Abstract
Cucurbitaceae comprises 800 species, the majority of which are known for their nutritive, economic, and health-promoting effects. This study aims at the metabolome profiling of cucumber (Cucumis sativus) and bottle gourd (Lagenaria siceraria) fruits in a comparative manner for the first time, considering that both species are reported to exhibit several in-common phytochemical classes and bioactivities. Nevertheless, bottle gourd is far less known and/or consumed than cucumber, which is famous worldwide. A multiplex approach, including HR-UPLC/MS/MS, GNPS networking, SPME, and GC/MS, was employed to profile primary and secondary metabolites in both species that could mediate for new health and nutritive aspects, in addition to their aroma profiling, which affects the consumers’ preferences. Spectroscopic datasets were analyzed using multivariate data analyses (PCA and OPLS) for assigning biomarkers that distinguish each fruit. Herein, 107 metabolites were annotated in cucumber and bottle gourd fruits via HR-UPLC/MS/MS analysis in both modes, aided by GNPS networking. Metabolites belong to amino acids, organic acids, cinnamates, alkaloids, flavonoids, pterocarpans, alkyl glycosides, sesquiterpenes, saponins, lignans, fatty acids/amides, and lysophospholipids, including several first-time reported metabolites and classes in Cucurbitaceae. Aroma profiling detected 93 volatiles presented at comparable levels in both species, from which it can be inferred that bottle gourds possess a consumer-pleasant aroma, although data analyses detected further enrichment of bottle gourd with ketones and esters versus aldehydes in cucumber. GC/MS analysis of silylated compounds detected 49 peaks in both species, including alcohols, amino acids, fatty acids/esters, nitrogenous compounds, organic acids, phenolic acids, steroids, and sugars, from which data analyses recognized that the bottle gourd was further enriched with fatty acids in contrast to higher sugar levels in cucumber. This study provides new possible attributes for both species in nutrition and health-care fields based on the newly detected metabolites, and further highlights the potential of the less famous fruit “bottle gourd”, recommending its propagation.
Keywords:
aroma; bottle gourd; chemometrics; cucumber; Cucumis sativus; Lagenaria siceraria; metabolomics; nutrients 1. Introduction
Family Cucurbitaceae comprises 130 genera and 800 species, including several crucial crops of high nutritive and medicinal values and increasing economic interest [1,2]. Roots and seeds of most Cucurbitaceae members share the presence of toxic triterpenes, cucurbitacins. In contrast, their fruits are safe, edible, nutritious, and possess health-promoting effects [1,3]. Cucumis sativus (cucumber) is a well-known species of Cucurbitaceae that is cultivated worldwide, and its fruit is edible either fresh or cooked [1,4]; in contrast, the less famous Lagenaria siceraria species (bottle gourd) is commonly used on the Indo-Pakistan subcontinent and few other countries in Europe and Africa for its nutritive and therapeutic values [5]. Aside from the macromorphological resemblance, both species are reported to exhibit anti-inflammatory, antioxidant, antimicrobial, anticancer, antihyperlipidemic, and cardioprotective activities, and are used in treatment of GIT disorders [1,4]; additionally, fruits’ extracts are involved in the treatment of skin disorders [3,4,6]. Previous phytochemical screening of L. siceraria and C. sativus indicated the presence of flavonoids, sterols, terpenes, phenolic acids, saponins, and carbohydrates in both fruits, whereas alkaloids were detected in C. sativus [3,4,7].
Despite that both bottle gourd and cucumber fruits share several common nutritional and medicinal purposes as well as their involvement in Ayurvedic and folk medicines [4,8], few studies have reported on their metabolome heterogeneity and how the bottle gourd compares to the most famous fruit in F. Cucurbitaceae, the cucumber, as analyzed using metabolomics and chemometrics. In this study, the first comparative metabolome profiling of L. siceraria and C. sativus fruit crude extracts is presented as measured via high resolution-ultrahigh performance liquid chromatography coupled to mass spectrometry (HR-UPLC/MS/MS) in both ionization modes for a comprehensive overview of metabolites. HR-UPLC/MS/MS is the analytical tool of choice for rapid and sensitive monitoring of secondary metabolites due to its robustness and selectivity [9]. Identification of metabolites was aided by Global Natural Products Social molecular networking (GNPS), that allowed rapid dereplication of compounds and extrapolating tentative identification into the unknown [9,10].
Solid phase microextraction (SPME) of volatile constituents alongside silylation of nonvolatile compounds followed by GC/MS analyses provided insight of their aroma and nutrients profile, respectively, which affect food value and consumer preferences, likewise for the first time [11]. Owing to the complexity of the obtained datasets via GC/MS analyses, multivariate data analyses using principal component analysis (PCA) and orthogonal projection to latent structures-discriminant supervised data analysis (OPLS) were applied for the first time for samples classification and assigning discriminating metabolites for each fruit.
The study provides new insights into the chemical composition of cucumber and the less investigated species, bottle gourd, in a comparative approach leading to the identification of several first-time-reported metabolites and classes in both species and a better rationalization of their health effects and potential application as nutraceuticals in the future, based on such chemical profiling.
2. Materials and Methods
2.1. Plant Material
The fresh fruits of cucumber (Cucumis sativus) and bottle gourd (Lagenaria siceraria var. siceraria) were obtained from Barrage Experimental Farm of Horticulture Research Station, El-Kanater El-Khyreia, Qalubia Governorate, Egypt. They were identified by Prof. Dr. Mahmoud Kotb Hatem; Breeding Research Department for Vegetable Crops, Medicinal, and Aromatic Plants, Horticulture Research Institute, Agricultural Research Center.
2.2. Chemicals and Fibers
SPME fiber of stableflex coated with divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS, 50/30 µm) was purchased by Supelco (Oakville, ON, Canada). All chemicals and standards were purchased from Sigma Aldrich (St. Louis, MO, USA). Acetonitrile and formic acid (LC–MS grade) were obtained from J. T. Baker (The Netherlands), and milliQ water was used for UPLC/PDA/ESI–qTOF-MS analysis.
2.3. Extracts Preparation for UPLC-MS Analysis
Freeze-dried samples of C. sativus and L. siceraria fruits (n = 3 each) were ground in liquid nitrogen (Sigma-Aldrich, St. Louis, MO, USA, purity ≥ 99.998%) using pestle and mortar. The extraction procedure was conducted as previously described in Hegazi, N.M. et al., [9]. About 150 of the powdered samples was mixed with 6 mL methanol containing 1 g/mL umbelliferone as an internal standard (Sigma-Aldrich, St. Louis, MO, USA, purity ≥ 98.0%) and homogenized with an Ultra-Turrax (IKA, Staufen, Germany) at 11,000 rpm, 5 × 60 s with 1 min break intervals. Extracts were then vortexed for 1 min, centrifuged at 3000× g for 30 min, and filtered through a 22 m pore size filter.
2.4. UPLC-MS Analysis
Chromatographic separation was performed on an ACQUITY UPLC system (Waters, Milford, MA, USA) equipped with a HSS T3 column (100 × 1.0 mm, particle size 1.8 µm; Waters). The analysis was carried out under exact conditions described in Hegazi, N.M. et.al., [9]. Characterization of compounds was performed by generation of the elemental formula (mass accuracy < 5 ppm) and considering RT, MS2 data and reference literature.
2.5. Molecular Networking
The molecular network (MN) was constructed using the UPLC-HRMS/MS data (in the negative and positive ion mode) from both fruit extracts as prepared in Section 2.3 following exact conditions mentioned in Hegazi, N.M. et al. [9]. The MN parameters were as follows: minimum cosine score 0.70; 0.1 Da parent mass tolerance, 0.5 Da as fragment ion tolerance to create consensus spectra, more than 6 matched peaks, and a minimum cluster size of 2. All of the matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 4 matched peaks. Cytoscape (ver. 3.8.2.) was used for network visualization and analysis.
2.6. Headspace Volatiles Analysis of C. sativus and L. siceraria
The sample was prepared and analyzed following the same procedure and conditions reported in Farag et al. [12]. GC–MS analysis was adopted on an Agilent 5977B GC/MSD (Santa Clara, CA, USA) equipped with a DB-5 column (30 m × 0.25 mm i.d. × 0.25 µm film thickness; Supelco, Bellefonte, PA, USA) and coupled to a quadrupole mass spectrometer following the exact conditions mentioned in Farag, M.A. et al., [12]. For assessment of replicates, three different samples for each fruit were analyzed under the same conditions. Blank runs were conducted during sample analyses. The mass spectrometer was adjusted to EI mode at 70 eV with a scan range set at m/z 40–500.
2.7. GC–MS Analysis of Silylated Primary Metabolites in C. sativus and L. siceraria Fruits
100 Mg of finely freeze-dried powdered sample (for both fruits) was extracted with 5 mL 100% methanol with sonication for 30 min using Branson CPX-952-518R set at 36 °C, (Branson Ultrasonics, Carouge, SA Switzerland.) and with regular shaking, followed by centrifugation (LC-04C 80-2C regen lab prp centrifuge, Zhejiang, China) at 12,000× g for 10 min to eliminate debris. For evaluation of biological replicates, 3 independent samples for each fruit was analyzed under the same conditions. Then, 100 µL of the methanol extract was kept in opened screw-cap vials and left to evaporate under stream of nitrogen gas until full dryness. For derivatization, 150 µL of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA), previously diluted 1/1 with anhydrous pyridine, was mixed with the dried methanol extract and incubated (Yamato Scientific DGS400 Oven, QTE TECHNOLOGIES, Hanoi, Vietnam) for 45 min at 60 °C previous analysis using GC–MS. Separation of silylated derivatives was completed on a Rtx-5MS Restek, Bellefonte, PA, USA (30-m length, 0.25-mm inner diameter and 0.25-m film). Analysis of these primary metabolites followed the exact protocol detailed in Sedeek, M.S. et al., and Farag, M.A. et al. [11,12].
