Characterization of a Galactose Oxidase from Fusarium odoratissimum and Its Application in the Modification of Agarose
Abstract
:1. Introduction
2. Materials and Methods
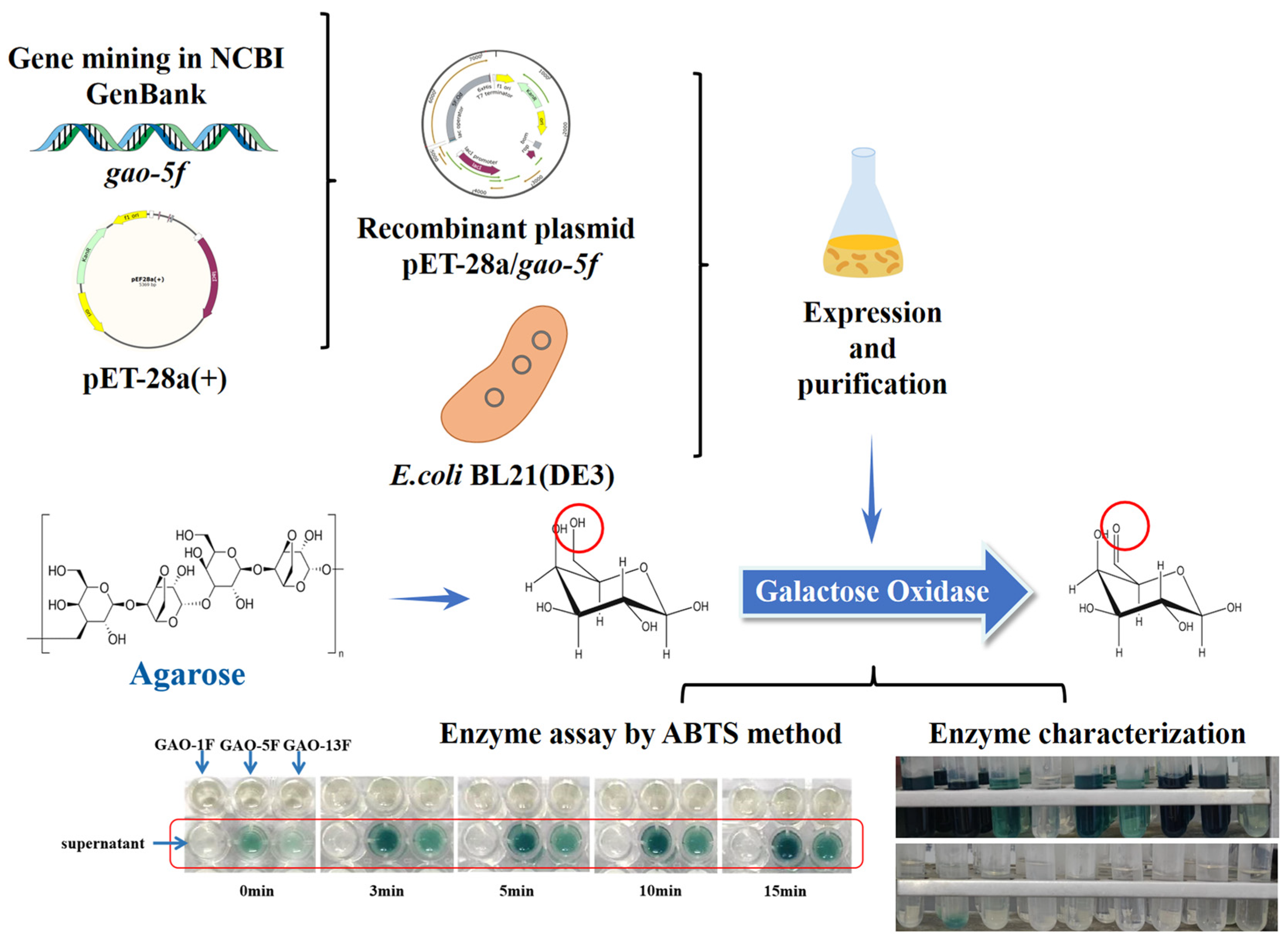
2.1. Schematic Overview of the Experimental Program
2.2. Materials, Strains and Culture Conditions
2.3. Gene Mining and Sequence Analysis
2.4. Expression of the Three Galactose Oxidase Genes in E. coli
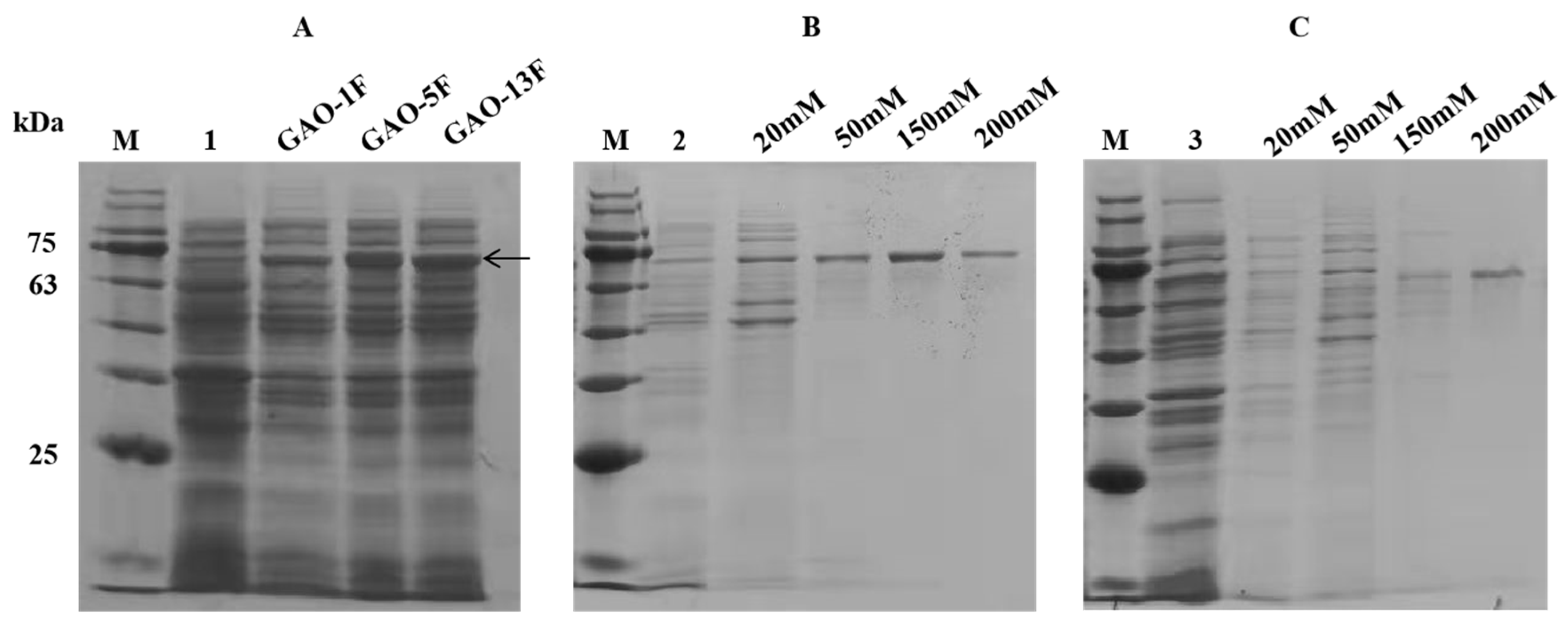
2.5. Purification of the Three Galactose Oxidases
2.6. Enzyme Assay
2.7. Enzyme Characterization
2.8. Oxidation of Agarose by GAO-5F
3. Results and Discussion
3.1. Sequences Analysis
3.2. Enzyme Expression, Purification, and Molecular Properties
3.3. Characterization of GAO-5F
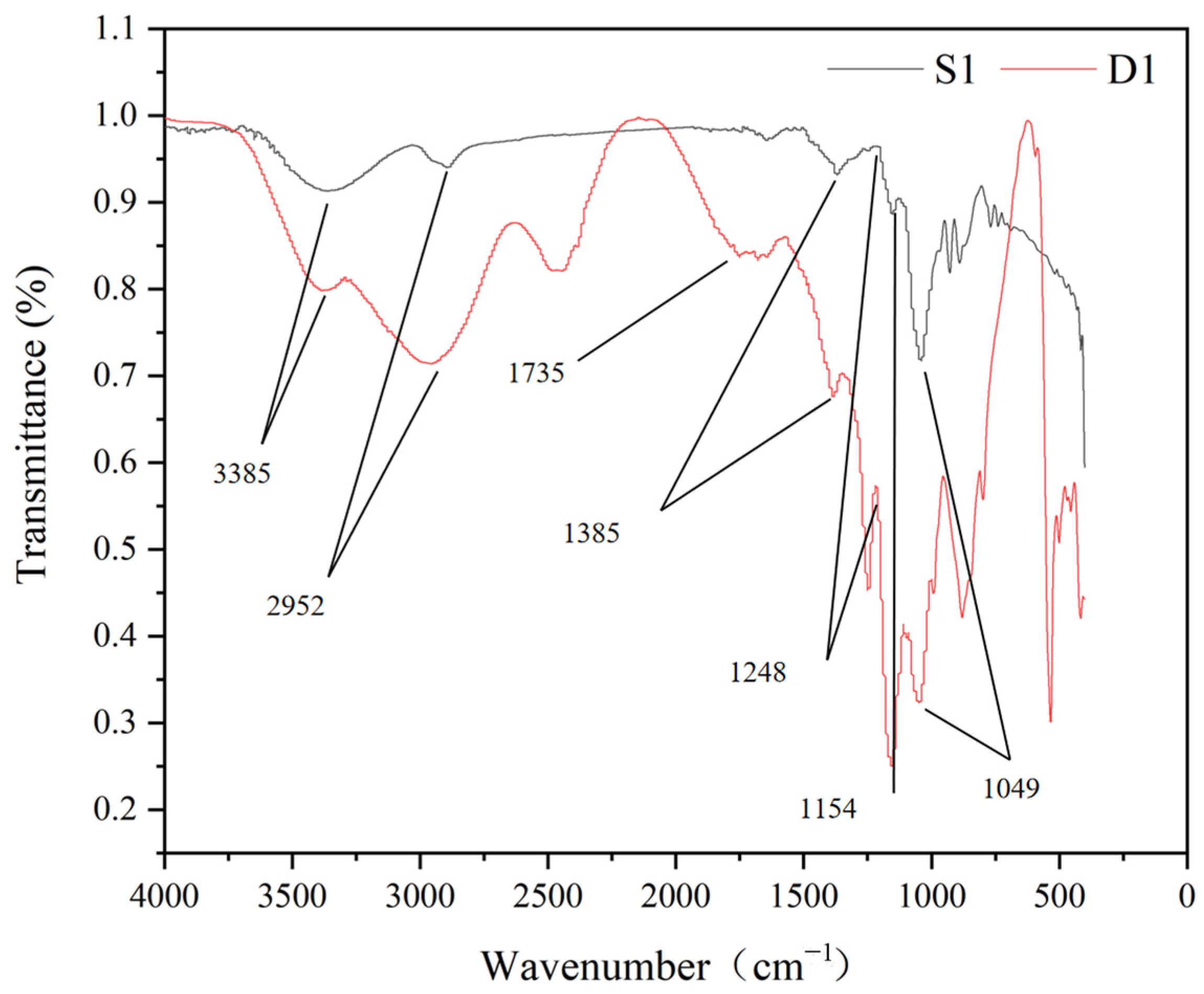
3.4. Oxidation of Agarose by GAO-5F
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Saha, T.; Hoque, M.E.; Mahbub, T. Chapter 13—Biopolymers for Sustainable packaging in Food, Cosmetics, and Pharmaceuticals. In Advanced Processing, Properties, and Applications of Starch and Other Bio-Based Polymers; Al-oqla, F.M., Sapuan, S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 197–214. [Google Scholar]
- Bharti, S.K.; Pathak, V.; Arya, A.; Alam, T.; Rajkumar, V.; Verma, A.K. Packaging potential of Ipomoea batatas and κ-carrageenan biobased composite edible film: Its rheological, physicomechanical, barrier and optical characterization. J. Food Process. Preserv. 2021, 45, e15153. [Google Scholar] [CrossRef]
- Galus, S.; Arik Kibar, E.A.; Gniewosz, M.; Kraśniewska, K. Novel materials in the preparation of edible films and coatings—A review. Coatings 2020, 10, 674. [Google Scholar] [CrossRef]
- Mohamed, S.A.A.; El-Sakhawy, M.; El-Sakhawy, M.A. Polysaccharides, protein and lipid-based natural edible films in food packaging: A review. Carbohyd. Polym. 2020, 238, 116178. [Google Scholar] [CrossRef]
