Detection Method for Tomato Leaf Mildew Based on Hyperspectral Fusion Terahertz Technology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Method
2.2. Cultivation of Samples
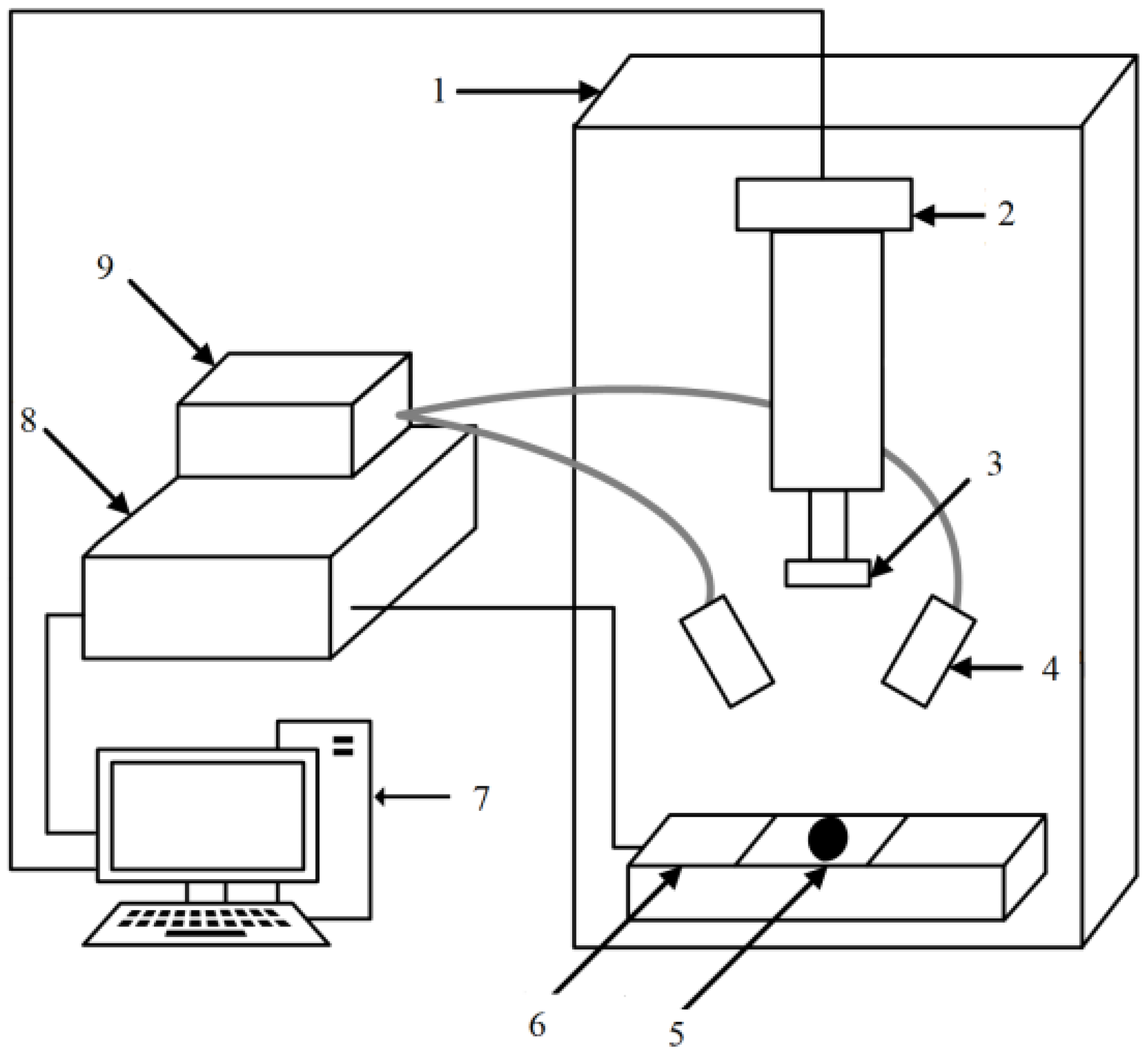
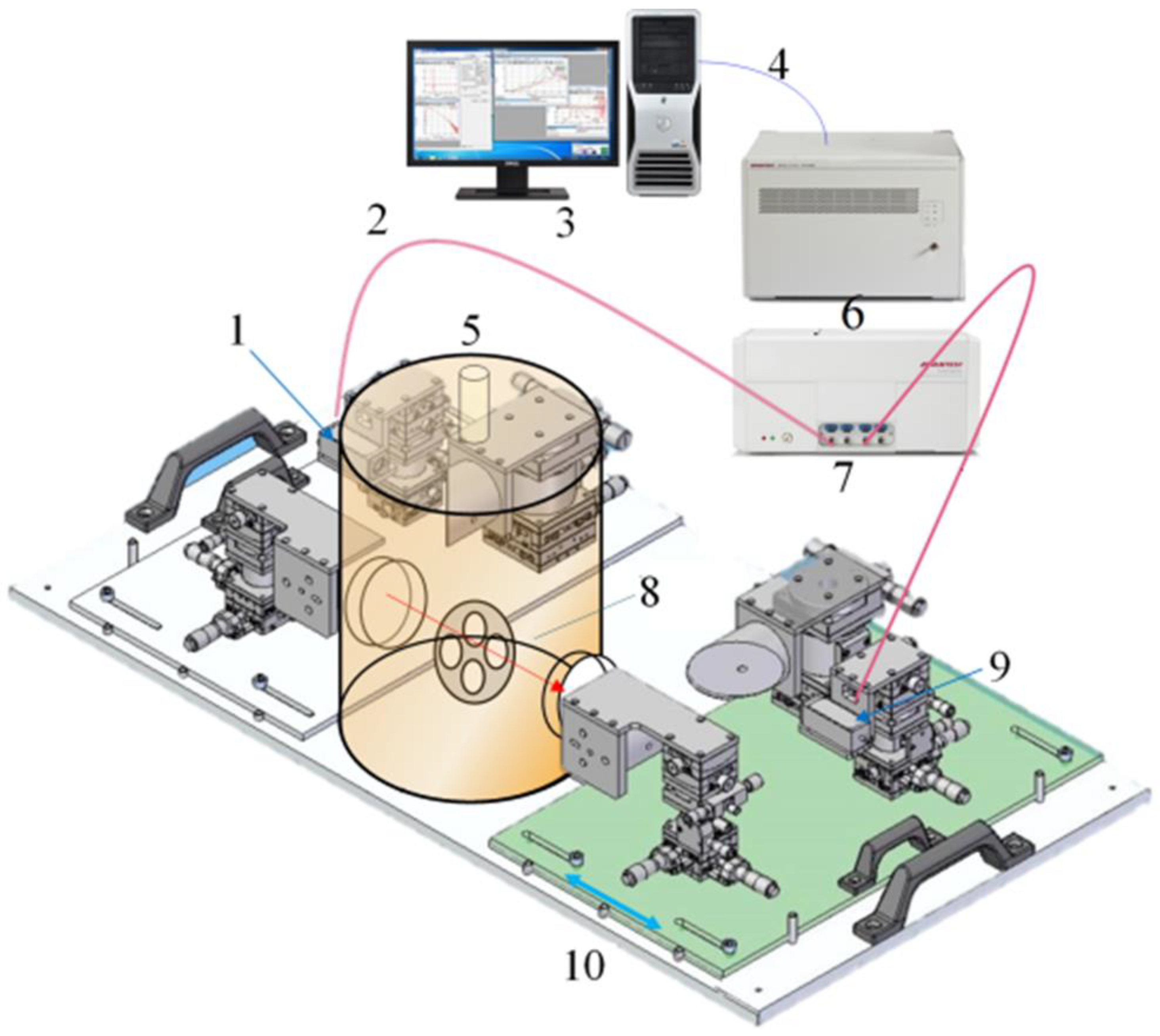
2.3. Equipment Used for Experiments
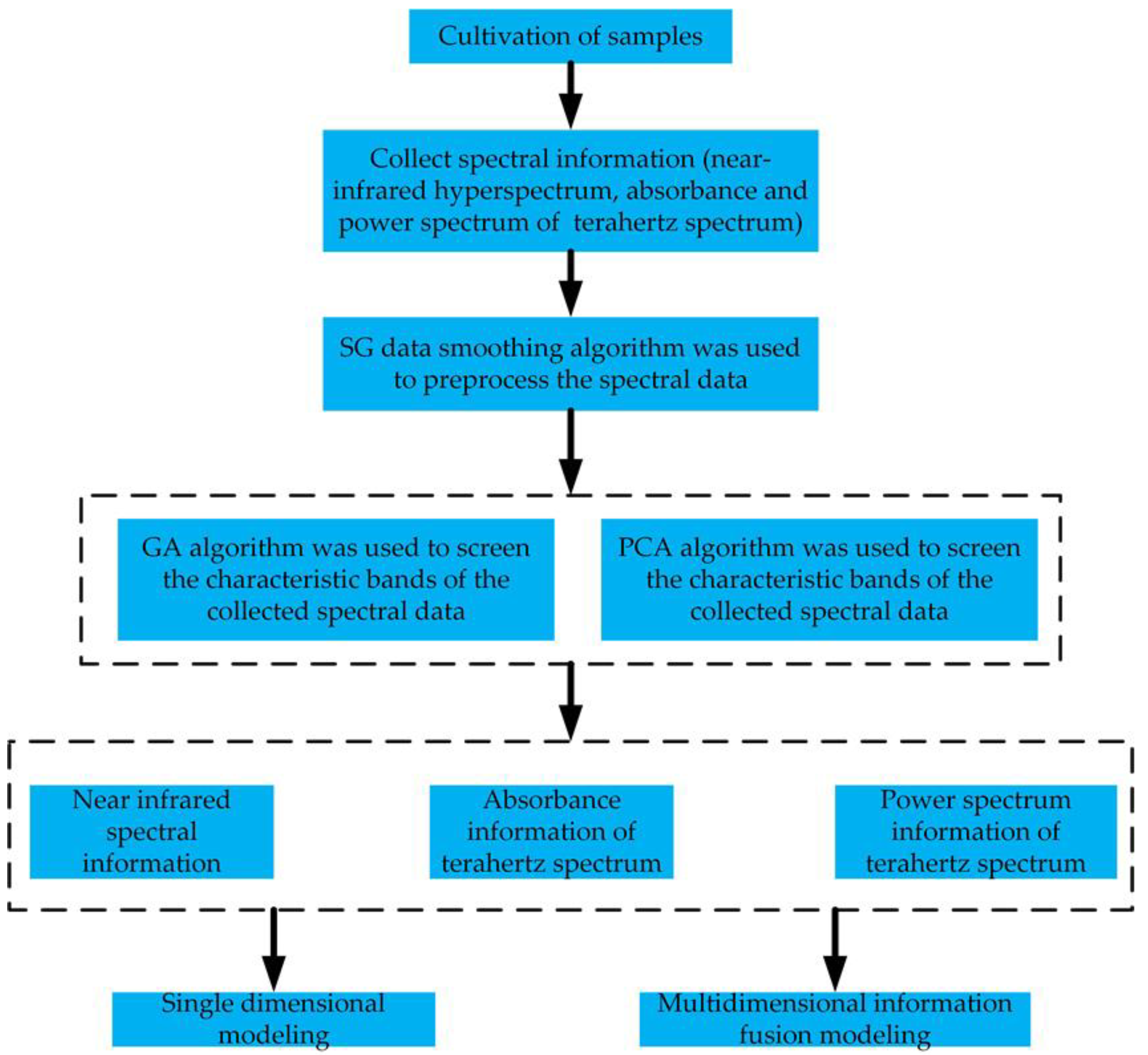
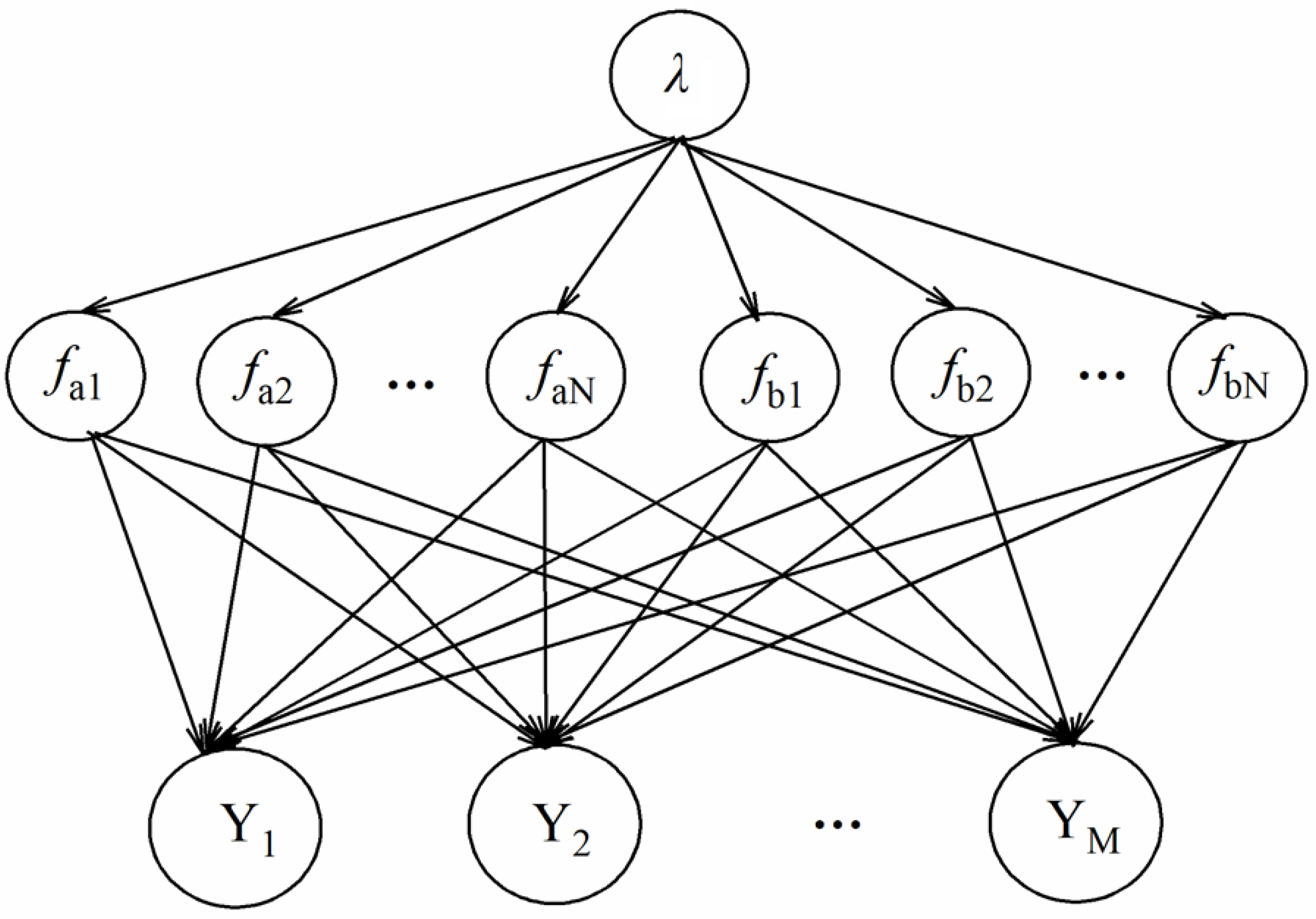
2.4. Data Processing
2.4.1. Data Smoothing
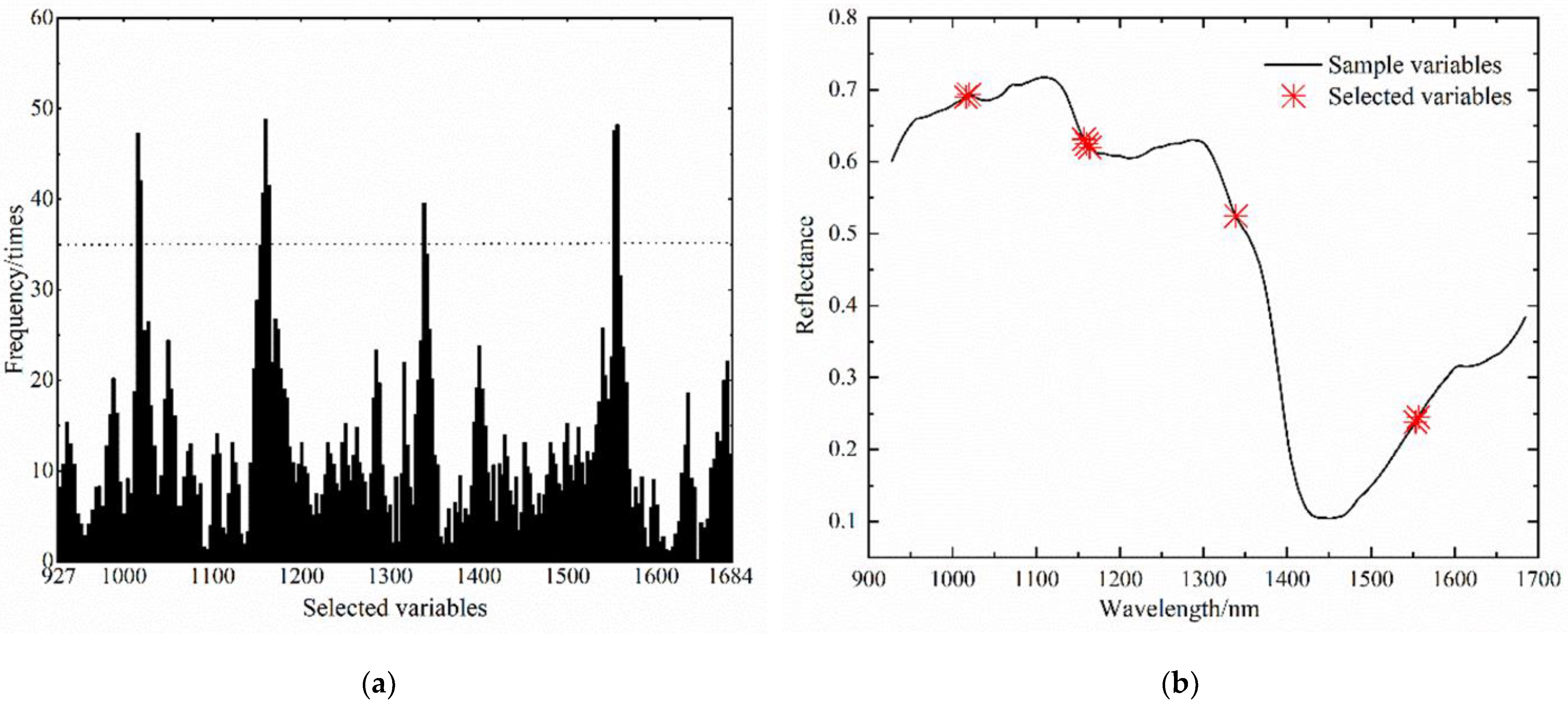
2.4.2. Characteristic Band screening
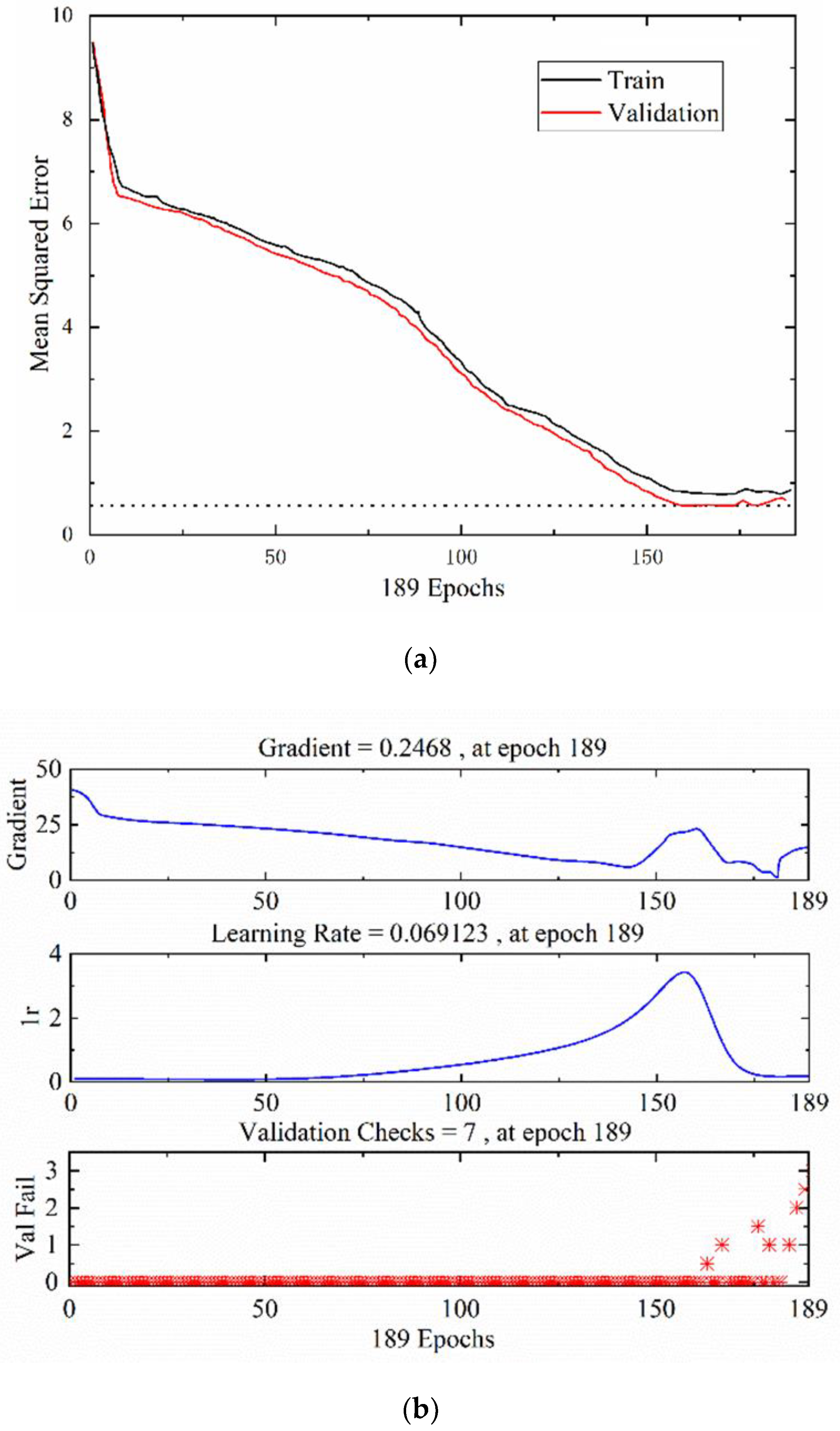
2.4.3. Establishment of the Model
3. Results and Discussion
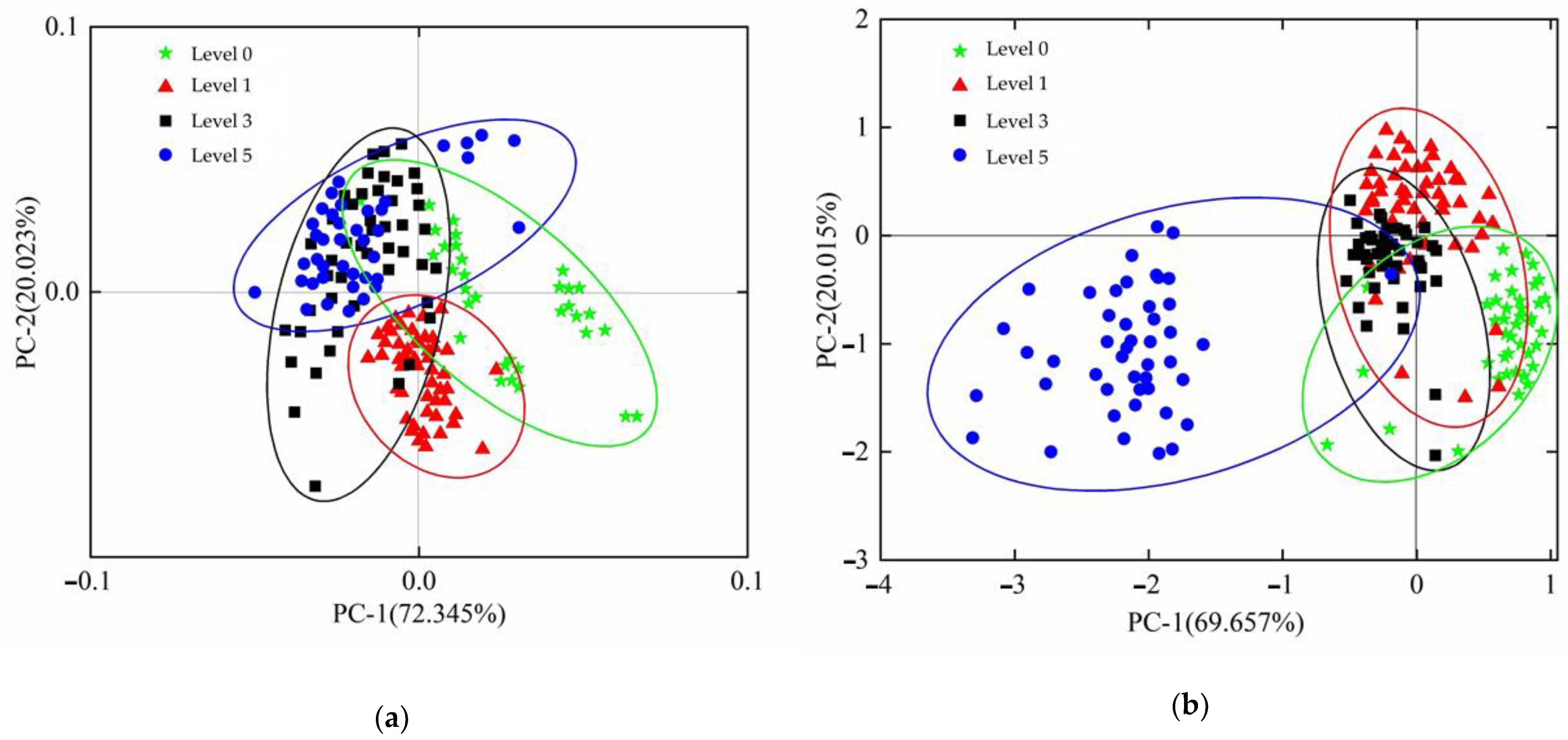
3.1. Screening of Near-Infrared Hyperspectral Characteristic Bands
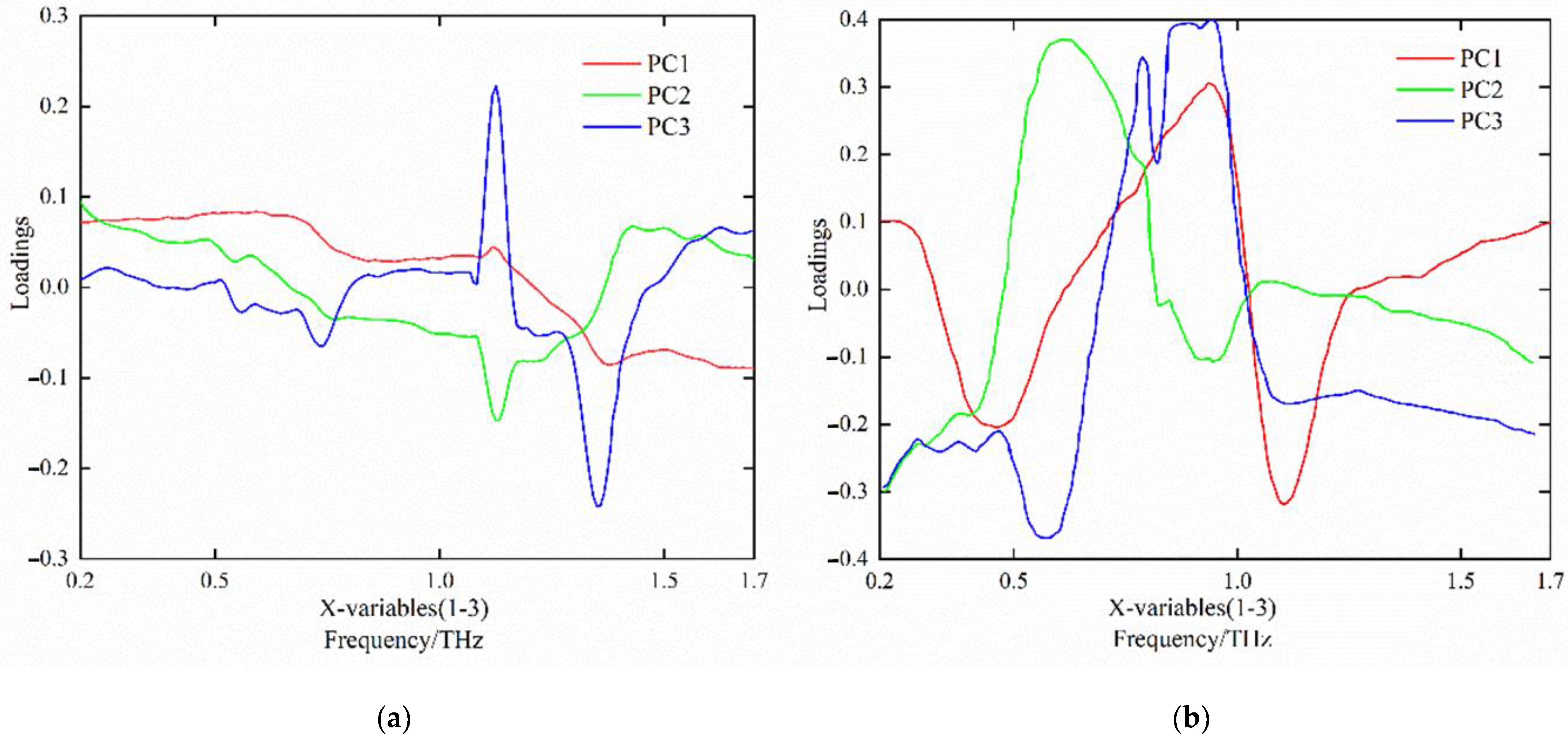
3.2. Terahertz Time-Domain Spectral Data Processing Results
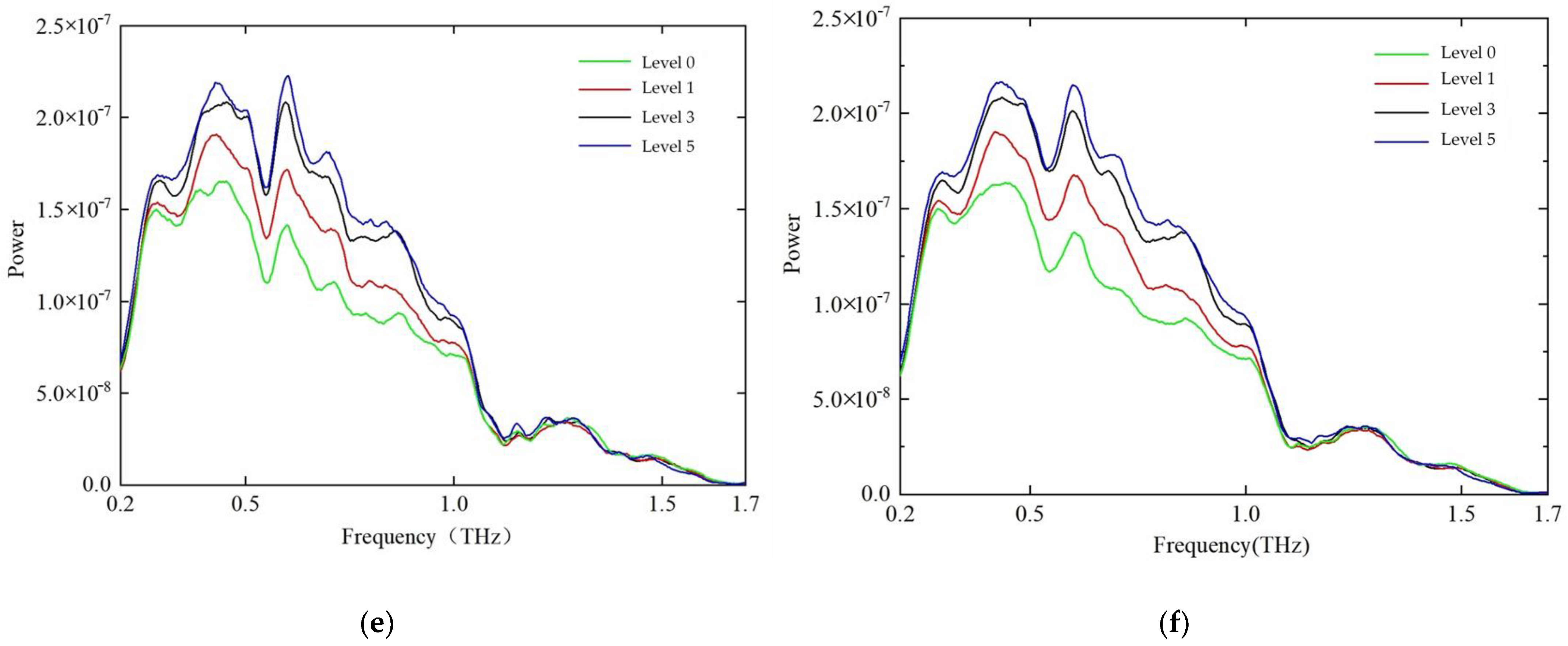
3.2.1. Terahertz Time-Domain Spectral Analysis
3.2.2. Screening of the Terahertz Time-Domain Spectrum Characteristic Frequency Band
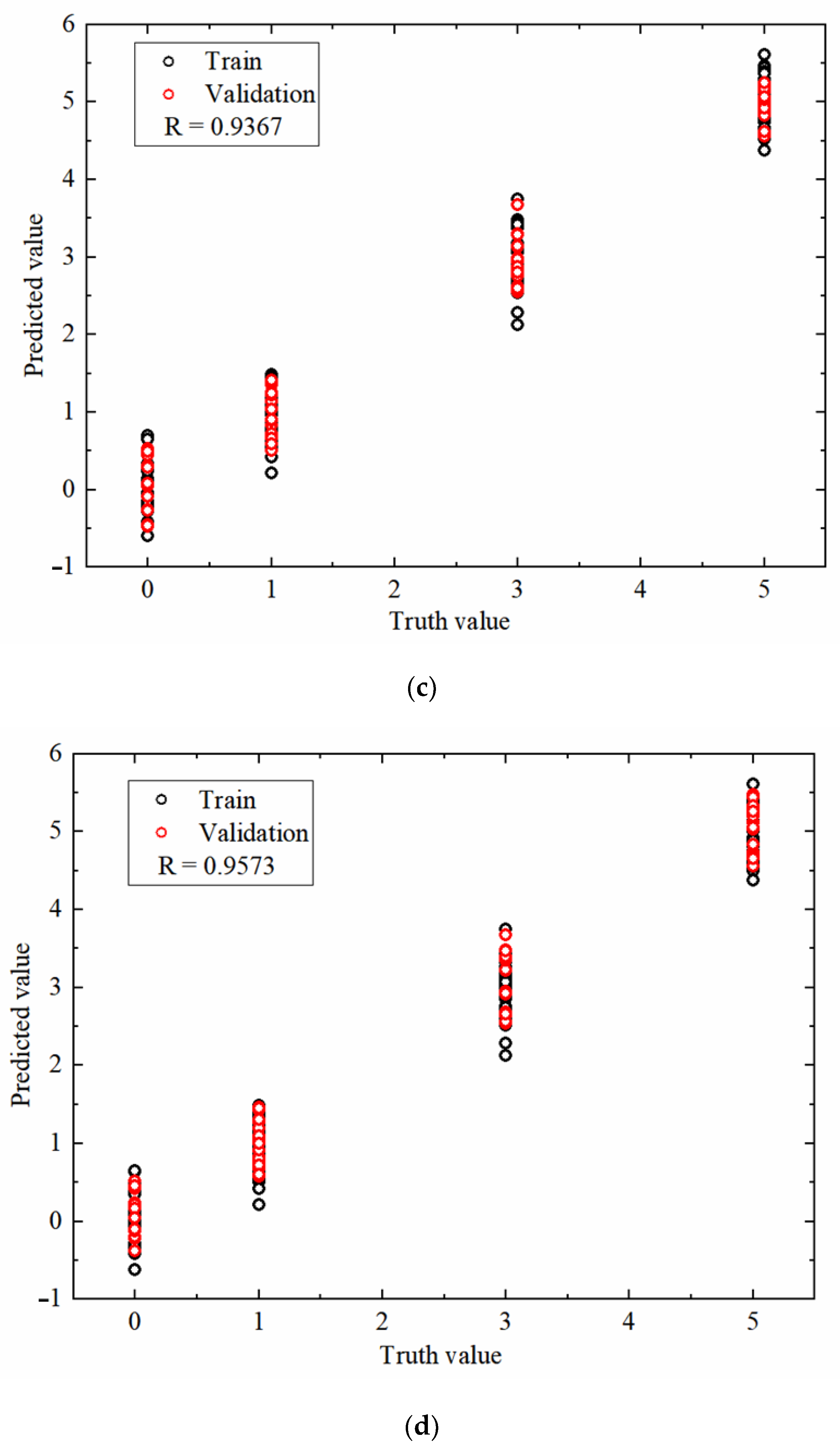
3.3. Single-Model Analysis
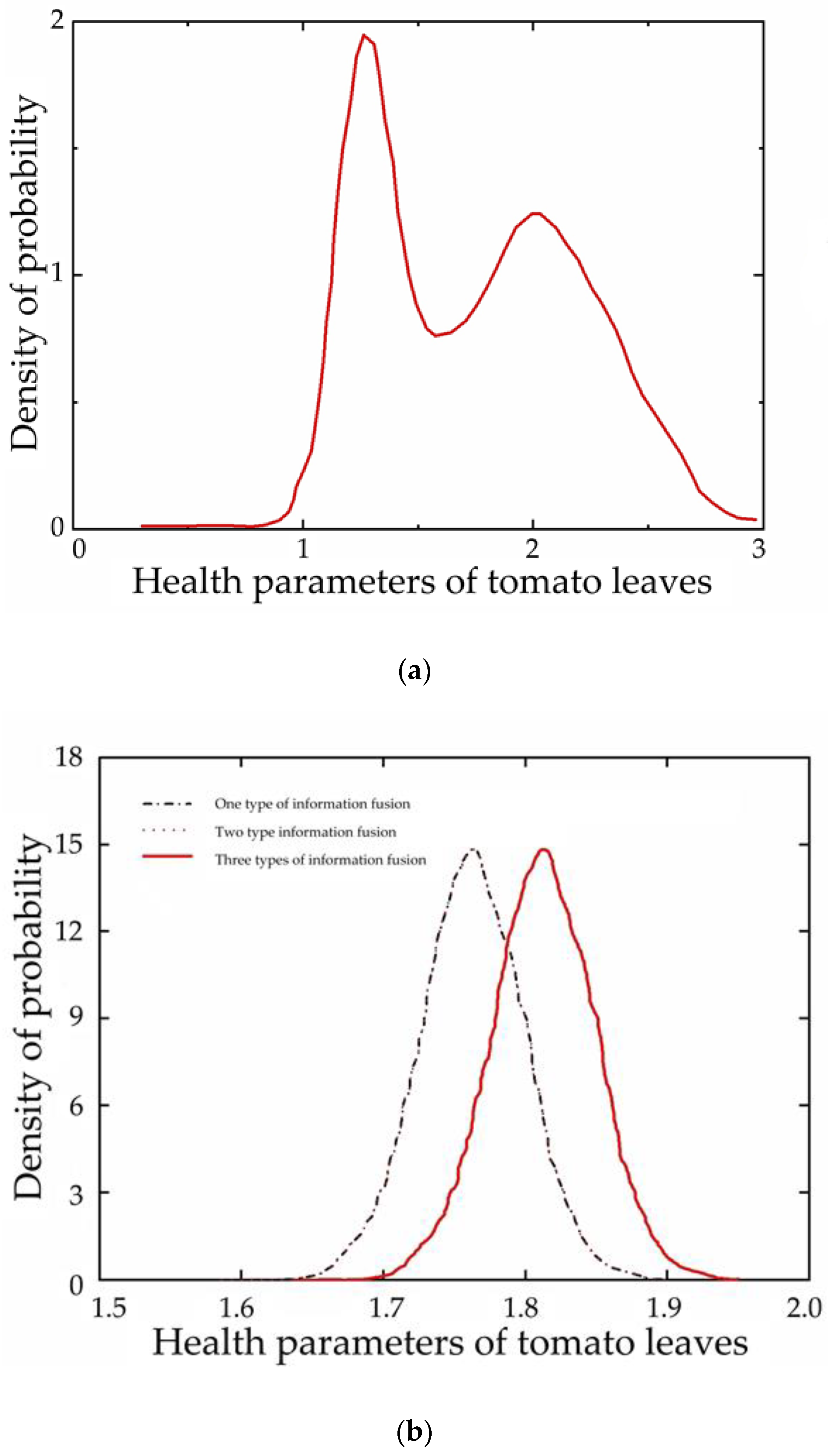