2.8. Metabolites Identification and Multivariate Data Analyses of Volatile and Non-Volatile Silylated Components Analyzed Using GC/MS
Identification was performed by comparing their retention indices (RI) in relation to n-alkanes (C8-C30), mass matching to NIST, WILEY library database and with standards if available. Peaks were first deconvoluted using AMDIS software (www.amdis.net, accessed on 23 April 2022) before mass spectral matching. Peak abundance data were exported for multivariate data analysis by extraction using MET-IDEA software (Broeckling, Reddy, Duran, Zhao, and Sumner, 2006). Data were then normalized to the amount of spiked internal standard (Z)-3-hexenyl acetate, pareto scaled and then subjected to principal component analysis (PCA), hierarchical clustering analysis (HCA) and partial least squares discriminant analysis (OPLS-DA) using SIMCA-P version 13.0 software package (Umetrics, Umeå, Sweden). All variables were mean-centered and scaled to Pareto variance.
3. Results and Discussion
3.1. Metabolome Profiling of L. siceraria and C. sativus Fruit Extracts via HR-UPLC/PDA/ESI-MS Based Molecular Networking
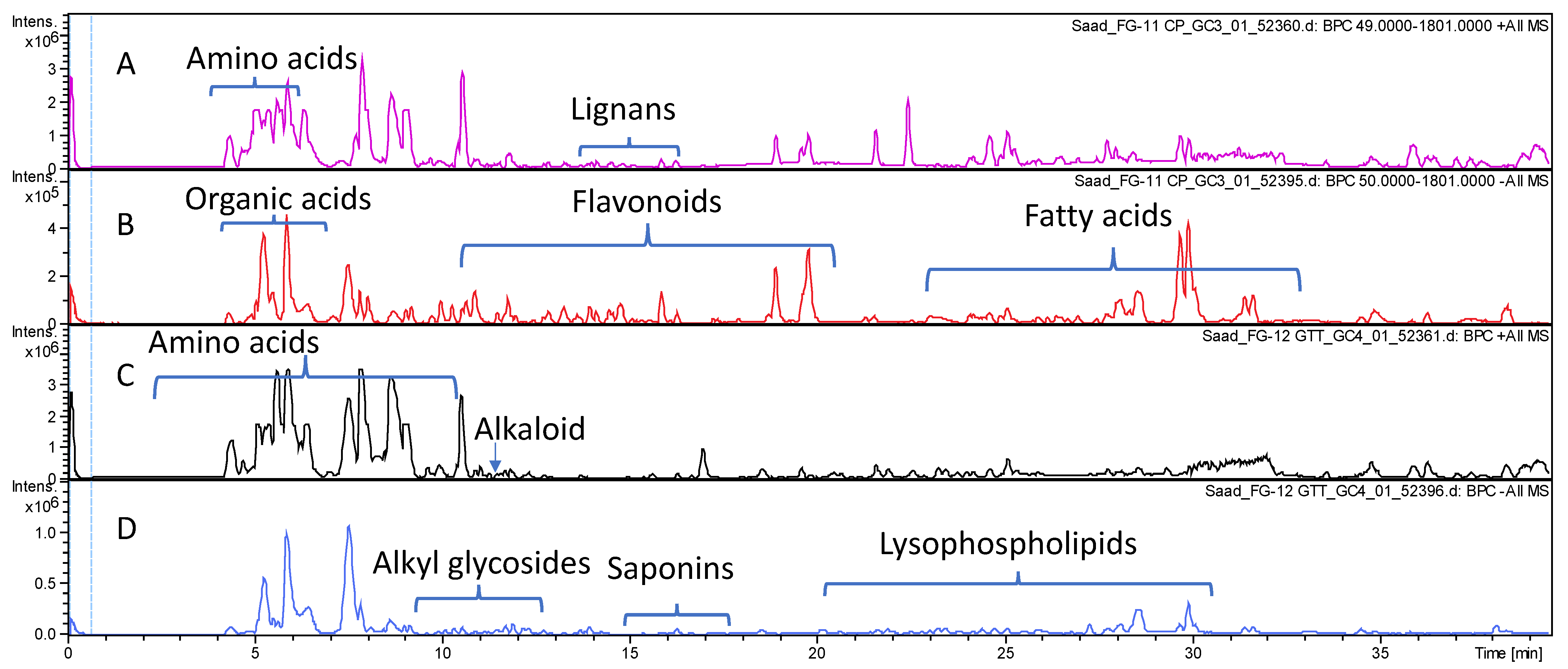
The comparative profiling of metabolites in bottle gourd and cucumber crude fruit methanol extracts was conducted via HR-UPLC/MS/MS in both negative and positive ionization modes (Section 2.4) for a comprehensive overview of metabolites that belong to different phytochemical classes. Gradient elution system of formic acid in water (0.1%): acetonitrile allowed for metabolites’ elution in order of their decreasing polarity. The obtained base peak chromatograms (BPCs) of each fruit extract in both ionization modes are presented in Figure 1.
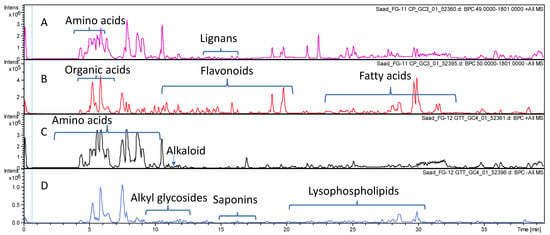
Figure 1.
Base peak chromatograms of Lagenaria siceraria (A,B) and Cucumis sativus (C,D) crude fruit extracts as analyzed via HR-UPLC/MS/MS in both positive (A,C) and negative (B,D) ionization modes.
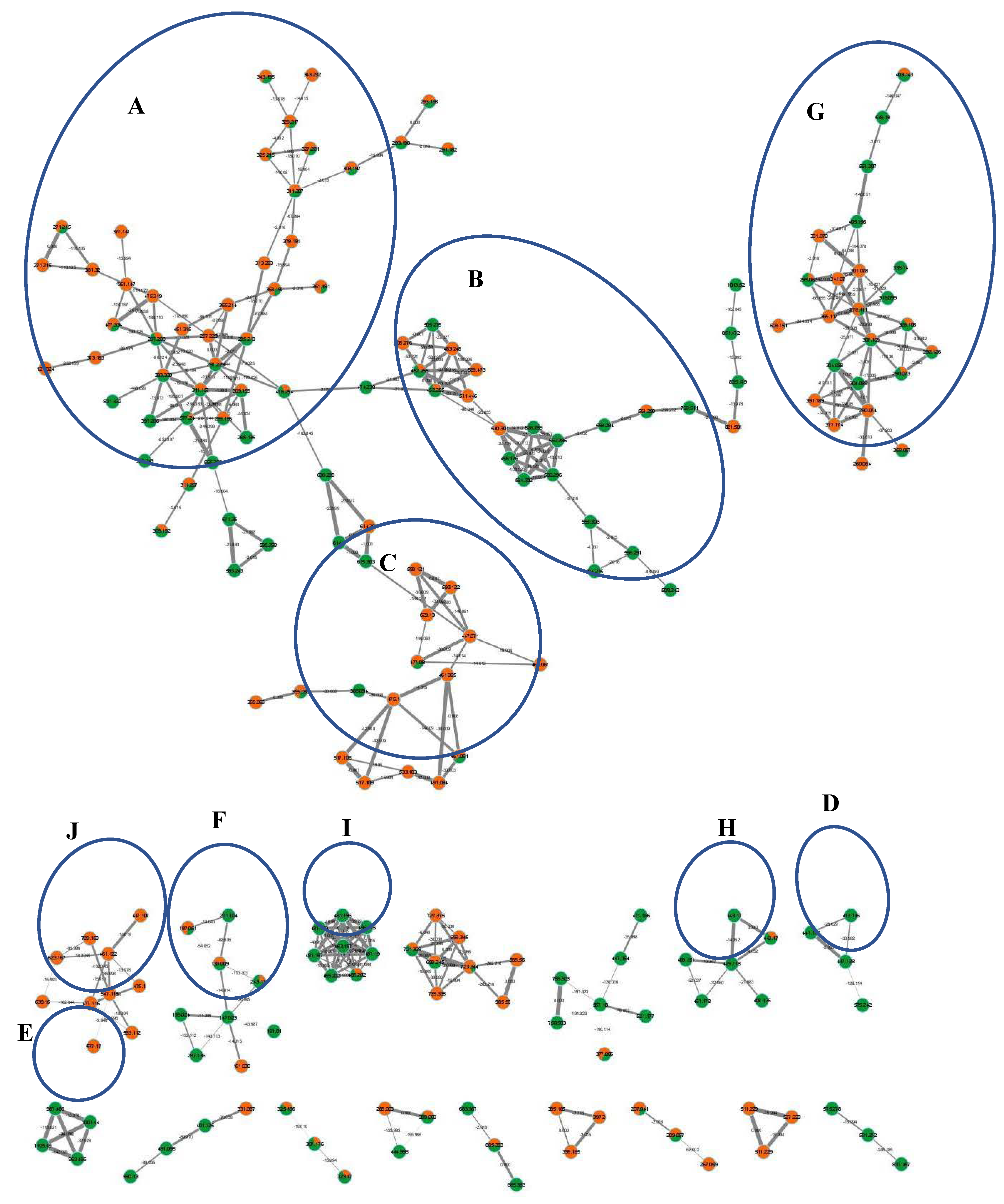
Furthermore, molecular networking was applied herein for the in-depth exploration and discrimination of samples guided by the Global Natural Products Social (GNPS) networking software and its spectral library that aided in peaks’ assignment and annotation based on analyzing the HR-tandem MS/MS data [9,10]. Two molecular networks (MN) were constructed from both ionization mode analyses that provided visual discrimination of samples based on metabolites’ abundance in each node, presented as a pie chart, allowing for rapid dereplication of known compounds (Figure 2 and Figure S1). Samples were coded with orange and green colors for bottle gourd and cucumber, respectively. All nodes were labelled with parent mass and edges were labelled with neutral loss values.
