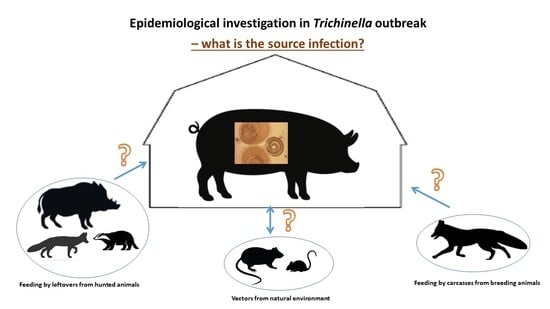
Scheme of Effective Epidemiological Investigations in Trichinella Outbreaks on Pig Farms
Abstract
:1. Introduction
1.1. Trichinellosis Infections in Europe
1.2. Trichinella in Pigs in EU
1.3. Epidemiologic Epidemiological Investigation on Farms with Trichinella Infected Pigs
1.3.1. Epidemiological Interview
1.3.2. Serological Investigations
1.3.3. Removing Other Infected Pigs from Farms
1.3.4. Species Identification of Detected Larvae
1.3.5. Differentiation of Isolates of the Same Species Using Available Molecular Epidemiology Tools
2. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dupouy-Camet, J. Trichinellosis: A worldwide zoonosis. Vet. Parasitol. 2000, 93, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Pozio, E. Trichinellosis in the European Union: Epidemiology, Ecology and Economic Impact. Parasitol. Today 1998, 14, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Webster, P.; Kapel, C.M.O. Studies on vertical transmission of Trichinella spp. in experimentally infected ferrets (Mustela putorius furo), foxes (Vulpes vulpes), pigs, guinea pigs and mice. Vet. Parasitol. 2005, 130, 255–262. [Google Scholar] [CrossRef]
- Pozio, E. The broad spectrum of Trichinella hosts: From cold- to warm-blooded animals. Vet. Parasitol. 2005, 132, 3–11. [Google Scholar] [CrossRef]
- Pozio, E.; Darwin Murrell, K. Systematics and Epidemiology of Trichinella. In Advances in Parasitology; Baker, J.R., Muller, R., Rollinson, D., Eds.; Academic Press: Cambridge, MA, USA, 2006; Volume 63, pp. 367–439. [Google Scholar]
- Pozio, E. World distribution of Trichinella spp. infections in animals and humans. Vet. Parasitol. 2007, 149, 3–21. [Google Scholar] [CrossRef]
- Pozio, E.; Rinaldi, L.; Marucci, G.; Musella, V.; Galati, F.; Cringoli, G.; Boireau, P.; La Rossa, G. Hosts and habitats of Trichinella spiralis and Trichinella britovi in Europe. Int. J. Parasitol. 2009, 39, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Vasilev, S.; Mitic, I.; Mirilovic, M.; Plavsa, D.; Milakara, E.; Plavsic, B.; Sofronic-Milosavljevic, L. Trichinella infectiob in Serbia from 2011 to 2020: A success story in the field of One Health. Epidemiol. Infect. 2023, 151, E20. [Google Scholar] [CrossRef] [PubMed]
- Różycki, M.; Korpysa-Dzirba, W.; Bełcik, A.; Pelec, T.; Mazurek, J.; Cencek, T. Analysis of a Trichinellosis Outbreak in Poland after Consumption of Sausage Made of Wild Boar Meat. J. Clin. Med. 2022, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Fichi, G.; Stefanelli, S.; Pagani, A.; Luchi, S.; De Gennaro, M.; Gómez-Morales, M.A.; Selmi, M.; Rovai, D.; Mari, M.; Fischetti, R.; et al. Trichinellosis outbreak caused by meat from a wild boar hunted in an italian region considered to be at negligible risk for Trichinella. Zoonoses Public Health 2015, 62, 285–291. [Google Scholar] [CrossRef]
- Gari-Toussaint, M.; Tieulié, N.; Baldin, J.L.; Dupouy-Camet, J.; Delaunay, P.; Fuzibet, J.G.; Le Fichoux, Y.; Pozio, E.; Marty, P. Human trichinellosis due to Trichinella britovi in southern France after consumption of frozen wild boar meat. Eurosurveill 2005, 10, 11–12. [Google Scholar] [CrossRef]
- Fosse, J.; Seegers, H.; Magras, C. Foodborne zoonoses due to meat: A quantitative approach for a comparative risk assessment applied to pig slaughtering in Europe. Vet. Res. 2008, 39, 1. [Google Scholar] [CrossRef] [Green Version]
- Pozio, E. Factors affecting the flow among domestic, synanthropic and sylvatic cycles of Trichinella. Vet. Parasitol. 2000, 93, 241–262. [Google Scholar] [CrossRef]
- Commission Implementing Regulation (EU) 2020/1478 of 14 October 2020 Amending Implementing Regulation (EU) 2015/1375 as Regards Sampling, the Reference Method for Detection and Import Conditions Related to Trichinella Control L338/7-L338/9. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32020R1478&from=EN (accessed on 8 February 2023).
