Molecular Recognition Patterns between Vitamin B12 and Proteins Explored through STD-NMR and In Silico Studies
Abstract
1. Introduction
2. Hypotheses
3. Materials and Methods
3.1. Reagents
3.2. Characterization of Cobalamin
3.3. Saturation Transfer Difference via Nuclear Magnetic Resonance (STD–NMR)
3.4. Molecular Docking
4. Results and Discussion
4.1. Cyanocobalamin Structure after the 1H Scan in NMR
4.2. Interaction between Vitamin B12 and Protein Identified Using STD–NMR
4.3. Binding Capability Judgment Based on Cyanocobalamin
4.4. Binding Conformation Simulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DMB | Dimethyl benzimidazole |
| STD–NMR | Saturation transfer difference by nuclear magnetic resonance |
| CNBL | Cyanocobalamin |
| OHBL | Aqua cobalamin |
| Kα | Binding constant |
| KD | Dissociation constant |
References
- Martens, J.-H.; Barg, H.; Warren, M.; Jahn, D. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 2002, 58, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Klug, G. Beyond catalysis: Vitamin B12 as a cofactor in gene regulation. Mol. Microbiol. 2014, 91, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Stabler, S.P. Vitamin B12 deficiency. N. Engl. J. Med. 2013, 368, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, F. Vitamin B12 sources and bioavailability. Exp. Biol. Med. 2007, 232, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Gille, D.; Schmid, A. Vitamin B12 in meat and dairy products. Nutr. Rev. 2015, 73, 106–115. [Google Scholar] [CrossRef]
- Matte, J.J.; Britten, M.; Girard, C.L. The importance of milk as a source of vitamin B12 for human nutrition. Anim. Front. 2014, 4, 32–37. [Google Scholar] [CrossRef]
- Fedosov, S.N.; Nexo, E.; Heegaard, C.W. Binding of aquocobalamin to bovine casein and its peptides via coordination to histidine residues. Int. Dairy J. 2018, 76, 30–39. [Google Scholar] [CrossRef]
- Fedosov, S.N.; Nexo, E.; Heegaard, C.W. Vitamin B12 and its binding proteins in milk from cow and buffalo in relation to bioavailability of B12. J. Dairy Sci. 2019, 102, 4891–4905. [Google Scholar] [CrossRef]
- Liu, Z.; Gong, Z.; Dong, X.; Tang, C. Transient protein–protein interactions visualized by solution NMR. Biochim. Biophys. Acta-Proteins Proteom. 2016, 1864, 115–122. [Google Scholar] [CrossRef]