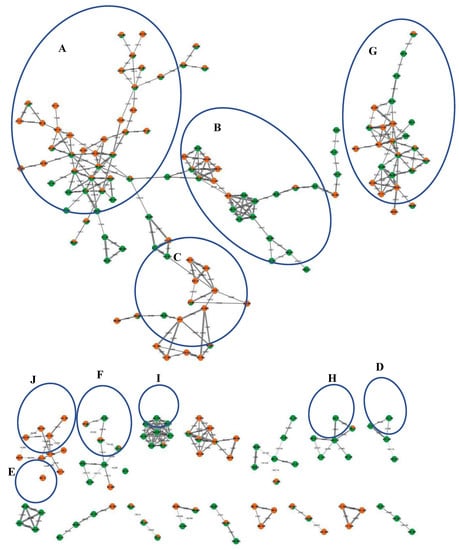
Figure 2.
Full molecular networking created using MS/MS data in negative ionization mode for L. siceraria (bottle gourd) and C. sativus (cucumber) crude fruit extracts showing 351 nodes and 449 edges. All nodes are labelled with parent mass and edges are labelled with neutral loss values. The network is displayed as a pie chart with orange and green colors representing distribution of the precursor ion intensity in the bottle gourd and cucumber extracts, respectively. Clusters annotation: (A): fatty acids/amides, (B): lysophospholipids, (C): flavonoids and ptercarpans, (D): alkyl glycosides, (E): saponins, (F): organic acids, (G): amino acids derivatives, (H): sesquiterpenes, (I): lysophosphatidic acid derivatives, and (J): cinnamates.
Identification was based on determining retention time (Rt. min.) for each metabolite, its mass spectral data—including molecular ion, daughter ions, their respective formulae and fragmentation pattern—and comparing the collective data with the reported literature and databases such as HMDB, PubChem, FooDB, the Phytochemical dictionary of natural products, and others, combined with GNPS library annotations. The clustering of metabolites in MN (minimum two connected nodes) was based on shared fragments and their fragmentation pattern; this allowed us to extrapolate the identification of annotated compounds to the unknown peaks aided by the generated formulae [10].
Overall, 107 peaks were annotated in both modes (Table 1), belonging to amino acids, organic acids, cinnamates, alkaloids, flavonoids, pterocarpans, alkyl glycosides, sesquiterpenes, saponins, lignans, fatty acids/amides, and lysophospholipids, including several first-time reported metabolites and classes, as explained in detail in the following subsections. The constructed MN from the HR- negative ESI-MS/MS analysis was composed of 351 nodes and 449 edges, in which the clusters of interest included cluster A: fatty acids/amides, cluster B: lysophospholipids, cluster C: flavonoids and pterocarpans, cluster D: alkyl glycosides, cluster E: saponins, cluster F: organic acids, cluster G: amino acids derivatives, cluster H: sesquiterpenes, cluster I: lysophosphatidic acid derivatives, and cluster J: cinnamates (Figure 2). The positive MN was composed of 1002 nodes and 1451 edges, in which the clusters of interest included cluster A: lignans, cluster B: flavonoids, cluster C: amino acid derivatives, singleton D: alkaloid, and cluster E: fatty acids/amides (Figure S1).

Table 1.
Metabolome profiling of Cucumis sativus and Lagenaria siceraria crude fruit extracts via HR-UPLC/MS/MS in both negative and positive modes of analyses.
The detection of alkaloids in positive mode versus phenolic acids in negative mode is expected considering the improved sensitivity for each class in respective mode and highlighting the importance of acquiring in different ionization types. Glycosides were detected based on the neutral loss of the attached O-sugar moieties at 162 amu (C6H10O5) for hexoses, 146 amu (C6H10O4) for rhamnose, 176 amu (C6H8O6) for glucuronide and 132 amu (C5H8O4) for pentoses. In some spectra, peaks detected at m/z 113 (C5H5O3−), m/z 101 (C4H5O3−), m/z 71 (C3H3O2−) and/or m/z 59 (C2H3O2−) correspond to hexose fragments [13]. Acylation of glycosides with acetyl or malonyl moieties was recognized by additional mass and/or neutral loss of 42 amu (C2H2O) or 86 amu (C3H2O3), respectively, while C-glycosides showed significant neutral losses of 90 amu and 120 amu resulting from 0,2 and 0,3-sugar ring cleavage [14].
3.1.1. Amino Acids and Amines
Several amino acids and amine derivatives were eluted early, as detected in chromatograms at Rt. 4–11 min. (Figure 1), and grouped in two major clusters, i.e., G and C, in negative and positive MNs, respectively (Figure 2 and Figure S1), from which 14 metabolites could be annotated in peaks 2–15 (Table 1). The identified metabolites included four amines and 10 derivatives of amino acids viz. lysine, iso/leucine, phenylalanine, and glutamine, in which decarboxylation (-CO2, 44 amu), demethylation (-CH2, 14 amu), deglycosilation, deamination (-NH2, 17 amu) and dehydration (-H2O, 18 amu) were the major fragmentation pathways matching the reported literature, GNPS library, and HMDB and MassBank databases. All identified metabolites were common between both fruits except for peaks 14 and 15 (Table 1), which were detected in L. siceraria at [M+H]+ 267.1337, C13H19N2O4+ and [M+H]+ 245.1859, C12H25N2O3+, identified as N-(methoxybenzyl)glutamine, which is previously reported in Cucurbitaceae [15] and Iso/Leucyl-Iso/Leucine dipeptide, respectively.
3.1.2. Organic Acids
6 Organic acids were identified in peaks 17–22 (Table 1) and grouped in cluster F in negative MN (Figure 2) based on decarboxylation and dehydration shared fragments. For example, homocitric acid in peak 18 (Table 1) was detected in L. siceraria for the first time at [M-H]−, C7H9O7− and fragmented into m/z 161, C6H9O5− (-COO, 44 amu), m/z 125, C6H5O3− [M-H-CO2-2H2O], and m/z 81, C5H5O− [M-H-2CO2-2H2O].
3.1.3. Alkaloids
Aside from previous studies that reported the presence of alkaloids in cucumber at moderate levels [3,16,17], peak 16 at [M+H]+ 247.1446, C14H19N2O2+ appeared as a singleton in positive MN (Figure S1) at Rt. 11.4 min. (Table 1) and was identified as N,N,N-trimethyltryptophan betaine, known as lenticin or hypaphorin alkaloid, which is reported herein for the first time in cucumber fruit. Identification was based on the presence of diagnostic daughter ions at m/z 188, C11H10NO2+, post the loss of N-trimethyl moiety, followed by decarboxylation at m/z 144 C10H10N+ or dehydration at m/z 170, C11H8NO+. The latter proceeded into ring cleavage and demethylation at m/z 146, C9H8NO+ and m/z 122 C7H8NO+, as explained in Figure S2, matching the reported literature [18] and HMDB spectrum (https://hmdb.ca/spectra/ms_ms/2947783, accessed on 11 January 2023). Lenticin is distributed in various vegetables and known to exhibit cardioprotective and neurological effects, and thus could be correlated with cucumbers’ cardioprotective reported effect for the first time using such a metabolomics approach [19]. It is believed that cucumber may encompass several other alkaloids, but a special extract targeting method with solvents of lower polarity is needed to enhance their detection.
3.1.4. Phenolics and Cinnamic Acid Derivatives
In total, 15 glycosylated and/or acylated derivatives of phenolic and cinnamic acids were eluted at Rt. 8–16 min. in peaks 23–38 (Table 1). MSn ions for cinnamic acid derivatives were evidenced at m/z 179, C9H7O4−, m/z 161, C9H5O3− and m/z 135, C8H7O2− for caffeoyl, m/z 193, C10H9O4− and m/z 175, C10H7O3− for feruloyl, m/z 223, C11H11O5− and m/z 205, C11H9O4− for sinapoyl moieties (Table 1). While sinapic acid derivatives were detected in cucumber only, bottle gourd extract was exclusively enriched with conjugates of phenyl and caffeoyl or feruloyl glycosides (Figure S3–S5) that appeared in negative MN grouped in cluster J (Figure 2) and were identified in peaks 28, 29, 31–33 and 35–38 (Table 1). Such conjugates are reported herein for the first time in the genus Lagenaria based on the diagnostic peaks at m/z 123, C7H7O2– and m/z 105, C7H5O− for hydroxymethylphenyl fragments or m/z 137, C7H5O3– and m/z 93, C6H5O− for hydroxybenzoic acid fragments, or m/z 139, C7H7O3− and m/z 121, C7H5O2− for dihydroxymethoxybenzene fragments (Figure S3–S5). Hexosyl moieties in peaks 32, 37, and 38 (Table 1) were additionally acylated with malonic acid (C3H2O3−, 86 amu), which is known to improve the biological effects of phenolics [20].
For example, peak 35 detected in bottle gourd at [M-H]− 461.1449, C23H25O10− was identified as hydroxymethylphenyl-O-feruloyl-O-hexoside fragmented into m/z 337, C16H17O8− post losing hydroxymethylphenyl moiety, m/z 193, C10H9O4− for ferulic acid and its dehydrated and demethylated fragments at m/z 175, C10H7O3− and m/z 160, respectively, whereas ions at m/z 123, C7H7O2− and m/z 105, C7H5O− were detected for hydroxymethlphenyl ion and its dehydrated form, respectively (Figures S3 and S4). The node of peak 35 in negative MN was directly connected to molecular ion [M-H]− 547.1452 in peak 38 at C26H27O13− having an extra C3H2O3− (malonyl, 86 amu), and thus identified as hydroxymethylphenyl-O-feruloyl-O-malonylhexoside, which, in turn, was connected to [M-H]− 709.1985, C32H37O18− having an additional C6H10O5− (hexosyl, 162 amu), and hence identified as hydroxymethylphenyl-O-hexosylferuloyl-O-malonylhexoside, aided by the necessary MSn ions and their predicted formulae to confirm their identification (Table 1). Little information is available on the bioactivities of such acylated conjugates and how they contribute to L. siceraria health effects; thus, further studies are required to explore their potential pharmacological effect.