3.4. Fusion Model Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mao, H.P.; Wang, Y.F.; Yang, N.; Liu, Y.; Zhang, X.D. Effects of nutrient solution irrigation quantity and powdery mildew infection on the growth and physiological parameters of greenhouse cucumbers. Int. J. Agric. Biol. Eng. 2022, 15, 68–74. [Google Scholar] [CrossRef]
- Bandamaravuri, K.B.; Nayak, A.K.; Bandamaravuri, A.S.; Samad, A. Simultaneous detection of downy mildew and powdery mildew pathogens on Cucumis sativus and other cucurbits using duplex-qPCR and HRM analysis. AMB Express 2020, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Saberi, Z.; Rezaei, B.; Khayamian, T. A fluorescent aptasensor for analysis of adenosine triphosphate based on aptamer–magnetic nanoparticles and its single-stranded complementary DNA labeled carbon dots. Luminescence 2018, 33, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Li, K.T. Analysis of PCR Kinetics inside a Microfluidic DNA Amplification System. Micromachines 2018, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Sireesha, Y.; Velazhahan, R. Rapid and specific detection of Peronosclerospora sorghi in maize seeds by conventional and real-time PCR. Eur. J. Plant Pathol. 2018, 150, 521–526. [Google Scholar] [CrossRef]
