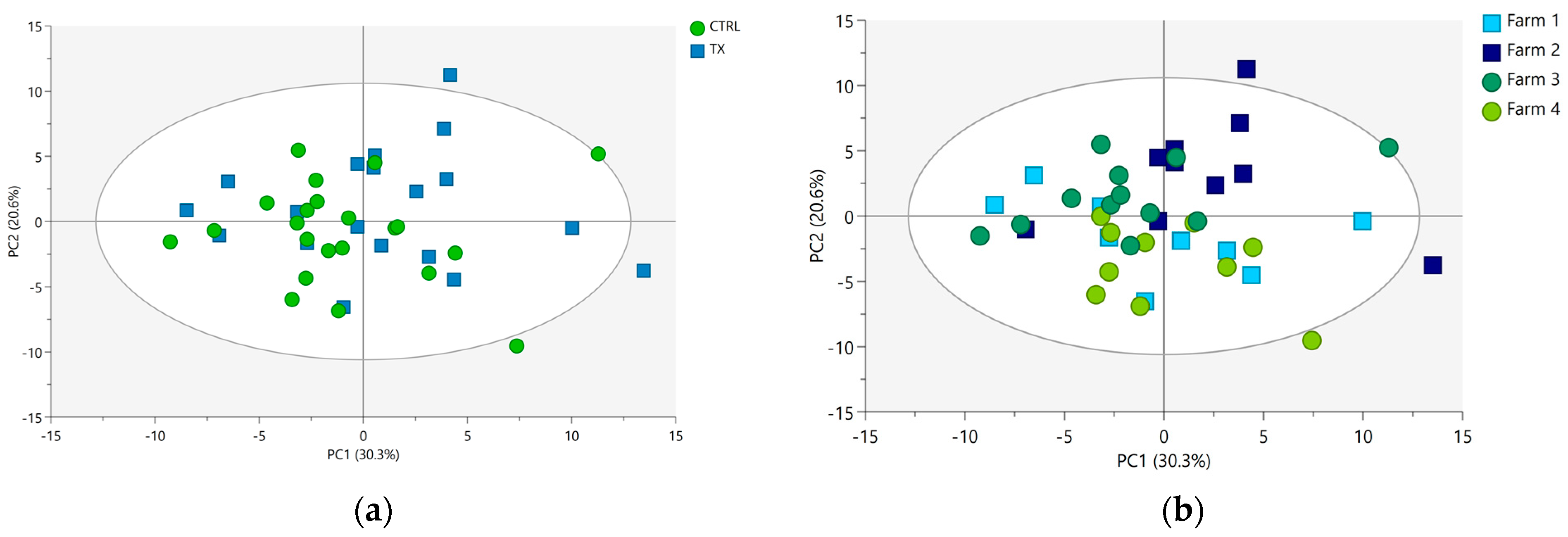
| Biochemical Category | N a | Metabolite Name | Assignment | δ ppm and Multiplicity b | Formula |
|---|---|---|---|---|---|
| AA and Derivatives | 1 | Isoleucine | γCH3 | 0.96 (t) | C6H13NO2 |
| δCH3 | 1.04 (d) | ||||
| αCH | 3.65 (d) | ||||
| 2 | Leucine | δCH3 | 0.96 (t) | C6H13NO2 | |
| βCH2 | 0.99 (d) | ||||
| αCH | 3.72 (m) | ||||
| γCH | 1.72 (m) | ||||
| 3 | Valine | γCH3 | 0.98 (d) | C5H11NO2 | |
| γCH3 | 1.04 (d) | ||||
| βCH2 | 2.25 (m) | ||||
| αCH | 3.61(d) | ||||
| 4 | Alanine | βCH3 | 1.49 (d) | C3H7NO2 | |
| 5 | Lysine | δCH2 | 1.72 (m) | C6H14N2O2 | |
| 8 | Taurine | N-CH2 | 3.25 (t) | C2H7NO3S | |
| 11 | Ornithine | ½ γCH2 | 1.72 (m) | C5H12N2O2 | |
| δCH2 | 3.04 (t) | ||||
| αCH | 3.77 (t) | ||||
| 12 | Proline | γCH2 | 2.06 (m) | C5H9NO2 | |
| 13 | Methionine | δCH3 | 2.13 (s) | C5H11NO2S | |
| αCH | 3.78 (m) | ||||
| 14 | Glutamate | βCH2 | 2.07 (m) | C5H8NO4− | |
| 2.13 (m) | |||||
| γCH2 | 2.35 (m) | ||||
| 16 | Creatine | N-CH3 | 3.04 (s) | C4H9N3O2 | |
| 18 | β-Alanine | CH2COOH | 2.56 (t) | C3H7NO2 | |
| N-CH2 | 3.18 (t) | ||||
| 21 | Tyrosine | C3H&C5H | 6.91 (d) | C9H11NO3 | |
| C2H&C6H | 7.20 (d) | ||||
| 22 | Phenylalanine | C3H &C5H | 7.33 (m) | C9H11NO2 | |
| C4H | 7.38 (m) | ||||
| C2H&C6H | 7.42 (m) | ||||
| 24 | Threonine | γCH3 | 1.33 (d) | C4H9NO3 | |
| αCH | 3.57 (d) | ||||
| 17 | Glutathione | αCH | 3.77 (m) | C10H17N3O6S | |
| 23 | Tryptophan | αCH | 4.06 (m) | C11H12N2O2 | |
| C1H | 7.33 (s) | ||||
| C3H | 7.55 (d) | ||||
| 26 | Glutamine | βCH2 | 2.13 (m) | C5H10N2O3 | |
| αCH | 3.77 (t) | ||||
| Organic Acids | 4 | Lactate | βCH3 | 1.33 (d) | C3H5O3− |
| αCH | 4.12 (q) | ||||
| 7 | Acetate | CH3 | 1.92 (s) | C2H3O2− | |
| 15 | Succinate | CH2 | 2.41 (s) | C4H4O4−2 | |