- Lepre, C.A.; Moore, J.M.; Peng, J.W. Theory and Applications of NMR-Based Screening in Pharmaceutical Research. Chem. Rev. 2004, 104, 3641–3676. [Google Scholar] [CrossRef]
- Walpole, S.; Monaco, S.; Nepravishta, R.; Angulo, J. Chapter Twelve—STD NMR as a Technique for Ligand Screening and Structural Studies. In Biological NMR Part B; Wand, J.A., Ed.; Academic Press: Cambridge, MA, USA, 2019; ISBN 0076-6879. [Google Scholar]
- Maity, S.; Gundampati, R.K.; Kumar, T.K.S. NMR Methods to Characterize Protein-Ligand Interactions. Nat. Prod. Commun. 2019, 14, 1934578X19849296. [Google Scholar] [CrossRef]
- Mayer, M.; Meyer, B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- Qin, J.; Gronenborn, A.M. Weak protein complexes: Challenging to study but essential for life. FEBS J. 2014, 281, 1948–1949. [Google Scholar] [CrossRef]
- Murray, C.W.; Blundell, T.L. Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol. 2010, 20, 497–507. [Google Scholar] [CrossRef]
- Harner, M.J.; Frank, A.O.; Fesik, S.W. Fragment-based drug discovery using NMR spectroscopy. J. Biomol. NMR 2013, 56, 65–75. [Google Scholar] [CrossRef]
- Hwang, T.L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson. Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Payne, K.A.P.; Quezada, C.P.; Fisher, K.; Dunstan, M.S.; Collins, F.A.; Sjuts, H.; Levy, C.; Hay, S.; Rigby, S.E.J.; Leys, D. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 2015, 517, 513–516. [Google Scholar] [CrossRef]
- Russell, H.J.; Jones, A.R.; Hay, S.; Greetham, G.M.; Towrie, M.; Scrutton, N.S. Protein motions are coupled to the reaction chemistry in coenzyme b12-dependent ethanolamine ammonia lyase. Angew. Chem. 2012, 124, 9440–9444. [Google Scholar] [CrossRef]
- Sun, Q.; Zhai, Y.; Wang, W.; Gan, N.; Zhang, S.; Suo, Z.; Li, H. Molecular recognition patterns between vitamin B12 and human serum albumin explored through STD–NMR and spectroscopic methods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 258, 119828. [Google Scholar] [CrossRef]
- Sun, Q.; Tang, P.; Zhao, L.; Pu, H.; Zhai, Y.; Li, H. Mechanism and structure studies of cinnamaldehyde/cyclodextrins inclusions by computer simulation and NMR technology. Carbohydr. Polym. 2018, 194, 294–302. [Google Scholar] [CrossRef]