- Wang, Y.F.; Mao, H.P.; Zhang, X.D.; Liu, Y.; Du, X.X. A Rapid Detection Method for Tomato Gray Mold Spores in Greenhouse Based on Microfluidic Chip Enrichment and Lens-Less Diffraction Image Processing. Foods 2022, 10, 3011. [Google Scholar] [CrossRef]
- Marin-Ortiz, J.C.; Gutierrez-Toro, N.; Botero-Fernandez, V.; Hoyos-Carvajal, L.M. Linking physiological parameters with visible/near-infrared leaf reflectance in the incubation period of vascular wilt disease. Saudi J. Biol. Sci. 2022, 27, 88–99. [Google Scholar] [CrossRef]
- Feng, X.P.; Yu, C.L.; Chen, Y.; Oeng, J.Y.; Ye, L.H.; Shen, T.T.; Wen, H.Y.; He, Y. Non-destructive Determination of Shikimic Acid Concentration in Transgenic Maize Exhibiting Glyphosate Tolerance Using Chlorophyll Fluorescence and Hyperspectral Imaging. Front. Plant Sci. 2018, 9, 468. [Google Scholar] [CrossRef] [PubMed]
- Fazari, A.; Pellicer-Valero, O.J.; Gomez-Sanchis, J.; Bernardi, B.; Cubero, S.; Benalia, S.; Zimbalatti, G.; Blasco, J. Application of deep convolutional neural networks for the detection of anthracnose in olives using VIS/NIR hyperspectral images. Comput. Electron. Agric. 2021, 187, 106252. [Google Scholar] [CrossRef]