- Luo, Q.; Hossen, M.A.; Zeng, Y.; Dai, J.; Li, S.; Qin, W.; Liu, Y. Gelatin-based composite films and their application in food packaging: A review. J. Food Eng. 2022, 313, 110762. [Google Scholar] [CrossRef]
- Fan, H.Y.; Duquette, D.; Dumont, M.; Simpson, B.K. Salmon skin gelatin-corn zein composite films produced via crosslinking with glutaraldehyde: Optimization using response surface methodology and characterization. Int. J. Biol. Macromol. 2018, 120, 263–273. [Google Scholar] [CrossRef]
- Dong, J.; Yu, D.; Yu, Z.; Zhang, L.; Xia, W. Thermally-induced crosslinking altering the properties of chitosan films: Structure, physicochemical characteristics and antioxidant activity. Food Packag. Shelf 2022, 34, 100948. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, L.; Liu, C.; Zheng, X.; Tang, K. Tuning structure and properties of gelatin edible films through pullulan dialdehyde crosslinking. LWT Food Sci. Technol. 2021, 138, 110607. [Google Scholar] [CrossRef]
- Parka, J.; Namb, J.; Yunc, H.; Jinb, H.; Kwakad, H. Aquatic polymer-based edible films of fish gelatin crosslinked with alginate dialdehyde having enhanced physicochemical properties. Carbohyd. Polym. 2021, 254, 117317. [Google Scholar] [CrossRef]
- Liu, L.; Huang, X.; Geng, F.; Huang, Q. Optimization of preparation process of egg white protein/κ-carrageenan composite film. J. Food Process. Preserv. 2021, 46, e16167. [Google Scholar] [CrossRef]
- Kumar, N.; Neeraj. Polysaccharide-based component and their relevance in edible film/coating: A review. Nutr. Food Sci. 2019, 49, 793–823. [Google Scholar] [CrossRef]
- Bäumgen, M.; Dutschei, T.; Bornscheuer, U.T. Marine polysaccharides: Occurrence, enzymatic degradation and utilization. Chembiochem 2021, 22, 2247–2256. [Google Scholar] [CrossRef]