3.1.5. Sesquiterpenes
The family Cucurbitaceae is known to accumulate di-/sesqui-/and triterpenes, to which various bioactivities are attributed [4,21] nevertheless, sesquiterpenes are the least reported in the literature. Cluster H in negative MN consists of five metabolites (Figure 2), from which peak 39 was detected in both fruit extracts and was identified as cymaroside A at [M-H]− 443.1918, C21H31O10− (Table 1). Cynaroside A is sesquiterpene lactone-O-hexoside, abundant in artichoke, that undergoes deglycosilation at m/z 281, C15H21O5−, decarboxylation at m/z 237, C14H21O3, followed by sequential demethylation, dehydration and ring cleavage fragmentation processes to yield several diagnostic daughter ions, as explained in detail in Figure S6, in accordance with databases and the other literature [22]. Despite being previously detected/isolated from other green vegetables [23], this is the first report of its presence in F. Cucurbitaceae, and whether these fruits could serve as source of that sesquiterpene, similar to artichoke, should be considered. The other four metabolites clustered with cymaroside A sesquiterpene shared ions corresponding to lactone ring cleavage followed by sequential demethylation and dehydration at m/z 119, m/z 101, m/z 89, m/z 71, m/z 59 (Figure S6), but could not be completely identified.
3.1.6. Alkyl Glycosides
From cluster D in negative MN (Figure 2), four metabolites were detected exclusively in cucumber as formate adducts in peaks 44–47 that belong to the alkyl glycosides class (Table 1). Alkyl glycosides are formed of fatty alcohols glycosylated with sugars. In detail, peak 44, [M-H]− detected at m/z 367.1608, C15H27O10− which was identified as butanol-O-pentosyl-hexoside and yielded fragment ions at m/z 235, C10H19O6− and m/z 73, post sequential loss of both sugars followed by demethylation at m/z 59, while MSn ions at m/z 161, 113, 101, 71 belonged to hexosyl fragments (Figure S7) in good accordance with the literature [24] and databases. Similarly, peaks 45 and 46 were identified as benzyl-O-pentosyl-hexoside and hexanol-O-pentosyl-hexoside, respectively. Such compounds were previously detected in several fruits as in apples, anise, cumin, and others [15,25,26,27]; however, according to our knowledge, this is the first report of their presence in Cucurbitaceae. The exact bioactivities of such class of compounds in these fruits have yet to be studied.
3.1.7. Flavonoids
Previous studies reported the presence of isoflavones and acylated/methoxylated apigenin and luteolin-O/C-glycosides in cucumber [28,29], while flavonoid-O/C-glycosides were reported in both species [3,4,30]. In this study, 24 flavonoids were detected and tentatively identified in both species, including flavones, flavonols, and isoflavones. C-glycosides were recognized by MSn ions corresponding to sugar ring cleavage, unlike O-glycosides, which showed intact release of the dehydrated sugar. Aglycones were differentiated based on their typical RDA fragments [31]. Identification was guided by GNPS-based networking, which allowed extrapolating peaks’ annotations to unknown compounds, as observed in clusters B and C, in positive and negative MNs, respectively (Figure S1 and Figure 2).
Acylated flavonoids were detected in peaks 52, 54, 61, 62, 66–68, and 71 (Table 1), in which cucumber was more abundant in aromatic, i.e., feruloyl (C10H8O3, 176 amu) or coumaroyl (C9H6O2, 146 amu), acylated flavones, whereas bottle gourd was enriched with flavone and flavonols acylated with aliphatic, i.e., malonyl (C3H2O3, 86 amu) or acetyl (C2H2O, 42 amu), moieties. These differential metabolomics results could aid in annotating acetyl transferase enzyme by correlating metabolite data with gene expression data. For example, peak 54 was identified as iso/vitexin-O-couamroylhexoside, which was previously reported in cucumber [28]. The precursor ion detected at [M+H]+ 741.2041, C36H37O17+ showed fragments at m/z 595, C27H31O15+ [loss of coumaroyl, 146 amu], m/z 433 for iso/vitexin, C21H21O10+ upon loss of O-hexosyl moiety, followed by MSn ions for C-hexosyl ring cleavage and aglycone RDA at m/z 415, m/z 397, m/z 313 and m/z 283 (Figure S8).
Methoxylated flavonoids were detected abundantly in bottle gourd fruit as in peaks 55, 58, 60, 62, 63, 64, 66–69, and 71 (Table 1), identified based on the neutral loss of one or more methyl (-CH2, 14 amu) / methoxyl (-OCH2, 30 amu) moieties. For example, peak 62 was identified as isorhamnetin-O-malonylhexoside at [M+H]+ 565.1186, C25H25O15+ and was reported for the first time in both species (Figure S9). The precursor ion yielded fragments at m/z 479 [M+H-malonyl]+, m/z 317, C16H13O7+ for isorhamnetin post deglycosylation followed by demethylation at m/z 303, C15H11O7, dehydration at m/z 285, C15H9O6+ and finally RDA fragments at m/z 229, m/z 151 and m/z 127 [32]. To our knowledge, this is the first report of di/methoxylated flavonoid-O-acylated hexoside in L. siceraria, as in peaks 64, 66–69, and 71 (Table 1).
Such a mixture of flavonoids promotes the various health effects of both species, since acylation and methoxylation are known to impart structural and functional modifications that improve flavonoids’ bioactivity by increasing their lipophilicity and intracellular bioavailability, thus exhibiting better receptor/ligand binding compared to their non-acylated/methoxylated analogues, as observed in cancer chemoprevention and anti-acetylcholine esterase activities [20,33,34,35].
3.1.8. Pterocarpans
Pterocarpans are derivatives of isoflavonoids de novo biosynthesized as a response to stress, mainly detected in legumes [36,37]. Herein, three pterocarpans were detected in cucumber grouped with flavonoids in negative MN cluster C (Figure 2) and identified in peaks 72–74 (Table 1). Identification was based on the diagnostic fragmentation pattern, showing sequential loss of two methyl groups (−2 × CH2, 14 amu) followed by decarbonylation (-CO, 28 amu) confirmed by generated formulae [38]. For example, peak 72 in Table 1 showed the precursor ion at [M-H]− 475.124, C23H23O11− that yielded MSn ions at m/z 313, C17H13O6− [M-H-C6H10O5, hexosyl 162 amu], followed by demethylation at m/z 298 and m/z 283, C15H7O6−, then loss of carbonyl at m/z 255, C14H7O5− and was thus identified as hedysarimpterocarpene A-O-hexoside (HPA-O-hexoside) in accordance with the reported literature [38] (Figure S10). Peak 72 node in cluster C was directly connected to [M-H]− 517.1345, C25H25O12−, having an additional 42 amu (-C2H2O−) and sharing the same fragments at m/z 313, m/z 298 and m/z 255, and was thus identified as hedysarimpterocarpene A-O-acetylhexoside (HPA-O-acetylhexoside). Similarly, peak 74 at [M-H]− 313.0715, C17H13O6− was identified as hedysarimpterocarpene (HPA) (Table 1). This is the first report of pterocarpans in Cucurbitaceae; further studies are required to confirm their biosynthesis in plants other than legumes and present them as potential sources of pterocarpans.
3.1.9. Lignans
Three lignans belonging to the dibenzylbutanediol subclass were grouped in cluster A in positive MN as detected in bottle gourd for the first time at [M+H]+ 379.1747, [M+H]+ 363.1795 and [M+H]+ 347.1829 in peaks 75–77 (Table 1), identified as pentahydroxy-dimethoxylignan, known as carinol and secoisolariciresinol, previously reported in Cucurbitaceae [39], and trihydroxy-dimethoxylignan, respectively. Elemental composition of peaks 76, C20H27O6+ and 77, C20H27O5+ showed one and two hydroxyl moieties fewer than peak 75, C20H27O7+, respectively. Identification was based on diagnostic fragments corresponding to sequential dehydration, demethylation, and demethoxylation prior to and/or after cleavage of the bis(benzylbutanediol) bonding (Figure S11) sharing MSn ions at m/z 137, C8H9O2+, m/z 121, C8H9O+ m/z 107, C7H7O+ and m/z 93, C7H9+, in accordance with references and databases [40].
3.1.10. Saponins
Previously, cucumber was reported for the presence of triterpenoid saponins [41]. In this study, five saponins in peaks 78–82 were detected in cucumber fruit (Table 1). Peak 82 detected in positive ionization mode at [M+H]+ 943.5275, C48H79O18+ was identified as soyasaponin I based on fragments at m/z 797, [M+H-rhm], m/z 635, C36H59O9 [(M+H)-rhm-hex], 617, C36H57O8 +, m/z 599, C36H55O7+, m/z 581, C36H53O6+ and m/z 459, C30H51O3+ for soyasapogenol B post loss of glucuronide moiety (C6H8O6,176 amu) followed by dehydration, demethylation and RDA fragmentation of sapogenin at m/z 441, C30H49O2+, m/z 423, C30H47O+, m/z 405, C30H45+, m/z 383, C27H43O+ and other fragments (Figure S12) matching references and databases [42]. On the other hand, all saponins were detected in negative ionization mode analysis as formate adducts (+46 amu, HCOOH), in which soyasaponin I showed few fragments indicative for the loss of hydroxymethyl, hydroxyl and ring A cleavage at m/z 397 and m/z 341, as predicted in HMDB spectra. In negative MN (Figure 2), cluster E consists of saponins correlated with soyasaponin I for their soyasapogenol B/E MSn ions at m/z 397 and m/z 341, in addition to other ions indicating terminal sugar loss or a fragment of hexose at m/z 113 (Table 1, Figure S13). From the aforementioned data, peaks 79 and 81 were annotated as soyasapogenol E-O-dihexosyl-O-glucuronide (soyasaponin Bd) and soyasapogenol B-O-rhamnosyl-O-pentosyl-O-hexosyl-O-glucuronide (melilotus saponin O1), respectively. The mixture of these saponins is reported in legumes [43], but this was the first time it was reported in cucumber. Other saponins maybe present in L. siceraria and C. sativus fruits that could be revealed upon using an extraction-targeting method instead of crude methanol extracts.