- Zhang, L.X.; Tian, X.; Li, Y.X.; Chen, Y.Q.; Chen, Y.Y.; Ma, J.C. Estimation of Disease Severity for Downy Mildew of Greenhouse Cucumber Based on Visible Spectral and Machine Learning. Spectrosc. Spectr. Anal. 2020, 40, 227–232. [Google Scholar] [CrossRef]
- Qin, L.F.; Zhang, X.; Zhang, X.Q. Early Detection of Cucumber Downy Mildew in Greenhouse by Hyperspectral Disease Differential Feature Extraction. Trans. Chin. Soc. Agric. Mach. 2022, 51, 212–220. [Google Scholar]
- Cao, Q.H.; Lin, H.M.; Zhou, W.; Li, Z.X.; Zhang, T.J.; Huang, H.Q.; Li, X.M.; Li, D.H. Water Quality Analysis Based on Terahertz Attenuated Total Reflection Technology. Spectrosc. Spectr. Anal. 2022, 42, 31–37. [Google Scholar] [CrossRef]
- Zhang, X.D.; Wang, P.; Wang, Y.F.; Hu, L.; Luo, X.W.; Mao, H.P. Cucumber powdery mildew detection method based on hyperspectra-terahertz. Front. Plant Sci. 2022, 13, 1035731. [Google Scholar] [CrossRef] [PubMed]