- Meyer, B.; Peters, T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. 2003, 42, 864–890. [Google Scholar] [CrossRef] [PubMed]
- Schneider, Z.; Stroinski, A. Comprehensive B12. In Comprehensive B12; de Gruyter: Berlin, Germany, 2011; ISBN 311008239X. [Google Scholar]
- Swain, B.C.; Rout, J.; Tripathy, U. Interaction of vitamin B12 with β-lactoglobulin: A computational study. J. Biomol. Struct. Dyn. 2022, 40, 2146–2155. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.; Gupta, J.; Rabbani, G.; Khan, R.H.; Saleemuddin, M. Behaviour of oleic acid-depleted bovine alpha-lactalbumin made LEthal to tumor cells (BAMLET). Mol. Biosyst. 2016, 12, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Reddy, R.R.; Shanmugam, G.; Madhan, B.; Kumar, B.V.N.P. Selective binding and dynamics of imidazole alkyl sulfate ionic liquids with human serum albumin and collagen–a detailed NMR investigation. Phys. Chem. Chem. Phys. 2018, 20, 9256–9268. [Google Scholar] [CrossRef]
- Liu, J.; Wang, R. Classification of Current Scoring Functions. J. Chem. Inf. Model. 2015, 55, 475–482. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, X.; Russell, P.; Li, J.; Pan, W.; Fu, J.; Zhang, A. Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock. Ecotoxicol. Environ. Saf. 2022, 233, 113323. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.; Hou, M.; Xiao, Z.-X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Harner, M.J.; Mueller, L.; Robbins, K.J.; Reily, M.D. NMR in drug design. Arch. Biochem. Biophys. 2017, 628, 132–147. [Google Scholar] [CrossRef]
- Monaco, S.; Tailford, L.E.; Juge, N.; Angulo, J. Differential Epitope mapping by STD NMR spectroscopy (DEEP-STD NMR) to reveal the nature of protein-ligand contacts. Angew. Chem. Int. Ed. 2017, 129, 15491–15495. [Google Scholar] [CrossRef]
- Ferrer-Gallego, R.; Hernández-Hierro, J.M.; Brás, N.F.; Vale, N.; Gomes, P.; Mateus, N.; De Freitas, V.; Heredia, F.J.; Escribano-Bailón, M.T. Escribanobailón, Interaction between Wine Phenolic Acids and Salivary Proteins by Saturation-Transfer Difference Nuclear Magnetic Resonance Spectroscopy (STD-NMR) and Molecular Dynamics Simulations. J. Agric. Food Chem. 2017, 65, 6434. [Google Scholar] [CrossRef]
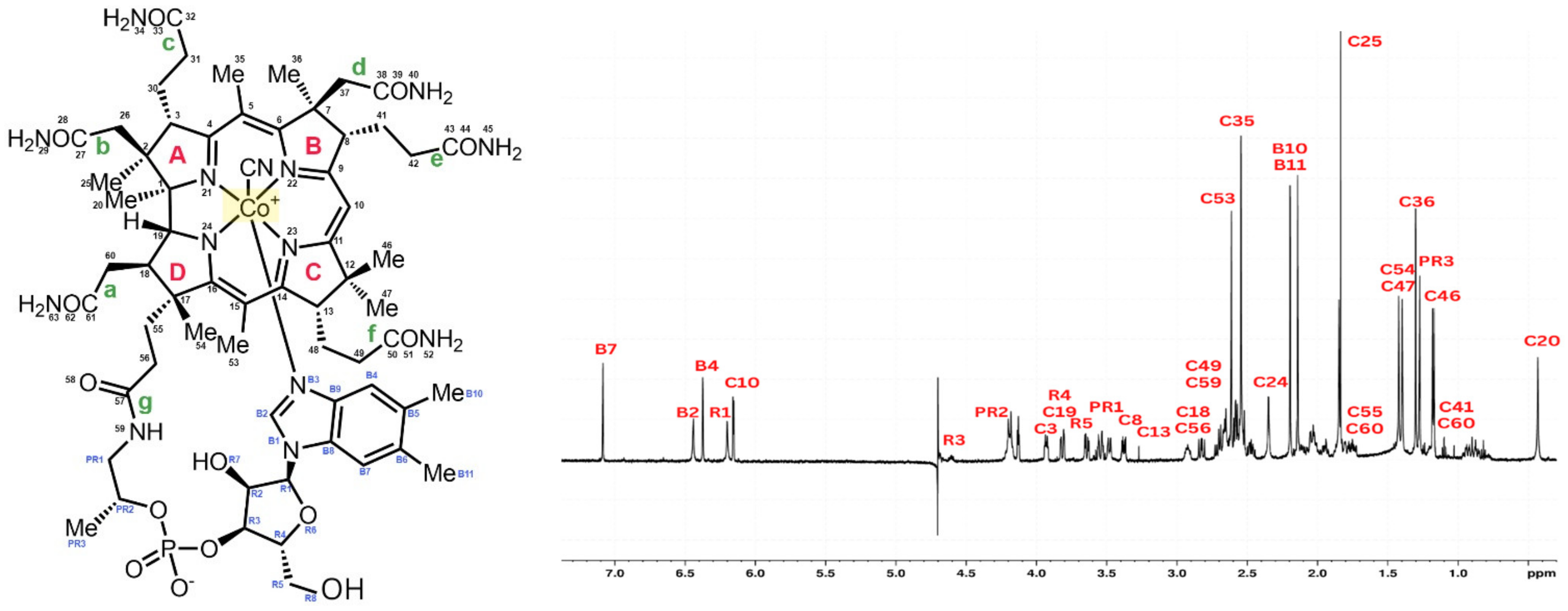
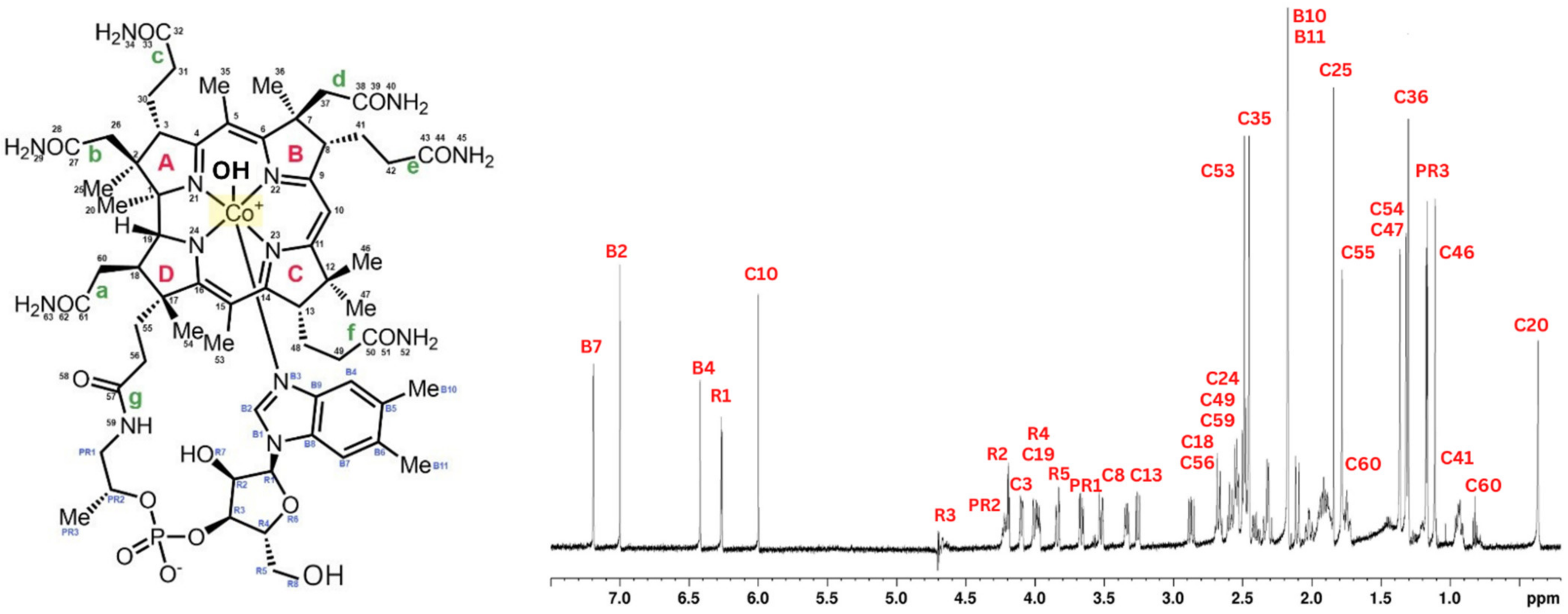