- Wong, T.; Brault, L.; Gasparotto, E.; Vallée, R.; Morvan, P.; Ferrières, V.; Nugier-Chauvin, C. Formation of amphiphilic molecules from the most common marine polysaccharides, toward a sustainable alternative? Molecules 2021, 26, 4445. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, T.; Xia, K.; Liu, X.; Zhang, X. Preparation and application of edible agar-based composite films modified by cellulose nanocrystals. Food Packag. Shelf 2022, 34, 100936. [Google Scholar] [CrossRef]
- Mostafavi, F.S.; Zaeim, D. Agar-based edible films for food packaging applications—A review. Int. J. Biol. Macromol. 2020, 159, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xue, Q.; Guo, Q.; Wang, G.; Yan, S.; Wu, Y.; Li, L.; Zhang, X.; Li, B. The covalent crosslinking of dialdehyde glucomannan and the inclusion of tannic acid synergistically improved physicochemical and functional properties of gelatin films. Food Packag. Shelf 2021, 30, 100747. [Google Scholar] [CrossRef]
- Chang, P.; Robyt, J. Oxidation of primary alcohol groups of natur1ally occurring polysaccharides with 2,2,6,6-Tetramethyl-1-piperidine oxoammonium ion. J. Carbohyd. Chem. 1996, 15, 819–830. [Google Scholar] [CrossRef]
- Shimizu, M.; Fukuzumi, H.; Saito, T.; Isogai, A. Preparation and characterization of TEMPO-oxidized cellulose nanofibrils with ammonium carboxylate groups. Int. J. Biol. Macromol. 2013, 59, 99–104. [Google Scholar] [CrossRef]
- Figueiredo, C.; De Lacey, A.L.; Pita, M. Electrochemical studies of galactose oxidase. Electrochem. Sci. Adv. 2022, 2, e2100171. [Google Scholar] [CrossRef]
- Li, J.; Davis, I.; Griffith, W.P.; Liu, A. Formation of monofluorinated radical cofactor in galactose oxidase through copper-mediated C-F bond scission. J. Am. Chem. Soc. 2020, 142, 18753–18757. [Google Scholar] [CrossRef]