- Di Girolamo, F.V.; Pagano, M.; Tredicucci, A.; Bitossi, M.; Paoletti, R.; Barzanti, G.P.; Benvenuti, C.; Roversi, P.F.; Toncelli, A. Detection of fungal infections in chestnuts: A terahertz imaging-based approach. Food Control 2021, 123, 107700. [Google Scholar] [CrossRef]
- Li, B.; Zhang, D.P.; Shen, Y. Study on Terahertz Spectrum Analysis and Recognition Modeling of Common Agricultural Diseases. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 243, 118820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Duan, C.H.; Gao, H.Y.; Shi, Q.; Wang, Y.F.; Shen, B.G.; Zhang, X. Tomato water stress state detection model by using terahertz spectroscopy technology. Trans. Chin. Soc. Agric. Eng. 2021, 37, 121–128. [Google Scholar]
- Jin, C.; Mao, H.P.; Chen, Y.; Shi, Q.; Wang, Q.R.; Ma, G.X.; Liu, Y. Engineering-oriented dynamic optimal control of a greenhouse environment using an improved genetic algorithm with engineering constraint rules. Comput. Electron. Agric. 2020, 177, 105698. [Google Scholar] [CrossRef]
- Zhang, F.; Cui, X.H.; Zhang, Y.K.; Wang, Y.X. Relationship Between Visible/Near Infrared Spectral Data and Fertilization Information at Different Positions of Hatching Eggs. Spectrosc. Spectr. Anal. 2021, 41, 3064–3068. [Google Scholar] [CrossRef]