| Binding Constant—Kα (L/mol) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C20 | C25 | C35 | C53 | C10 | B10/11 | B4 | B2 | B7 | ||||||||||
| Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | |
| Gluten | 2.88 × 104 | 3.21 × 104 | 3.03 × 104 | 3.59 × 104 | 3.7 × 103 | 2.78 × 103 | 1.99 × 104 | 4.65 × 103 | 2.77 × 103 | 2.83 × 103 | 3.02 × 104 | 3.46 × 104 | 3.16 × 103 | 3.65 × 103 | 3.77 × 103 | 3.09 × 103 | 2.76 × 103 | 2.40 × 103 |
| Myoglobin | 3.16 × 103 | 4.04 × 104 | 4.82 × 104 | 3.04 × 104 | 4.34 × 104 | 4.39 × 104 | 3.16 × 104 | 3.51 × 104 | 4.33 × 104 | 3.86 × 104 | 3.96 × 104 | 4.67 × 104 | 3.74 × 104 | 4.86 × 104 | 3.05 × 104 | 4.37 × 104 | 2.48 × 104 | 4.20 × 104 |
| Casein | 4.24 × 104 | 3.04 × 104 | 4.88 × 104 | 2.71 × 104 | 2.47 × 104 | 4.27 × 104 | 3.42 × 104 | 5.52 × 103 | 3.01 × 104 | 3.11 × 104 | 3.19 × 104 | 4.72 × 104 | 2.29 × 104 | 3.75 × 104 | 2.92 × 104 | 3.11 × 104 | 3.16 × 104 | 3.04 × 104 |
| Egg albumin | 2.35 × 104 | 3.73 × 104 | 3.04 × 104 | 3.20 × 104 | 2.33 × 104 | 3.44 × 104 | 2.43 × 104 | 2.66 × 104 | 1.87 × 104 | 3.28 × 104 | 1.61 × 104 | 2.60 × 104 | 2.48 × 104 | 4.16 × 104 | 1.60 × 104 | 6.46 × 104 | 2.41 × 104 | 8.03 × 104 |
| Pea | 7.29 × 103 | 4.10 × 104 | 2.96 × 104 | 1.48 × 104 | 1.13 × 104 | 3.06 × 104 | 3.85 × 104 | 3.04 × 104 | 1.08 × 104 | 3.21 × 104 | 7.65 × 103 | 3.80 × 104 | 1.28 × 104 | 4.54 × 104 | 1.09 × 104 | 4.54 × 104 | 1.01 × 104 | 4.97 × 104 |
| Rice | 3.13 × 104 | 3.43 × 104 | 3.84 × 104 | 2.46 × 104 | 3.52 × 104 | 2.46 × 104 | 2.21 × 104 | 3.91 × 104 | 2.44 × 104 | 2.60 × 104 | 2.49 × 104 | 4.28 × 104 | 2.01 × 104 | 2.41 × 104 | 2.31 × 103 | 2.56 × 103 | 4.81 × 104 | 2.95 × 104 |
| KD (mM) | Kα (L/mol)-Binding Constant | |||
|---|---|---|---|---|
| Cyanocobalamin | Aqua Cobalamin | Cyanocobalamin | Aqua Cobalamin | |
| Gluten | 0.23469 | 0.25545 | 4261.01 | 3914.70 |
| Myoglobin | 0.02911 | 0.02315 | 34,353.06 | 43,189.09 |
| Casein | 0.03401 | 0.02876 | 29,399.73 | 34,772.23 |
| Egg albumin | 0.04926 | 0.02527 | 20,301.11 | 39,573.24 |
| Pea | 0.10247 | 0.02554 | 9758.70 | 39,159.15 |
| Rice | 0.09200 | 0.08544 | 10,869.40 | 11,703.72 |
| Cyanocobalamin | Aqua Cobalamin | |||
|---|---|---|---|---|
| Vina Score | Amino Acids Interacting | Vina Score | Amino Acids Interacting | |