- Mollerup, F.; Parikka, K.; Vuong, T.V.; Tenkanen, M.; Master, E. Influence of a family 29 carbohydrate binding module on the activity of galactose oxidase from Fusarium graminearum. BBA Gen. Subj. 2016, 1860, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Koschorreck, K.; Alpdagtas, S.; Urlacher, V.B. Copper-radical oxidases a diverse group of biocatalysts with distinct properties and a broad range of biotechnological applications. Eng. Microbiol. 2022, 2, 10003. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, M.E.; Mathieu, Y.; Ribeaucourt, D.; Haon, M.; Mulyk, P.; Hein, J.E.; Lafond, M.; Berrin, J.; Brumer, H. A survey of substrate specificity among Auxiliary Activity Family 5 copper radical oxidases. Cell. Mol. Life Sci. 2021, 78, 8187–8208. [Google Scholar] [CrossRef] [PubMed]
- Parikka, K.; Master, E.; Tenkanen, M. Oxidation with galactose oxidase- Multifunctional enzymatic catalysis. J. Mol. Catal. B Enzym. 2015, 120, 47–59. [Google Scholar] [CrossRef]
- Parikka, K.; Leppänen, A.; Pitkänen, L.; Reunanen, M.; Willför, S.; Tenkanen, M. Oxidation of polysaccharides by galactose oxidase. J. Agric. Food Chem. 2010, 58, 262–272. [Google Scholar] [CrossRef]
- Xu, S.; Zheng, J.; Xiao, H.; Wu, R. Simultaneously identifying and distinguishing glycoproteins with O-GlcNAc and O-GalNAc (the Tn antigen) in human cancer cells. Anal. Chem. 2022, 94, 3343–3351. [Google Scholar] [CrossRef]
- Duke, J.A.; Paschall, A.V.; Glushka, J.; Lees, A.; Moremen, K.W.; Avci, F.Y. Harnessing galactose oxidase in the development of a chemoenzymatic platform for glycoconjugate vaccine design. J. Biol. Chem. 2022, 298, 101453. [Google Scholar] [CrossRef]
- Lee, S.; Jeong, D.; Lee, S.; Nam, S.; Lee, W.Y. A highly sensitive amperometric galactose biosensor based on graphene-doped sol-gel-derived titania-nafion composite films. Electroanal 2022, 34, 629–639. [Google Scholar] [CrossRef]