- Directive 2003/99/EC of the European Parliament and of the Council of 17 November 2003 on the Monitoring of Zoonoses and Zoonotic Agents, Amending Council Decision 90/424/EEC and Repealing Council Directive 92/117/EEC, L325/31-L325/40. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2003:325:0031:0040:en:PDF (accessed on 8 February 2023).
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar] [CrossRef] [Green Version]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015, 13, 3991. [Google Scholar] [CrossRef] [Green Version]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015, 13, 4329. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016, 16, 4634. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef] [Green Version]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 5926. [Google Scholar] [CrossRef] [Green Version]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, 6406. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union One Health 2020 Zoonoses. EFSA J. 2021, 19, 6971. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention Control. Trichinellosis. In ECDC. Annual Epidemiological Report for 2020; ECDC: Stockholm, Sweden, 2022. [Google Scholar]
- Pannwitz, G.; Mayer-Scholl, A.; Balicka-Ramisz, A.; Nöckler, K. Increased prevalence of Trichinella spp., northeastern Germany, 2008. Emerg. Infect. Dis. 2010, 16, 936–942. [Google Scholar] [CrossRef]
- ISO 18743; Microbiology of the Food Chain—Detection of Trichinella Larvae in Meat by Artificial Digestion Method. ISO: Geneva, Switzerland, 2015.
- EFSA (European Food Safety Authority); ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, 7666. [Google Scholar]
- Marquer, P.; Rabade, T.; Forti, R. Pig farming in the European Union: Considerable variations from one Member State to another. Stat. Focus 2014, 15, 1–12. [Google Scholar]
- EFSA (European Food Safety Authority). Scientific Opinion on the public health hazards to be covered by inspection of meat (swine). EFSA J. 2011, 10, 2351. [Google Scholar] [CrossRef] [Green Version]
- Regulation (EU) 2017/625 of the European Parliament and of the Council of 15 March 2017 on Official Controls and Other Official Activities Performed to Ensure the Application of Food and Feed Law, Rules on Animal Health and Welfare, Plant Health and Plant Protection Products, Amending Regulations (EC) No 999/2001, (EC) No 396/2005, (EC) No 1069/2009, (EC) No 1107/2009, (EU) No 1151/2012, (EU) No 652/2014, (EU) 2016/429 and (EU) 2016/2031 of the European Parliament and of the Council, Council Regulations (EC) No 1/2005 and (EC) No 1099/2009 and Council Directives 98/58/EC, 1999/74/EC, 2007/43/EC, 2008/119/EC and 2008/120/EC, and Repealing Regulations (EC) No 854/2004 and (EC) No 882/2004 of the European Parliament and of the Council, Council Directives 89/608/EEC, 89/662/EEC, 90/425/EEC, 91/496/EEC, 96/23/EC, 96/93/EC and 97/78/EC and Council Decision 92/438/EEC (Official Controls Regulation). Off. J. Eur. Union 2017, 7, 1–142.
- Ustawa z dnia 29 stycznia 2004 r. o Inspekcji Weterynaryjnej, Dz.U. 2004 nr 33 poz. 287.2004. Available online: https://isap.sejm.gov.pl/isap.nsf/DocDetails.xsp?id=WDU20040330287 (accessed on 8 February 2023).