| Casein | −5.9 | ASP184, VAL186, ILE187, ARG189, PRO214, THR219, ALA220, GLU223, ASP284, ILE351, LYS358, TYR359, PRO389, ASP390, LEU393, ASP394 | −4.4 | SER344, VAL345, ARG346, VAL349, ARG353, VAL373, GLN377, GLY629, ASN633, ASP634, GLU635, GLU792, GLU793, ARG794, PHE795, GLU796, GLN797, ASN798 |
| myoglobin | −5.1 | PHE44, LYS46, PHE47, ASP61, HIS65, THR68, ALA72, ALA85, HIS89, HIS98 | −5.2 | ARG32, LEU33, GLY36, HIS37, LYS103, GLU106, PHE107, ILE108, ASP110, ALA111, ILE113, HIS114, HIS117, GLN129 |
| Egg Albumin | −6.5 | VAL222, LYS223, TYR246, MET319, GLU320, GLU322, PHE323, LYS457, LYS464, LEU468, ARG472, ALA475, TYR480, ILE483, VAL484 | −5.6 | GLU43, GLU44, LYS47, MET51, TYR63, SER67, LYS68, VAL70, LYS71, ASP75, GLN78, ASN158, VAL160, SER161, HIS165 |
| Gluten | −7.1 | PRO46, ASP174, VAL177, LEU46, ARG123, VAL168, LEU170, PRO187, GLN188, PRO189, LEU190, LYS191 | −7.5 | HIS10, GLN44, LEU101, PHE103, PRO120, ARG123, ASP166, HIS167, VAL168, GLU169, LEU170, SER171, GLU178, VAL179, HIS180, SER181, GLY182, VAL183, CYS184, THR185, ASP186, PRO187, GLN188, PRO189, LEU203, TYR228 |
| Rice | −8.1 | ARG199, HIS200, ARG201, PHE203, PHE204, ILE211, LEU215, GLU219, ASP222, SER224, ASN226, VAL227 | −6.7 | TRP34, GLN35, SER36, SER37, ARG38, ARG39, GLY40, SER41, GLU44, CYS45, ARG46, PHE47, CYS78, THR79, TYR190, ILE371, ASN372, HIS374, ASN391, GLN411, HIS412, HIS413 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghosh, R.; Thomas, D.S.; Arcot, J. Molecular Recognition Patterns between Vitamin B12 and Proteins Explored through STD-NMR and In Silico Studies. Foods 2023, 12, 575. https://doi.org/10.3390/foods12030575
Ghosh R, Thomas DS, Arcot J. Molecular Recognition Patterns between Vitamin B12 and Proteins Explored through STD-NMR and In Silico Studies. Foods. 2023; 12(3):575. https://doi.org/10.3390/foods12030575
Chicago/Turabian StyleGhosh, Ruchira, Donald S. Thomas, and Jayashree Arcot. 2023. "Molecular Recognition Patterns between Vitamin B12 and Proteins Explored through STD-NMR and In Silico Studies" Foods 12, no. 3: 575. https://doi.org/10.3390/foods12030575
APA StyleGhosh, R., Thomas, D. S., & Arcot, J. (2023). Molecular Recognition Patterns between Vitamin B12 and Proteins Explored through STD-NMR and In Silico Studies. Foods, 12(3), 575. https://doi.org/10.3390/foods12030575