- Kanyong, P.; Krampa, F.D.; Aniweh, Y.; Awandare, G.A. Enzyme-based amperometric galactose biosensors: A review. Microchim. Acta 2017, 184, 3663–3671. [Google Scholar] [CrossRef]
- Mckenna, S.M.; Leimkühler, S.; Herter, S.; Turner, N.J.; Carnell, A.J. Enzyme cascade reactions: Synthesis of furandicarboxylic acid (FDCA) and carboxylic acids using oxidases in tandem. Green Chem. 2015, 17, 3271–3275. [Google Scholar] [CrossRef]
- Paukner, R.; Staudigl, P.; Choosri, W.; Haltrich, D.; Leitner, C. Expression, purification, and characterization of galactose oxidase of Fusarium sambucinum in E. coli. Protein Expr. Purif. 2015, 108, 73–79. [Google Scholar] [CrossRef]
- Paukner, R.; Staudigl, P.; Choosri, W.; Sygmund, C.; Halada, P.; Haltrich, D.; Leitner, C. Galactose oxidase from Fusarium oxysporum—Expression in E. coli and P. pastoris and biochemical characterization. PLoS ONE 2014, 9, e100116. [Google Scholar] [CrossRef] [PubMed]
- Faria, C.B.; de Castro, F.F.; Martim, D.B.; Abe, C.A.L.; Prates, K.V.; de Oliveira, M.A.S.; Barbosa-Tessmann, I.P. Production of galactose oxidase inside the Fusarium fujikuroi species complex and recombinant expression and characterization of the galactose oxidase GaoA protein from Fusarium subglutinans. Mol. Biotechnol. 2019, 61, 633–649. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Leitner, C.; Volc, J.; Haltrich, D. Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor. Appl. Environ. Microbiol. 2001, 67, 3636–3644. [Google Scholar] [CrossRef]
- Bron, A.J.; Stevens, C.; Wilmot, C.; Seneviratne, K.D.; Blakeley, V.; Dooley, D.M.; Philips, S.E.; Knowles, P.F.; McPherson, M.J. Structure and mechanism of galactose oxidase. The free radical site. J. Biol. Chem. 1994, 269, 25095–25105. [Google Scholar] [CrossRef]
- Whittaker, M.M.; Whittaker, J.W. Expression of recombinant galactose oxidase by Pichia pastoris. Protein Expr. Purif. 2000, 20, 105–111. [Google Scholar] [CrossRef]