- Pozio, E. Searching for Trichinella: Not all pigs are created equal. Trends Parasitol. 2014, 30, 4–11. [Google Scholar] [CrossRef]
- Pozio, E. Trichinella pseudospiralis an elusive nematode. Vet. Parasitol. 2016, 231, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Nockler, K.; Serrano, F.J.; Boireau, P.; Kapel, C.M.O.; Pozio, E. Experimental studies in pigs on Trichinella detection in different diagnostic matrices. Vet. Parasitol. 2005, 132, 85–90. [Google Scholar] [CrossRef]
- Bilska-Zając, E.; Różycki, M.; Korpysa-Dzirba, W.; Bełcik, A.; Ziętek-Barszcz, A.; Włodarczyk-Ramus, M.; Gontarczyk, A.; Cencek, T. Trichinella outbreaks on pig farms in Poland in 2012–2020. Pathogens 2021, 10, 1504. [Google Scholar] [CrossRef]
- Bliska-Zajac, E.; Różycki, M.; Antolak, E.; Belcik, A.; Gradziel-Krukowska, K.; Karamon, J.; Sroka, J.; Zdybel, J.; Cencek, T. Occurrence of Trichinella spp. in rats on pig farms. Ann. Agric. Environ. Med. 2018, 25, 4. [Google Scholar] [CrossRef] [PubMed]
- Stojcevic, D.; Zivicnjak, T.; Marinculic, A.; Marucci, G.; Andelko, G.; Brstilo, M.; Pavo, L.; Pozio, E. The epidemiological investigation of Trichinella infection in brown rats (Rattus norvegicus) and domestic pigs in Croatia suggests that rats are not a reservoir at the farm level. J. Parasitol. 2004, 90, 666–670. [Google Scholar] [CrossRef]
- Bilska-Zając, E.; Franssen, F.; Różycki, M.; Swart, A.; Karamon, J.; Sroka, J.; Zdybel, J.; Ziętek-Barszacz, A.; Cencek, T. Intraspecific genetic variation in Trichinella spiralis and Trichinella britovi populations circulating in different geographical regions of Poland. Int. J. Parasitol Parasites Wildl. 2019, 10, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Leiby, D.A.; Duffy, C.H.; Murrell, K.D.; Schad, G.A. Trichinella spiralis in an agricultural ecosystem: Transmission in the rat population. J. Parasitol. 1990, 76, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Bandi, C.; La Rosa, G.; Comincini, S.; Damiani, G.; Pozio, E. Random amplified polymorphic DNA technique for the identification of Trichinella species. Parasitology 1993, 107, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Gondek, M.; Bień, J.; Nowakowski, Z. Use of ELISA and Western blot for serological detection of antibodies to E-S antigens of Trichinella spiralis muscle larvae in sera of swine experimentally infected with Trichinella spiralis. Vet. Immunol. Immunopathol. 2018, 203, 13–20. [Google Scholar] [CrossRef]
- Kořínková, K.; Kovařčík, K.; Pavlíčková, Z.; Svoboda, M.; Koudela, B. Serological detection of Trichinella spiralis in swine by ELISA (enzyme-linked immunosorbent assay) using an excretory–secretory (E/S) antigen. Parasitol. Res. 2008, 102, 1317–1320. [Google Scholar] [CrossRef]
- Smith, H.J. Evaluation of the ELISA for the serological diagnosis of trichinosis in Canadian swine. Can. J. Vet. Res. 1987, 51, 194–197. [Google Scholar]
- Zarlenga, D.S.; Chute, M.B.; Martin, A.; Kapel, C.M.O. A multiplex PCR for unequivocal differentiation of all encapsulated and non-encapsulated genotypes of Trichinella. Int. J. Parasitol. 1999, 29, 1859–1867. [Google Scholar] [CrossRef]
- Sun, G.-G.; Wang, Z.-Q.; Liu, C.-Y.; Jiang, P.; Liu, R.-D.; Wen, H.; Qi, X.; Wang, L.; Cui, J. Early serodiagnosis of trichinellosis by ELISA using excretory–secretory antigens of Trichinella spiralis adult worms. Parasites Vectors 2015, 8, 484. [Google Scholar] [CrossRef] [Green Version]