- Wang, Y.F.; Zhang, X.D.; Ma, G.X.; Du, X.X.; Shaheen, N.; Mao, H.P. Recognition of weeds at asparagus fields using multi-feature fusion and backpropagation neural network. Int. J. Agric. Biol. Eng. 2021, 14, 190–198. [Google Scholar] [CrossRef]
- Wang, Z.F.; He, X.Q.; Shen, H.L.; Fan, S.J.; Zeng, Y.L. Multi-source information fusion to identify water supply pipe leakage based on SVM and VMD. Inf. Process. Manag. 2022, 59, 102819. [Google Scholar] [CrossRef]
- Li, L.; Zhu, F.; Sun, H.; Hu, Y.Y.; Yang, Y.Y.; Jin, D.W. Multi-source information fusion and deep-learning-based characteristics measurement for exploring the effects of peer engagement on stock price synchronicity. Inf. Fusion 2021, 69, 1–21. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Cheng, Q.Q.; Fang, X.M.; Yu, H.H.; Li, D.L. Principal component analysis and long short-term memory neural network for predicting dissolved oxygen in water for aquaculture. Trans. Chin. Soc. Agric. Eng. 2018, 34, 183–191. [Google Scholar] [CrossRef]
- Wang, Y.F.; Mao, H.P.; Xu, G.L.; Zhang, X.D.; Zhang, Y.K. A Rapid Detection Method for Fungal Spores from Greenhouse Crops Based on CMOS Image Sensors and Diffraction Fingerprint Feature Processing. J. Fungi 2022, 8, 374. [Google Scholar] [CrossRef] [PubMed]


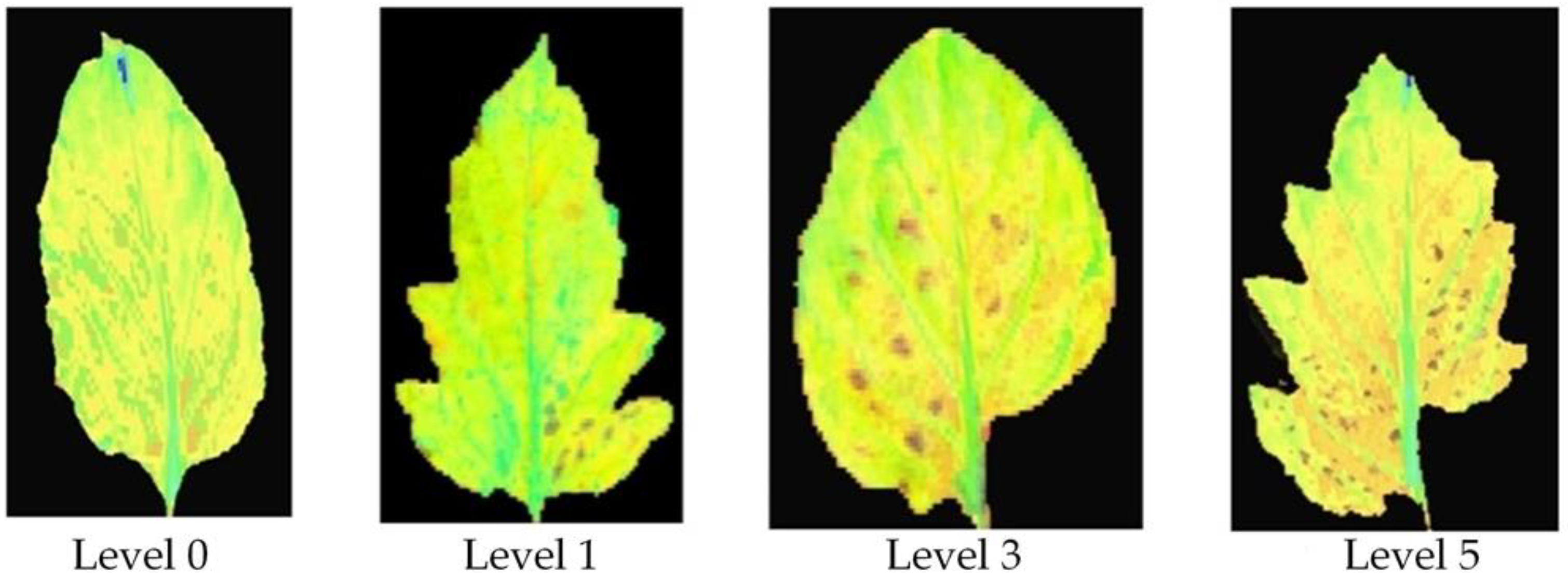



| Disease Level | Number | Training Set | Prediction Set |
|---|---|---|---|
| Level 0 (healthy samples) | 42 | 28 | 14 |
| Level 1 (disease spot area < 5%) | 76 | 51 | 25 |
| Level 3 (6% < disease spot area < 10%) | 65 | 43 | 22 |
| Level 5 (11% < disease spot area < 25%) | 57 | 38 | 19 |
| Total samples | 240 | 160 | 80 |
| Principal Component/Cumulative Contribution Rate (%) | PC1 | PC2 | PC3 |
|---|---|---|---|
| absorbance | 72.345 | 92.368 | 94.522 |
| power spectrum | 69.657 | 89.672 | 93.914 |
| Dimensions | Model | Number of Characteristic Variables | Prediction Accuracy (%) | ||||
|---|---|---|---|---|---|---|---|
| Level 0 | Level 1 | Level 3 | Level 5 | Total | |||
| Near-infrared hyperspectrum | GA-BPNN | 8 | 100 | 96 | 90.90 | 94.74 | 95 |
| THz power spectrum | PCA-BPNN | 5 | 100 | 96 | 95.45 | 94.74 | 96.67 |
| THz absorbance | PCA-BPNN | 6 | 100 | 92 | 95.45 | 94.74 | 95 |