- Alberton, D.; De Oliveira, L.S.; Peralta, R.M.; Barbosa-Tessmann, I.P. Production, purification, and characterization of a novel galactose oxidase from Fusarium acuminatum. J. Basic Microbiol. 2007, 47, 203–212. [Google Scholar] [CrossRef]
- Solomon, E.I.; Heppner, D.E.; Johnston, E.M.; Ginsbach, J.W.; Cirera, J.; Qayyum, M.; Kieber-Emmons, M.T.; Kjaergaard, C.H.; Hadt, R.G.; Tian, L. Copper active sites in biology. Chem. Rev. 2014, 114, 3659–3853. [Google Scholar]
- Hu, Z.; Hong, P.; Liao, M.; Kong, S.; Huang, N.; Ou, C.; Li, S. Preparation and characterization of chitosan-agarose composite films. Materials 2016, 9, 816. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wu, C.; Huang, J.; Peng, X.; Chen, P.; Tang, S. Synthesis and characterization of a degradable composite agarose/HA hydrogel. Carbohyd. Polym. 2012, 88, 1445–1452. [Google Scholar] [CrossRef]
- Gómez-Ordóñez, E.; Rupérez, P. FTIR-ATR spectroscopy as a tool for polysaccharide identification in edible brown and red seaweeds. Food Hydrocoll. 2011, 25, 1514–1520. [Google Scholar] [CrossRef]
- Trivedi, T.J.; Kumar, A. Efficient extraction of agarose from red algae using ionic liquids. Green Sustain. Chem. 2014, 4, 190–201. [Google Scholar] [CrossRef]
- Lee, H.; You, J.; Jin, H.J.; Kwak, H.W. Chemical and physical reinforcement behavior of dialdehyde nanocellulose in PVA composite film: A comparison of nanofiber and nanocrystal. Carbohyd. Polym. 2020, 232, 115771. [Google Scholar] [CrossRef] [PubMed]


| Enzyme | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) |
|---|---|---|---|
| GAO-5F | 0.13 | 8.17 | 62.84 ± 0.021 |
| GAO-13F | 0.11 | 4.25 | 38.64 ± 0.023 |
| Compound | Concentration (mM) | Relative Activity (%) a |
|---|---|---|
| CuCl2·2H2O | 5 | 142 ± 3.59 |
| Tween 80 | 0.25% | 142 ± 1.58 |
| None | - | 100 ± 1.57 |
| NaCl | 5 | 87.9 ± 2.34 |
| MgCl2·6H2O | 5 | 85.1 ± 2.03 |