3.1.11. Fatty Acids/Amides
In total, 16 fatty acids and one fatty acyl amide were annotated in peaks 83–99, equally distributed in both species (Table 1) and grouped in the major clusters in both negative and positive MNs, A and E, respectively (Figure 2 and Figure S1). This included mono-, di-, tri-, and tetrahydroxylated fatty acids, as evidenced by the loss of water (H2O,18 amu) and carboxylate (CO2, 44 amu) moieties. Six saturated fatty acids were detected in peaks 83, 84, 86, 87, 98, and 99. For example, peak 86—detected in both species— was identified as azelaic acid at [M-H]− 187.0975, C9H15O4− a dicarboxylic fatty acid, which showed fragment ions at m/z 169, C9H13O3− [M-H-H2O], m/z 143, C8H15O2− [M-H-CO2], m/z 125, C8H13O− [M-H-H2O-CO2], m/z 97, C6H9O−, m/z 80, m/z 71, m/z 69 C4H5O− and m/z 57, matching the literature [44]. Azelaic acid is incorporated into skin products for treatment of alopecia, as well as its reported cytotoxic and anti-inflammatory effects [44,45]. This could account for the popular usage of cucumber in skin preparations [3]. In contrast, 10 unsaturated fatty acids were detected in peaks 85, 88, and 90–97 (Table 1). Peak 85 was identified as octadecenamide at [M+H]+ 282.2796, C18H36NO+ showed fragment at m/z 265 C18H33O+ upon loss of ammonia (NH3, 17 amu). The herein identified fatty acids matched reported literature on Cucurbitaceae seed oil contents [15].
3.1.12. Lysophospholipids
In negative MN (Figure 2), cluster B and cluster I grouped lysophospholipids and lysophosphatidic acids, respectively, from which peaks 100–107 were detected (Table 1). Metabolites detected at [M-H]− 481.2203, C21H38O10P−, [M-H]− 497.2156, C21H38O11P−, [M-H]− 463.2100, C21H36O9P− and [M-H]− 431.2202, C21H36O7P− are lysophophatidic acid derivatives, detected in cucumber only in peaks 100–102, and 104 (Table 1)
Overall, metabolome profiling of cucumber and bottle gourd based on GNPS-networking showed that both fruits share the abundance of amino acids, organic acids, flavones-C-glycosides, fatty acids and lysophospholipids in a comparable manner. By contrast, triterpenoid saponins, alkaloids, flavones-C-glycosides acylated with ferulic or coumaric acids, isoflavones alkyl glycosides and pterocarpans were exclusively detected in cucumber, and the latter two classes were reported for the first time in Cucurbitaceae. On the other hand, bottle gourd was further distinguished by the presence of dimethoxylated flavonoids acylated with malonic or acetic acids, lignans and conjugates of phenyl compounds with caffeic/ferulic acid-O-glycosides, all of which are reported for the first time in genus Lagenaria. Sesquiterpenes were more abundant in bottle gourd. except for cynaroside A, which was detected in both fruits for the first time as well. Successive extraction of fruits with solvents of different polarities is recommended to further reveal phytochemical classes that require targeting, such as alkaloids and saponins. The herein identified metabolites rationalized several of the reported pharmacological effects in both species, and further promoted them for new in vitro and in vivo medicinal research. Cucumber shared several phytochemicals that are characteristic to legumes (Fabaceae), e.g., soyasaponins, isoflavones, and pterocarpans, thus supporting the idea of an in-depth study of its biosynthetic pathways and gene expression data [37,43].
3.2. Aroma Profiling of L. siceraria and C. sativus Fruits Using SPME Coupled to GC-MS
SPME is well suited for the profiling of aroma for low-strength aroma food matrices, and also at low temperature for collection, compared to steam distillation, providing the true composition of volatile blends, [11] and this is the first time it has been reported in both species.
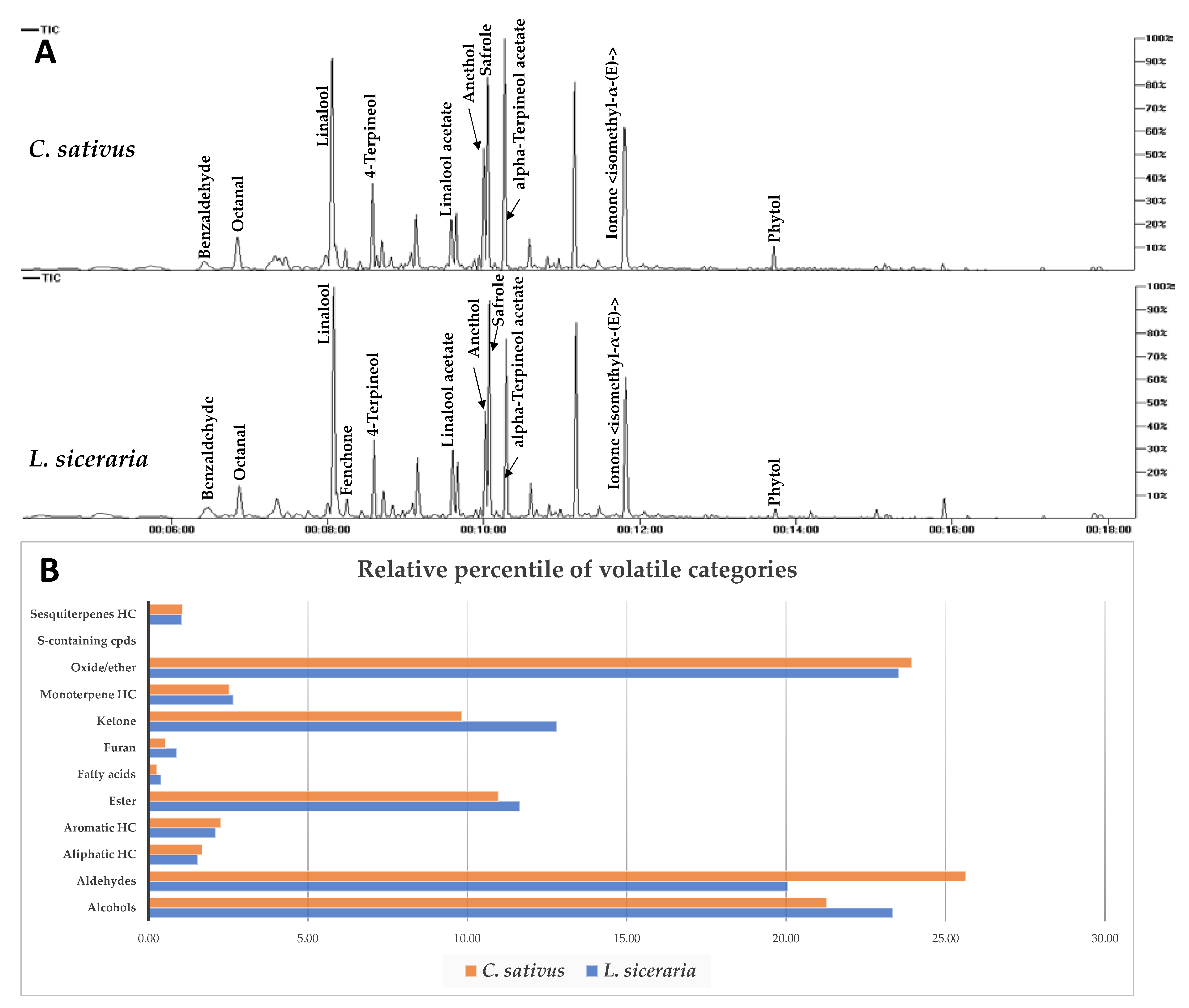
In this study, three independent replicates were analyzed under the same conditions via SPME-GC/MS and represented by total ion chromatograms (Figure 3A). 93 volatile constituents were detected in L. siceraria and C. sativus fruits and categorized into alcohols (11), aldehydes (16), ketones (14), fatty acids (3), oxides/ethers (7), S-containing compounds (1), monoterpenes (3), sesquiterpenes (5), esters (22), furans (1), and aliphatic (7) and aromatic (3) hydrocarbons (Table S1). Both species showed comparable aroma composition, as seen in the relative percentile of each class in Figure 3B.
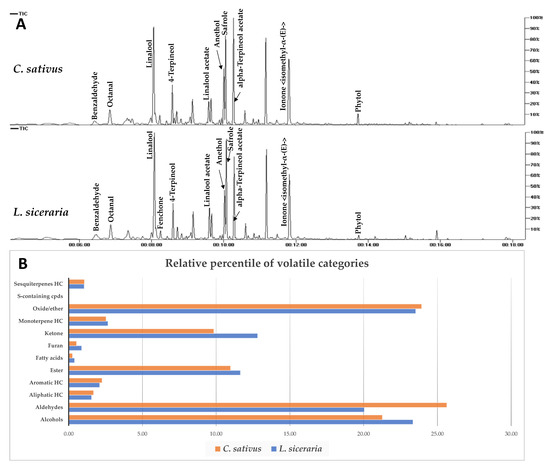
Figure 3.
(A) Total ion chromatogram of C. sativus and L. siceraria aroma constituents as analyzed via SPME GC/MS. Assigned compounds names follow those shown in Supplementary Table S1. (B) Relative percentile of different volatile categories detected via SPME-GC/MS analysis in C. sativus and L. siceraria fruits.
In L. siceraria, alcohols and ethers were the major volatile classes, detected at 23.34% and 23.52%, respectively, whereas aldehydes were the most abundant in C. sativus (25.63%), followed by ethers (23.93 %) (Figure 3B, Table S1). Sesquiterpenes were detected in both fruits at low levels, i.e., 1.05% and 1.07% in bottle gourd and cucumber, respectively. Linalool is the major volatile constituent detected in both fruits at 18.76 % and 17.70 %, followed by safrole (11.4%, 10.5%), anethol (9.43%, 11.2%), and octanol (9.18%, 11.1%) in bottle gourd and cucumber, respectively.