- Gamble, H.R.; Anderson, W.R.; Graham, C.E.; Murrell, K.D. Diagnosis of swine trichinosis by enzyme-linked immunosorbent assay (ELISA) using an excretory-secretory antigen. Vet. Parasitol. 1983, 13, 349–361. [Google Scholar] [CrossRef]
- Kociecka, W.; Bruschi, F.; Marini, C.; Mrozewicz, B.; Pielok, L. Clinical appraisal of patients and detection of serum antibodies by ELISA and CIA tests in late periods of Trichinella sp. invasion. Parasite 2001, 8, S147–S151. [Google Scholar] [CrossRef] [Green Version]
- Gondek, M.; Bien, J.; Nowakowski, Z. Detection of experimental swine trichinellosis using commercial ELISA test. Pol. J. Vet. Sci. 2017, 20, 445–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottstein, B.; Pozio, E.; Nöckler, K. Epidemiology, diagnosis, treatment, and control of Trichinellosis. Clin. Microbiol. Rev. 2009, 22, 127–145. [Google Scholar] [CrossRef] [Green Version]
- Nöckler, K.; Reckinger, S.; Broglia, A.; Mayer-Scholl, A.; Bahn, P. Evaluation of a Western Blot and ELISA for the detection of anti-Trichinella-IgG in pig sera. Vet. Parasitol. 2009, 163, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Gondek, M.; Grzelak, S.; Pyz-Łukasik, R.; Knysz, P.; Ziomek, M.; Bień-Kalinowska, J. Insight into Trichinella britovi infection in pigs: Effect of various infectious doses on larvae density and spatial larvae distribution in carcasses and comparison of the detection of Anti-T. britovi IgG of Three Different Commercial ELISA Tests and Immunoblot Assay. Pathogens 2022, 11, 735. [Google Scholar] [CrossRef] [PubMed]
- Pozio, E.; Merialdi, G.; Licata, E.; Della Casa, G.; Fabiani, M.; Amati, M.; Cherchi, S.; Ramini, M.; Faeti, V.; Interisano, M.; et al. Differences in larval survival and IgG response patterns in long-lasting infections by Trichinella spiralis, Trichinella britovi and Trichinella pseudospiralis in pigs. Parasites Vectors 2020, 13, 520. [Google Scholar] [CrossRef]
- van der Leek, M.L.; Dame, J.B.; Adams, C.L.; Gillis, K.D.; Littell, R.C. Evaluation of an enzyme-linked immunosorbent assay for diagnosis of trichinellosis in swine. Am. J. Vet. Res. 1992, 53, 877–882. [Google Scholar] [PubMed]
- Gamble, H.R. Detection of Trichinellosis in pigs by artificial digestion and Enzyme Immunoassay. J. Food Prot. 1996, 59, 295–298. [Google Scholar] [CrossRef]
- Gamble, H.R.; Solomon, M.B.; Long, J.B. Effects of Hydrodynamic Pressure on the viability of Trichinella spiralis in pork. J. Food Prot. 1998, 61, 637–639. [Google Scholar] [CrossRef]
- Kapel, C.M.O.; Webster, P.; Lind, P.; Pozio, E.; Henriksen, S.A.; Murrell, K.D.; Nansen, P. Trichinella spiralis, T. britovi, and T. nativa: Infectivity, larval distribution in muscle, and antibody response after experimental infection of pigs. Parasitol. Res. 1998, 84, 264–271. [Google Scholar] [CrossRef]
- Ljungström, I.; Hammarström, L.; Kociecka, W.; Smith, C.I. The sequential appearance of IgG subclasses and IgE during the course of Trichinella spiralis infection. Clin. Exp. Immunol. 1988, 74, 230–235. [Google Scholar]
- Reiterová, K.; Dubinsky, P.; Tomasovicova, O.; Dvoroznakova, E.; Klimenko, V.V. Comparison of Trichinella spiralis larva antigens for the detection of specific antibodies in pigs. Vet. Med-Czech 1999, 44, 1–5. [Google Scholar]