| Number of Characteristic Variables | Prediction Accuracy (%) | ||||
|---|---|---|---|---|---|
| Level 0 | Level 1 | Level 2 | Level 3 | Total | |
| 19 | 99.36 | 95.57 | 96.20 | 97.35 | 97.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, Y.; Zhou, Z.; Zhang, Y.; Wang, X. Detection Method for Tomato Leaf Mildew Based on Hyperspectral Fusion Terahertz Technology. Foods 2023, 12, 535. https://doi.org/10.3390/foods12030535
Zhang X, Wang Y, Zhou Z, Zhang Y, Wang X. Detection Method for Tomato Leaf Mildew Based on Hyperspectral Fusion Terahertz Technology. Foods. 2023; 12(3):535. https://doi.org/10.3390/foods12030535
Chicago/Turabian StyleZhang, Xiaodong, Yafei Wang, Zhankun Zhou, Yixue Zhang, and Xinzhong Wang. 2023. "Detection Method for Tomato Leaf Mildew Based on Hyperspectral Fusion Terahertz Technology" Foods 12, no. 3: 535. https://doi.org/10.3390/foods12030535
APA StyleZhang, X., Wang, Y., Zhou, Z., Zhang, Y., & Wang, X. (2023). Detection Method for Tomato Leaf Mildew Based on Hyperspectral Fusion Terahertz Technology. Foods, 12(3), 535. https://doi.org/10.3390/foods12030535