Interestingly, hexenal, nonenal, and nonadienal (cucumber aldehyde) aldehydes that were reported to account for cucumber’s pleasant aroma [46] are herein detected in bottle gourd as well for the first time, at comparable levels except for 2,6-nonadienal (peak 19), which is more abundant in cucumber (Table S1).
3.3. Multivariate Data Analysis of Volatiles Dataset Acquired from SPME-GC/MS
Although notable differences in volatile constituents were observed visually between both fruits, multivariate data analyses were employed to classify both fruits in an untargeted manner using PCA and OPLS modelling (Figure S14). Samples segregation was observed along PC1 and PC2 to account for more than 80% of the total variance (Figure S14A,B). Supervised OPLS was further applied for its superiority in class separation [47] to obtain a model with good variance and prediction power (R2 = 0.88, Q2 = 0.82) (Figure S14C,D). The loading plot and S-loading plots identified metabolites mediating for samples separation as denoted by Mol. ion/Rt and labelled with their peak numbers in accordance with Table S1 (Figure S14B,D).
Examination of the PCA loading plot revealed that aldehydes were abundant in cucumber, represented by benzaldhyde (peak 14), octanal (peak 15), benzenacetaldehyde, (peak 17) and nonadienal (peak 19), in addition to anethol (peak 83), whereas bottle gourd was further enriched with ketones and esters, i.e., fenchone (peak 67) and methyl hexadecanoate (peak 58) (Table S1, Figure S14A,B).
Furthermore, OPLS score plot showed better discrimination of samples (Figure S14C). The S-loading plot derived from OPLS model (Figure S14D) confirmed aldehydes’ abundance in cucumber as discriminating metabolites, i.e., octanal (peak 15), benzenacetaldehyde (peak 17), and nonadienal (peak 19), versus anethol ether (peak 83) fenchone (peak 67), and methyl hexadecanoate (peak 58) in bottle gourd fruit.
The overall aroma of bottle gourd and cucumber is manifested by a mixture of characteristic volatiles. Linalool, that is, the major detected constituent in both fruits, is known to impart an orange oil-like aroma, whereas fenchone in bottle gourd possesses a pleasant camphor-like aroma and is incorporated in food as a perfumery flavor [48]. On the other hand, aldehydes in cucumber, i.e., nonadienal, nonenal, and hexenal are known to mediate for the pleasant characteristic aroma of fresh cucumber that is further enhanced upon chewing, while benzaldhyde has an almond-like odor, all of which are shared in bottle gourd as well at variable concentrations. [49,50,51]. In addition to their role in taste and odor, such volatiles are known to mediate for antimicrobial activity [51,52]. The study infers that bottle gourd possesses a consumer-pleasant aroma.
3.4. Unsupervised PCA Data Analysis of C. sativus and L. siceraria Primary Metabolites
To provide insight into C. sativus and L. siceraria fruits’ primary metabolome mediating for their nutritive value, GC–MS was employed, leading to the detection of 49 peaks. Chromatograms displayed a representative profile of C. sativus and L. siceraria fruits’ nutrient primary metabolites (Figure S15).
Metabolites were categorized into alcohols (6), amino acids (2), fatty acids/esters (12), nitrogenous compounds (1), organic acids (12), phenolic acids (1), steroids (2), sugars (9), sugar acids (1), and sugar alcohols (3), as shown in Table 2.

Table 2.
Primary metabolite analyzed using GC-MS of different C. sativus and L. siceraria with results expressed as relative percentile of the total peak area (n = 3).
In C. sativus, sugars represented the most abundant class at ca. 63% of detected primary metabolites, mostly represented by monosaccharides at 61% and trace disaccharides at ca. 2%. Fructose, glucose, and galactose were the main identified monosaccharides, amounting to ca. 27, 19, and 14%, respectively, to account for the mild sweet taste of C. sativus (Table 2). The sugars in L. siceraria were detected at half that in C. sativus (ca. 33%), likewise represented by fructose, glucose, and galactose, though at lower levels than in C. sativus at 8–11%. Both samples encompassed trace levels of sugar acids and sugar alcohols (Table 2). Myo-inositol was the major detected sugar alcohol detected at 1%, which is in agreement with what was previously published for cucumber fruits [53].
12 Fatty acids/esters were identified in both fruit samples, i.e., cucumber and bottle gourd, amounting for ca. 15 and 36%, respectively (Table 2), mostly represented by saturated fatty acids detected at 13 and 29% in both fruit samples, respectively. The main identified fatty acids were palmitic, caprylic, and stearic acids (Table 2). At the cellular and tissue levels, palmitic acid performs a variety of important biological roles [54]. Caprylic acid is a medium-chain saturated fatty acid that exerts strong anti-atherosclerotic, anti-inflammatory, antifungal, and antibacterial effects [55,56]. Only three unsaturated fatty acids were identified, i.e., linoleic acid, oleic acid, and α-linolenic acid, amounting to 2 and 7% in cucumber and bottle gourd, respectively.
12 Organic acids were identified in both samples amounting to 13 and 17% in C. sativus and L. siceraria, respectively. Glutamic, malic, hydroxy butanoic, and lactic acids were the major identified organic acids in both fruits (Table 2). Organic acids are known to exert bactericidal activity acting as food acidulant preservatives [57].
Azelaic acid one of the organic acids identified, though at small levels, in both samples at 0.3%, and is well-reported for the treatment of a variety of skin conditions owing to its many skin-healing properties [58]. Whether L. siceraria has potential application in skin products comparable to that of C. sativus, with its well-reported anti-irritant, anti-acne, and skin-whitening properties has yet to be examined.
Amino acids were represented by two amino acids, viz. glycine and alanine, amounting to 2–3% of the total primary metabolites. Only one non-amino acid nitrogenous, i.e., urea, was identified in both fruits at 3–5% (Table 2).
3.5. Supervised Multivariate OPLS-DA Analysis of C. sativus and L. siceraria Primary Metabolites
(OPLS-DA) was further employed to confirm samples’ classification and primary metabolites’ marker determination obtained by GC-MS data-based PCA analysis. Both samples were modelled against each other (Figure S16C,D), showing clear separation as shown in the derived score plot, with R2 = 0.99 (variance coverage) and a prediction goodness parameter Q2 = 0.93 (Figure S16C). The corresponding S-plot (Figure S16D) showed that cucumber fruits were rich in sugars, i.e., glucose, galactose, and fructose, as listed in Table 2, found at a level twice that found in L. siceraria fruit, which accounts for its better recognition as fresh food, owing to its better taste. By contrast, the bottle gourd fruits were rich in fatty acids (Figure S16D), which confirmed the same markers obtained by PCA classification.
4. Conclusions
This study provided the first comparative metabolite profiling of cucumber and bottle gourd fruits via HR-UPLC/MS/MS based on GNPS-networking in both modes, allowing for annotation of 107 metabolites, i.e., amino acids (14), organic acids (6), phenolic/cinnamic acid derivatives (15), alkaloids (1), flavonoids (24), pterocarpans (3), alkyl glycosides (4), sesquiterpenes (5), saponins (5), lignans (3), fatty acids/amides (16), and lysophospholipids (8). Overall, both fruits share an abundance of amino acids, organic acids, flavones-C-glycosides, fatty acids and lysophospholipids in a comparable manner. However, triterpenoid saponins, alkaloids, flavones-C-glycosides acylated with ferulic or p-coumaric acids, alkyl glycosides, and pterocarpans were exclusively detected in cucumber, and the latter two classes were reported in this study for the first time to be present in Cucurbitaceae. On the other hand, bottle gourd was further distinguished by the presence of dimethoxylated flavonoids acylated with malonic or acetic acids, lignans, and conjugates of phenyl compounds with caffeic/ferulic acid-O-glycosides, all of which were reported for the first time in genus Lagenaria. Sesquiterpenes were more abundant in bottle gourd, except for cynaroside A, which was detected in both fruits for the first time. The herein-identified metabolites rationalized several of the reported pharmacological effects of both species and further promoted them for new in vitro and in vivo medicinal research. Several of the herein-detected phytochemicals require further bioactivity studies to study their potential health effects, e.g., pterocarpans, alkyl glycosides, and phenyl-cinnamic acids conjugates. Aroma profiling via SPME-GC/MS analysis detected 93 volatiles at comparable levels in both species, responsible for the pleasant aroma of bottle gourd, despite its enrichment with ketones and esters. 49 Silylated primary metabolites were detected in both species via GC/MS analysis at comparable levels, of which bottle gourd was further enriched with fatty acids, and cucumber with sugars.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods12040771/s1, Table S1: Relative percentile of volatile constituents detected in C. sativus and L. siceraria fruits via SPME/GCMS; Figure S1: Full molecular networking created using MS/MS data in positive ionization mode for L. siceraria (bottle gourd) and C. sativus (cucumber) crude fruit extracts showing 1002 nodes and 1451 edges. All nodes are labeled with parent mass and edges are labelled with neutral loss values. The network is displayed as pie chart with orange and green colors representing distribution of the precursor ion intensity in the pumpkin and cucumber extracts respectively; Figure S2: Tandem MS/MS spectrum of N,N,N-Trimethyltryptophan betaine (or Lenticin) alkaloid (peak 16) detected in C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in positive ionization mode; Figure S3: Tandem MS/MS spectrum of hydroxymethylphenyl-O-hexosylferuloyl-O-hexoside (peak 29) detected in L. siceraria fruit crude extract via HR-UPLC/MS/MS analysis in negative ionization mode; Figure S4: Tandem MS/MS spectrum of hydroxymethylphenyl -O-caffeoyl-O-hexoside (peak 31) detected in L. siceraria fruit crude extract via HR-UPLC/MS/MS analysis in negative ionization mode; Figure S5: Tandem MS/MS spectrum of hydroxybenzoic acid -O-feruloyl-O-hexoside (peak 36) detected in L. siceraria fruit crude extract via HR-UPLC/MS/MS analysis in negative ionization mode; Figure S6: Tandem MS/MS spectrum of cynaroside A sesquiterpene (peak 39) detected in both L. siceraria & C. sativus fruit crude extracts via HR-UPLC/MS/MS analysis in negative ionization mode; Figure S7: Tandem MS/MS spectrum of butanol-O-pentosyl-hexoside (peak 44) detected in C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in negative ionization mode; Figure S8: Tandem MS/MS spectrum of iso/vitexin-O-couamroylhexoside (peak 54) detected in C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in positive ionization mode; Figure S9: Tandem MS/MS spectrum of isorhamnetin-O-malonylhexoside (peak 62) detected in L. siceraria & C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in positive ionization mode; Figure S10: Tandem MS/MS spectrum of hedysarimpterocarpene A-O-hexoside or HPA-O-hexoside (peak 72) detected in C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in negative ionization mode; Figure S11: Tandem MS/MS spectrum of pentahydroxy-dimethoxylignan (peak 75) detected in C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in positive ionization mode; Figure S12. Tandem MS/MS spectrum of soyasaponin I (peak 82) detected in C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in positive ionization mode; Figure S13: Tandem MS/MS spectrum of melilotussaponin O1(peak 81) detected as formate adducts in C. sativus fruit crude extract via HR-UPLC/MS/MS analysis in negative ionization mode and identified based on GNPS networking with soyasaponin I in cluster E; Figure S14: Principal component analysis (PCA) and orthogonal projection to latent structures-discriminant analysis (OPLS) supervised data analysis of modelling C. sativus and L. siceraria fruit specimens analyzed via SPME GC-MS for their volatile metabolites (n = 3). PCA score (A) and loading plot (B) with PC1 = 61% and PC2 = 22%; OPLS-DA score plot (C) and loading S-plot (D). Variables labelled with peak numbers (as in Table S1) correspond to discriminating metabolites for each sample identified by their Mol.wt/Rt. Benzaldhyde (peak 14), octanal (peak 15), benzenacetaldehyde (peak 17), nonadienal (peak 19), anethol ether (peak 83), fenchone (peak 67) and methyl hexadecanoate (peak 58) are the discriminating biomarkers; Figure S15: Representative GC-MS chromatograms of C. sativus and L. siceraria fruit specimens’ silylated primary metabolites. Assigned peak numbers follow those shown in Table 2; Figure S16: Principal component analysis (PCA) and orthogonal projection to latent structures-discriminant analysis (OPLS) supervised data analysis of modelling C. sativus and L. siceraria fruit specimens analyzed via GC-MS for their silylated primary metabolites (n = 3). PCA score (A) and loading plot (B) with PC1 = 73% and PC2 = 21%; OPLS-DA score plot (C) and loading S-plot (D). Variables labelled with their names follow those listed in Table 2.