- Zarlenga, D.; Thompson, P.; Pozio, E. Trichinella species and genotypes. Res. Vet. Sci. 2020, 133, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Pozio, E.; Hoberg, E.; La Rosa, G.; Zarlenga, D.S. Molecular taxonomy, phylogeny and biogeography of nematodes belonging to the Trichinella genus. Infect. Genet. Evol. 2009, 9, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Pozio, E.; Zarlenga, D.S. Recent advances on the taxonomy, systematics and epidemiology of Trichinella. Int. J. Parasitol. 2005, 35, 1191–1204. [Google Scholar] [CrossRef]
- Yang, Y.-C.; Li, X.-M.; Yi, O.-Y.; Xie, Z.-Y. Study on 5S rDNA sequence of two isolates of Trichinella from Guangxi. Chin. J. Parasitol. Parasit. Dis. 2007, 25, 222–224. [Google Scholar]
- Wang, H.; Zhang, Y.; Lao, W.; Wu, Z. Restriction fragment length polymorphism (RFLP) analysis of genomic DNA of 5 strains of Trichinella spiralis in China. Chin. Med. Sci. J. 1995, 10, 131–135. [Google Scholar]
- Mayer-Scholl, A.; Broglia, A.; Reckinger, S.; Nöckler, K. Polymerase chain reaction—Restriction fragment length polymorphism analysis for the differentiation of Trichinella nativa and Trichinella britovi. Vet. Parasitol. 2014, 203, 247–249. [Google Scholar] [CrossRef]
- Perteguer, M.J.; Rodríguez, E.; García-Sánchez, R.N.; Nogal-Ruiz, J.J.; Bolas-Fernández, F.; Martínez-Fernández, A.R.; Garate, T. Identification of Spanish Trichinella isolates by ISSR-PCR: Intra-specific variability of Trichinella britovi. Vet. Parasitol. 2009, 159, 206–209. [Google Scholar] [CrossRef]
- Karadjian, G.; Bilska-Zając, E.; Bahn, P.; Py, J.-S.; Johne, A.; Gassilloud, B.; Rózycki, M.; Cencek, T.; Mayer-Scholl, A.; Vallée, I. Species identification of Trichinella originated from various host and different geographical location by MALDI-TOF. Exp. Parasitol. 2020, 213, 107890. [Google Scholar] [CrossRef] [PubMed]
- Järvis, T.; Miller, I.; Pozio, E. Trichinella britoviin Domestic Pig—As Case Report. Acta Vet. Scand. 2002, 43, 131. [Google Scholar] [CrossRef]
- Malakauskas, A.; Paulauskas, V.; Järvis, T.; Keidans, P.; Eddi, C.; Kapel, C.M.O. Molecular epidemiology of Trichinella spp. in three Baltic countries: Lithuania, Latvia, and Estonia. Parasitol. Res. 2007, 100, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Blanco, M.; García-Cabañas, A.; Guerra-Peguero, F.; Ramos-Aceitero, J.M.; Herrera-Guibert, D.; Martínez-Navarro, J.F. Outbreak of trichinellosis in Cáceres, Spain, December 2001–February 2002. Eurosurveillance 2002, 7, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Garcia, V.; Hernandez-Quero, J.; Rodriguez-Osorio, M. Short report: Human infection with Trichinella britovi in Granada, Spain. Am. J. Trop. Med. Hyg. 2003, 68, 463–464. [Google Scholar] [CrossRef] [PubMed]
- Dubinský, P.; Antolová, D.; Reiterová, K. Human Trichinella infection outbreaks in Slovakia, 1980–2008. Acta Parasitol. 2016, 61, 205–211. [Google Scholar] [CrossRef]
- Pozio, E.; Cossu, P.; Marucci, G.; Amati, M.; Ludovisi, A.; Morales, M.A.; La Rossa, G.; Firinu, T. The birth of a Trichinella britovi focus on the Mediterranean island of Sardinia (Italy). Vet. Parasitol. 2009, 159, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Beck, R.; Beck, A.; Lučinger, S.; Florijančić, T.; Bošković, I.; Marinculić, A. Trichinella pseudospiralis in pig from Croatia. Vet. Parasitol. 2009, 159, 304–307. [Google Scholar] [CrossRef]