| Carbohydrates | 20 | Alpha-glucose | C4H | 3.41 (t) | C6H12O6 |
| C2H | 3.54 (m) | ||||
| C3H | 3.72 (t) | ||||
| C1H | 5.24 (d) | ||||
| Beta-Glucose | C2H | 3.24 (m) | |||
| C5H | 3.83 (m) | ||||
| C6H | 3.90 (dd) | ||||
| C1H | 4.65 (d) | ||||
| Choline and Derivatives | 9 | Choline | N-(CH3)3 | 3.21 (s) | C5H14NO+ |
| βCH2 | 3.53 (dd) | ||||
| αCH2 | 4.07 (m) | ||||
| 10 | Glycerophosphocholine | N-(CH3)3 | 3.23 (s) | C8H21NO6P+ | |
| 19 | Phosphorylcholine | N-(CH3)3 | 3.22 (s) | C5H14NO4P | |
| Alcohol | 27 | Glycerol | CH | 3.77 (m) | C3H8O3 |
| ½ CH2 | 3.57 (m) | ||||
| ½ CH2 | 3.65 (m) | ||||
| Nucleoside | 25 | Adenosine | Ring protons | 8.25 (s) | C10H13N5O4 |
| Biochemical Category | N a | Metabolite Name | Assignment | δ ppm and Multiplicity b |
|---|---|---|---|---|
| Cholesterol | α | Total cholesterol | C18H3 | 0.61 (s) |
| β | C26H3, C27H3 | 0.84 (d) | ||
| δ | C21H3 | 0.82 (d) | ||
| ε | Esterified cholesterol | C19H3 | 0.93 (s) | |
| Fatty Acids | γ | Fatty acid residues | ꙍ-CH3 | 0.90 (t) |
| ζ | (CH2)n | 1.19 (m) | ||
| η | COCH2–CH2 | 1.52 (m) | ||
| 1.61 (m) | ||||
| θ | CH2–CH=(MUFA and PUFA) | 1.97 (m) | ||
| 2.04 (m) | ||||
| κ | –CO–CH2 | 2.23 (m) | ||
| λ | –CH=CH–CH2–CH=CH– of linoleic acid | 2.68 (t) | ||
| ν | –CH=CH– | 5.28 (m) | ||
| σ | (–CH2 all FA except ARA and EPA) | 1.61 (m) | ||
| μ | FA, PUFA | CH=CH–CH2–(CH=CH–CH2)n | 2.75 (m) | |
| MAG and TAG | ι | Monoglycerides (MAG) | FA, RH–CH2–CO–O–C2 | 2.20 (m) |
| φ | Triglycerides (TAG) | C1H and C3H of glycerol backbone | 4.22 (dd) | |
| C1H and C3H of glycerol backbone | 4.26 (dd) | |||
| Phospholipids | ξ | Sphingomyelin | (–CH2–N–(CH3)3) head group | 3.27 (s) |
| ο | Phosphatidylcholine (PC) | (–CH2–N–(CH3)3) head group | 3.31 (s) | |
| π | Phosphatidylethanolamine (PE) | –CH2–CH2–NH2 | 3.13 (s) | |