Author Contributions
Conceptualization, M.A.F.; methodology, M.A.F.; investigation, M.A.F., R.H.E.-A. and M.G.S.E.-D.; writing—original draft preparation, R.H.E.-A. and M.G.S.E.-D.; writing—review and editing, M.A.F., R.H.E.-A. and M.G.S.E.-D.; supervision, M.A.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding, and The APC was funded by Mohamed A. Farag.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in article and supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rolnik, A.; Olas, B. Vegetables from the Cucurbitaceae family and their products: Positive effect on human health. Nutrition 2020, 78, 110788. [Google Scholar] [CrossRef]
- Saboo, S.S.; Thorat, P.K.; Tapadiya, G.G.; Khadabadi, S. Ancient and recent medicinal uses of cucurbitaceae family. Int. J. Ther. Appl. 2013, 9, 11–19. [Google Scholar]
- Mukherjee, P.K.; Nema, N.K.; Maity, N.; Sarkar, B.K. Phytochemical and therapeutic potential of cucumber. Fitoterapia 2013, 84, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Rajasree, R.; Sibi, P.; Francis, F.; William, H. Phytochemicals of Cucurbitaceae family—A review. Int. J. Pharmacogn. Phytochem. Res. 2016, 8, 113–123. [Google Scholar]
- Ahmed, D.; Fatima, M.; Saeed, S. Phenolic and flavonoid contents and anti-oxidative potential of epicarp and mesocarp of Lagenaria siceraria fruit: A comparative study. Asian Pac. J. Trop. Med. 2014, 7, S249–S255. [Google Scholar] [CrossRef]
- Kumar, A.; Partap, S.; Sharma, N.K. Phytochemical, ethnobotanical and pharmacological profile of Lagenaria siceraria: A review. J. Pharmacogn. Phytochem. 2012, 1, 24–31. [Google Scholar]
- Shah, B.; Seth, A.; Desai, R. Phytopharmacological profile of Lagenaria siceraria: A review. Asian J. Plant Sci. 2010, 9, 152–157. [Google Scholar] [CrossRef]
- Tyagi, N.T.N.; Sharma, G.N.S.G.N.; Shrivastava, B.S.B. Medicinal value of Lagenaria siceraria: An overview. Int. J. Indig. Herbs Drugs 2017, 2, 36–43. [Google Scholar]
- Hegazi, N.M.; Khattab, A.R.; Frolov, A.; Wessjohann, L.A.; Farag, M.A. Authentication of saffron spice accessions from its common substitutes via a multiplex approach of UV/VIS fingerprints and UPLC/MS using molecular networking and chemometrics. Food Chem. 2022, 367, 130739. [Google Scholar] [CrossRef]
- Abdel Shakour, Z.T.; El-Akad, R.H.; Elshamy, A.I.; El Gendy, A.E.-N.G.; Wessjohann, L.A.; Farag, M.A. Dissection of Moringa oleifera leaf metabolome in context of its different extracts, origin and in relationship to its biological effects as analysed using molecular networking and chemometrics. Food Chem. 2023, 399, 133948. [Google Scholar] [CrossRef]
- Sedeek, M.S.; Afifi, S.M.; Mansour, M.K.; Hassan, M.; Mehaya, F.M.; Naguib, I.A.; Abourehab, M.A.S.; Farag, M.A. Unveiling Antimicrobial and Antioxidant Compositional Differences between Dukkah and Za’atar via SPME-GCMS and HPLC-DAD. Molecules 2022, 27, 6471. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Ramadan, N.S.; Shorbagi, M.; Farag, N.; Gad, H.A. Profiling of Primary Metabolites and Volatiles in Apricot (Prunus armeniaca L.) Seed Kernels and Fruits in the Context of Its Different Cultivars and Soil Type as Analyzed Using Chemometric Tools. Foods 2022, 11, 1339. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- El-Akad, R.H.; Abou Zeid, A.H.; El-Rafie, H.M.; Kandil, Z.A.-A.; Farag, M.A. Comparative metabolites profiling of Caryota mitis & Caryota urens via UPLC/MS and isolation of two novel in silico chemopreventive flavonoids. J. Food Biochem. 2021, 45, e13648. [Google Scholar] [CrossRef]
- Yannai, S. Dictionary of Food Compounds with CD-ROM; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Foong, F.H.N.; Mohammad, A.; Ichwan, S.J.A. Biological properties of cucumber (Cucumis sativus L.) extracts. Malays. J. Anal. Sci. 2015, 19, 1218–1222. [Google Scholar]
- Uzuazokaro, M.-M.A.; Okwesili, F.C.N.; Chioma, A.A. Phytochemical and proximate composition of cucumber (Cucumis sativus) fruit from Nsukka, Nigeria. Afr. J. Biotechnol. 2018, 17, 1215–1219. [Google Scholar] [CrossRef]
- Bondia-Pons, I.; Martinez, J.A.; de la Iglesia, R.; Lopez-Legarrea, P.; Poutanen, K.; Hanhineva, K.; Zulet, M.d.l.Á. Effects of short-and long-term Mediterranean-based dietary treatment on plasma LC-QTOF/MS metabolic profiling of subjects with metabolic syndrome features: The Metabolic Syndrome Reduction in Navarra (RESMENA) randomized controlled trial. Mol. Nutr. Food Res. 2015, 59, 711–728. [Google Scholar] [CrossRef]
- Wahid, M.; Saqib, F.; Chicea, L.; Ahmedah, H.T.; Sajer, B.H.; Marc, R.A.; Pop, O.L.; Moga, M.; Gavris, C. Metabolomics analysis delineates the therapeutic effects of hydroethanolic extract of Cucumis sativus L. seeds on hypertension and isoproterenol-induced myocardial infarction. Biomed. Pharmacother. 2022, 148, 112704. [Google Scholar] [CrossRef] [PubMed]
- Essa, A.F.; Teleb, M.; El-Kersh, D.M.; El Gendy, A.E.-N.G.; Elshamy, A.I.; Farag, M.A. Natural acylated flavonoids: Their chemistry and biological merits in context to molecular docking studies. Phytochem. Rev. 2022, 1–40. [Google Scholar] [CrossRef]
- Montesano, D.; Rocchetti, G.; Putnik, P.; Lucini, L. Bioactive profile of pumpkin: An overview on terpenoids and their health-promoting properties. Curr. Opin. Food Sci. 2018, 22, 81–87. [Google Scholar] [CrossRef]
- Shao, H.; Xiao, M.; Zha, Z.; Olatunji, O.J. UHPLC-ESI-QTOF-MS2 analysis of Acacia pennata extract and its effects on glycemic indices, lipid profile, pancreatic and hepatorenal alterations in nicotinamide/streptozotocin-induced diabetic rats. Food Sci. Nutr. 2022, 10, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Ishihara, N.; Umehara, K.; Miyase, T.; Ueno, A. Sesquiterpene glycosides and saponins from Cynara cardunculus L. Chem. Pharm. Bull. 1988, 36, 2466–2474. [Google Scholar] [CrossRef]
- Rivera-Mondragón, A.; Tuenter, E.; Ortiz, O.; Sakavitsi, M.E.; Nikou, T.; Halabalaki, M.; Caballero-George, C.; Apers, S.; Pieters, L.; Foubert, K. UPLC-MS/MS-based molecular networking and NMR structural determination for the untargeted phytochemical characterization of the fruit of Crescentia cujete (Bignoniaceae). Phytochemistry 2020, 177, 112438. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Ohtani, K.; Kasai, R.; Yamasaki, K.; Nguyen, M.D. n-Alkyl glycosides and p-hydroxybenzoyloxy glucose from fruits of Crescentia cujete. Phytochemistry 1998, 47, 259–263. [Google Scholar] [CrossRef]
- Takayanagi, T.; Ishikawa, T.; Kitajima, J. Sesquiterpene lactone glucosides and alkyl glycosides from the fruit of cumin. Phytochemistry 2003, 63, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, J.; Ishikawa, T.; Tanaka, Y. Water-soluble constituents of fennel. I. Alkyl glycosides. Chem. Pharm. Bull. 1998, 46, 1643–1646. [Google Scholar] [CrossRef]
- Abou-Zaid, M.M.; Lombardo, D.A.; Kite, G.C.; Grayer, R.J.; Veitch, N.C. Acylated flavone C-glycosides from Cucumis sativus. Phytochemistry 2001, 58, 167–172. [Google Scholar] [CrossRef]