| SDS | 5 | 81.6 ± 1.98 |
| KCl | 5 | 80.5 ± 4.48 |
| EDTA | 5 | 76.7 ± 1.85 |
| CTAB | 5 | 14.8 ± 3.69 |
| Substrate | Concentration (mM) | Relative Activity (%) |
|---|---|---|
| D-Galactose | 25 | 100 ± 3.57 |
| Methyl-α-D-galactoside | 25 | 99.7 ± 1.19 |
| D-(+)-Raffinose | 25 | 95.9 ± 2.93 |
| D-(+)-Melibiose | 25 | 57.6 ± 3.40 |
| Agarose | 0.1% | 12.6 ± 1.04 |
| κ-Carrageenan | 0.1% | 9.33 ± 0.52 |
| Lactose monohydrate | 25 | 4.56 ± 1.26 |
| D-Glucose | 25 | 0 |
| Sucrose | 25 | 0 |
| Maltose | 25 | 0 |
| Glyoxal aqueous 40% solution | 25 | 0 |
| Ethylene glycol | 25 | 0 |
| D-(+)-Galacturonic acid monohydrate | 25 | 0 |
| Soluble starch | 1% | 0 |
| Fructooligosaccharides | 1% | 0 |
| Guar gum | 1% | 0 |
| Gum arabic | 1% | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, N.; Xia, G.; Sun, H.; Zhao, L.; Cao, R.; Jiang, H.; Mao, X.; Liu, Q. Characterization of a Galactose Oxidase from Fusarium odoratissimum and Its Application in the Modification of Agarose. Foods 2023, 12, 603. https://doi.org/10.3390/foods12030603
Cao N, Xia G, Sun H, Zhao L, Cao R, Jiang H, Mao X, Liu Q. Characterization of a Galactose Oxidase from Fusarium odoratissimum and Its Application in the Modification of Agarose. Foods. 2023; 12(3):603. https://doi.org/10.3390/foods12030603
Chicago/Turabian StyleCao, Na, Guangli Xia, Huihui Sun, Ling Zhao, Rong Cao, Hong Jiang, Xiangzhao Mao, and Qi Liu. 2023. "Characterization of a Galactose Oxidase from Fusarium odoratissimum and Its Application in the Modification of Agarose" Foods 12, no. 3: 603. https://doi.org/10.3390/foods12030603
APA StyleCao, N., Xia, G., Sun, H., Zhao, L., Cao, R., Jiang, H., Mao, X., & Liu, Q. (2023). Characterization of a Galactose Oxidase from Fusarium odoratissimum and Its Application in the Modification of Agarose. Foods, 12(3), 603. https://doi.org/10.3390/foods12030603