| τ | Total phospholipids | Glycerol (C3H2) of phospholipids | 3.92 (m) 4.02 (m) |
| Metabolite | VIP Score a | FC b |
|---|---|---|
| Polar extract of pig liver | ||
| Glucose | 3.30 | 1.80 |
| Glutathione | 2.55 | 1.10 |
| Proline | 2.49 | 1.13 |
| Tryptophan | 2.29 | 1.13 |
| Choline | 1.30 | 1.23 |
| Lactate | 1.20 | 1.20 |
| Non-polar extract of pig liver | ||
| Fatty Acid | 4.50 | 0.95 |
| Phospholipids | 2.92 | 1.52 |
| Phosphatidylcholine | 1.94 | 1.25 |
| Total Cholesterol | 2.88 | 0.89 |
| Unsaturated Fatty Acid | 1.08 | 0.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabrile, M.P.; Ghidini, S.; Caligiani, A.; Scali, F.; Varrà, M.O.; Lolli, V.; Alborali, G.L.; Ianieri, A.; Zanardi, E. 1H NMR Metabolomics on Pigs’ Liver Exposed to Antibiotics Administration: An Explorative Study. Foods 2023, 12, 4259. https://doi.org/10.3390/foods12234259
Fabrile MP, Ghidini S, Caligiani A, Scali F, Varrà MO, Lolli V, Alborali GL, Ianieri A, Zanardi E. 1H NMR Metabolomics on Pigs’ Liver Exposed to Antibiotics Administration: An Explorative Study. Foods. 2023; 12(23):4259. https://doi.org/10.3390/foods12234259
Chicago/Turabian StyleFabrile, Maria Pia, Sergio Ghidini, Augusta Caligiani, Federico Scali, Maria Olga Varrà, Veronica Lolli, Giovanni Loris Alborali, Adriana Ianieri, and Emanuela Zanardi. 2023. "1H NMR Metabolomics on Pigs’ Liver Exposed to Antibiotics Administration: An Explorative Study" Foods 12, no. 23: 4259. https://doi.org/10.3390/foods12234259
APA StyleFabrile, M. P., Ghidini, S., Caligiani, A., Scali, F., Varrà, M. O., Lolli, V., Alborali, G. L., Ianieri, A., & Zanardi, E. (2023). 1H NMR Metabolomics on Pigs’ Liver Exposed to Antibiotics Administration: An Explorative Study. Foods, 12(23), 4259. https://doi.org/10.3390/foods12234259