- Khan, A.L.; Hamayun, M.; Waqas, M.; Kang, S.-M.; Kim, Y.-H.; Kim, D.-H.; Lee, I.-J. Exophiala sp. LHL08 association gives heat stress tolerance by avoiding oxidative damage to cucumber plants. Biol. Fertil. Soils 2012, 48, 519–529. [Google Scholar] [CrossRef]
- Gangwal, A.; Parmar, S.; Sheth, N. Triterpenoid, flavonoids and sterols from Lagenaria siceraria fruit. Der Pharm. Lett. 2010, 2, 307–317. [Google Scholar]
- Fabre, N.; Rustan, I.; de Hoffmann, E.; Quetin-Leclercq, J. Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar] [CrossRef]
- Li, K.; Gao, C.; Li, W. Study on fragmentation of vitexin and isorhamnetin-3-o-beta-D-rutinoside using electrospray quadrupole time of flight mass spectrometry. Zhongguo Zhong Yao Za Zhi 2011, 36, 180–184. [Google Scholar]
- El-Kersh, D.M.; Abou El-Ezz, R.F.; Fouad, M.; Farag, M.A. Unveiling Natural and Semisynthetic Acylated Flavonoids: Chemistry and Biological Actions in the Context of Molecular Docking. Molecules 2022, 27, 5501. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, P.A.; Stephenson, K.K.; Wade, K.L.; Liu, H.; Fahey, J.W. Structure-activity analysis of flavonoids: Direct and indirect antioxidant, and antiinflammatory potencies and toxicities. Nutr. Cancer 2013, 65, 1014–1025. [Google Scholar] [CrossRef]
- Xie, H.; Wang, J.-R.; Yau, L.-F.; Liu, Y.; Liu, L.; Han, Q.-B.; Zhao, Z.; Jiang, Z.-H. Quantitative Analysis of the Flavonoid Glycosides and Terpene Trilactones in the Extract of Ginkgo biloba and Evaluation of Their Inhibitory Activity towards Fibril Formation of β-Amyloid Peptide. Molecules 2014, 19, 4466–4478. [Google Scholar] [CrossRef]
- Uchida, K.; Akashi, T.; Aoki, T. The Missing Link in Leguminous Pterocarpan Biosynthesis is a Dirigent Domain-Containing Protein with Isoflavanol Dehydratase Activity. Plant Cell Physiol. 2017, 58, 398–408. [Google Scholar] [CrossRef]
- Jiménez-González, L.; Álvarez-Corral, M.; Muñoz-Dorado, M.; Rodríguez-García, I. Pterocarpans: Interesting natural products with antifungal activity and other biological properties. Phytochem. Rev. 2008, 7, 125–154. [Google Scholar] [CrossRef]
- Yang, M.; Wang, W.; Sun, J.; Zhao, Y.; Liu, Y.; Liang, H.; Guo, D.A. Characterization of phenolic compounds in the crude extract of Hedysarum multijugum by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. Int. J. Devoted Rapid Dissem. Up Minute Res. Mass Spectrom. 2007, 21, 3833–3841. [Google Scholar] [CrossRef] [PubMed]
- Sicilia, T.; Niemeyer, H.B.; Honig, D.M.; Metzler, M. Identification and Stereochemical Characterization of Lignans in Flaxseed and Pumpkin Seeds. J. Agric. Food Chem. 2003, 51, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 4, D608–D617. [Google Scholar] [CrossRef]
- Subandi; Wijaya, M.; Sudarmo, T.P.B.; Suarsini, E. Saponin isolates from cucumber (Cucumis sativus L.) fruit mesocarp and their activity as pancreatic lipase inhibitor. In Proceedings of the AIP Conference Proceedings, Online, 17 October 2018; p. 070016. [Google Scholar]
- Daveby, Y.D.; Åman, P.; Betz, J.M.; Musser, S.M. Effect of storage and extraction on ratio of soyasaponin I to 2, 3-dihydro-2, 5-dihydroxy-6-methyl-4-pyrone-conjugated soyasaponin I in dehulled peas (Pisum sativum L.). J. Sci. Food Agric. 1998, 78, 141–146. [Google Scholar] [CrossRef]
- Al-Snafi, A.E. Chemical constituents and pharmacological effects of Melilotus Officinalis-A review. IOSR J. Pharm. 2020, 10, 26–36. [Google Scholar]
- Lu, X.; Zheng, Y.; Wen, F.; Huang, W.; Chen, X.; Ruan, S.; Gu, S.; Hu, Y.; Teng, Y.; Shu, P. Study of the active ingredients and mechanism of Sparganii rhizoma in gastric cancer based on HPLC-Q-TOF–MS/MS and network pharmacology. Sci. Rep. 2021, 11, 1905. [Google Scholar] [CrossRef]
- Garelnabi, M.; Litvinov, D.; Parthasarathy, S. Evaluation of a gas chromatography method for azelaic acid determination in selected biological samples. N. Am. J. Med. Sci. 2010, 2, 397. [Google Scholar] [CrossRef]
- Schieberle, P.; Ofner, S.; Grosch, W. Evaluation of potent odorants in cucumbers (Cucumis sativus) and muskmelons (Cucumis melo) by aroma extract dilution analysis. J. Food Sci. 1990, 55, 193–195. [Google Scholar] [CrossRef]
- Farrag, A.R.H.; Abdallah, H.M.I.; Khattab, A.R.; Elshamy, A.I.; Gendy, A.E.-N.G.E.; Mohamed, T.A.; Farag, M.A.; Efferth, T.; Hegazy, M.-E.F. Antiulcer activity of Cyperus alternifolius in relation to its UPLC-MS metabolite fingerprint: A mechanistic study. Phytomedicine 2019, 62, 152970. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Maroto, M.C.; Díaz-Maroto Hidalgo, I.J.; Sánchez-Palomo, E.; Pérez-Coello, M.S. Volatile components and key odorants of Fennel (Foeniculum vulgare Mill.) and Thyme (Thymus vulgaris L.) oil extracts obtained by simultaneous distillation-extraction and supercritical fluid extraction. J. Agric. Food Chem. 2005, 53, 5385–5389. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Qian, M.C. Identification of aroma compounds in Chinese ‘Yanghe Daqu’ liquor by normal phase chromatography fractionation followed by gas chromatography [sol] olfactometry. Flavour Fragr. J. 2006, 21, 333–342. [Google Scholar] [CrossRef]
- Kula, J.; Sadowska, H. Unsaturated aliphatic C9-aldehydes as natural flavorants. Perfum. Flavorist 1993, 18, 23–25. [Google Scholar]
- Maamoun, A.A.; El-Akkad, R.H.; Farag, M.A. Mapping metabolome changes in Luffa aegyptiaca Mill fruits at different maturation stages via MS-based metabolomics and chemometrics. J. Adv. Res. 2021, 29, 179–189. [Google Scholar] [CrossRef]
- Park, S.-N.; Lim, Y.K.; Freire, M.O.; Cho, E.; Jin, D.; Kook, J.-K. Antimicrobial effect of linalool and α-terpineol against periodontopathic and cariogenic bacteria. Anaerobe 2012, 18, 369–372. [Google Scholar] [CrossRef]
- Clements, R.; Darnell, B. Myo-inositol content of common foods: Development of a high-myo-inositol diet. Am. J. Clin. Nutr. 1980, 33, 1954–1967. [Google Scholar] [CrossRef] [PubMed]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef]
- Nair, M.K.; Joy, J.; Vasudevan, P.; Hinckley, L.; Hoagland, T.A.; Venkitanarayanan, K.S. Antibacterial effect of caprylic acid and monocaprylin on major bacterial mastitis pathogens. J. Dairy Sci. 2005, 88, 3488–3495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xue, C.; Xu, Q.; Zhang, Y.; Li, H.; Li, F.; Liu, Y.; Guo, C. Caprylic acid suppresses inflammation via TLR4/NF-κB signaling and improves atherosclerosis in ApoE-deficient mice. Nutr. Metab. 2019, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Gómez-García, M.; Sol, C.; de Nova, P.J.G.; Puyalto, M.; Mesas, L.; Puente, H.; Mencía-Ares, Ó.; Miranda, R.; Argüello, H.; Rubio, P.; et al. Antimicrobial activity of a selection of organic acids, their salts and essential oils against swine enteropathogenic bacteria. Porc. Health Manag. 2019, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Chaudhery, R.; Ahmed, D.; Liaqat, I.; Dar, P.; Shaban, M. Study of bioactivities of lipid content of fresh Lagenaria siceraria seeds pulp and identification of its chemical constituents. J. Med. Plant Res. 2014, 8, 1014–1020. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).