- La Rosa, G.; Marucci, G.; Rosenthal, B.M.; Pozio, E. Development of a single larva microsatellite analysis to investigate the population structure of Trichinella spiralis. Infect. Genet. Evol. 2012, 12, 369–376. [Google Scholar] [CrossRef]
- La Rosa, G.; Vallée, I.; Marucci, G.; Casabianca, F.; Bandino, E.; Galati, F.; Boireau, P.; Pozio, E. Multilocus genotype analysis outlines distinct histories for Trichinella britovi in the neighboring Mediterranean islands of Corsica and Sardinia. Parasites Vectors 2018, 11, 353. [Google Scholar] [CrossRef] [Green Version]
- Bilska-Zajac, E.; Tonanzi, D.; Pozio, E.; Rozycki, M.; Cencek, T.; Thompson, P.C.; Rosenthal, B.M.; La Rossa, G. Genetic evidence substantiates transmission of Trichinella spiralis from one swine farm to another. Parasites Vectors 2021, 14, 359. [Google Scholar] [CrossRef]
- Bilska-Zając, E.; Rosenthal, B.; Thompson, P. Trich-tracker—A practical tool to trace Trichinella spiralis transmission based on rapid, cost-effective sampling of genome-wide genetic variation. Int. J. Parasitol. 2022, 52, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Bohling, J.; Small, M.; Von Bargen, J.; Louden, A.; DeHaan, P. Comparing inferences derived from microsatellite and RADseq datasets: A case study involving threatened bull trout. Conserv. Genet. 2019, 20, 329–342. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, D.B. Microsatellites. In Evolution and Application; Goldstein, D.B., Schlötterer, C., Eds.; Oxford University Press: Oxford, UK, 1999; pp. 1–368. [Google Scholar]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 2016, 17, 81–92. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.X.; Hewitt, G.M. Nuclear DNA analyses in genetic studies of populations: Practice, problems and prospects. Mol. Ecol. 2003, 12, 563–584. [Google Scholar] [CrossRef]
- Sunde, J.; Yıldırım, Y.; Tibblin, P.; Forsman, A. Comparing the performance of microsatellites and RADseq in population genetic studies: Analysis of data for pike (Esox lucius) and a synthesis of previous studies. Front. Genet. 2020, 11, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Year | 2017 | 2018 | 2019 | 2020 | 2021 |
|---|---|---|---|---|---|
| Total number of confirmed cases | 168 | 66 | 97 | 117 | 77 |
| Total number of confirmed cases/100,000 population (notification rates) | 0.03 | 0.01 | 0.02 | 0.03 | 0.02 |
| Number of reporting countries | 27 | 27 | 27 | 26 | 26 |
| Infections acquired in the EU | 81 | 18 | 26 | 99 | 29 |
| Infections acquired outside the EU | 2 | 1 | 2 | 2 | 2 |
| Unknown travel status or unknown country of infection | 85 | 47 | 69 | 16 | 46 |
| Number of outbreak-related cases | 199 | 114 | 44 | 119 | 2 |
| Total number of outbreaks | 11 | 10 | 5 | 6 | 1 |
| Year | 2017 | 2018 | 2019 | 2020 | 2021 |
|---|---|---|---|---|---|
| Domestic pigs raised under control housing condition | |||||
| Number of tested samples | 55,177,802 | 55,989,292 | 73,633,900 | 77,794,786 | 72,227,074 |
| % of positive samples (N) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Domestic pigs not raised under control housing condition | |||||
| Number of tested samples | 124,689,434 | 152,922,322 | 145,213,445 | 139,637,631 | 161,129,635 |
| % of positive samples (N) | 0.0002 (224) | 0.0003 (384) | 0.00015 (219) | 0.0001 (179) | 0.0001 (120) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bilska-Zając, E.; Korpysa-Dzirba, W.; Bełcik, A.; Karamon, J.; Sroka, J.; Cencek, T. Scheme of Effective Epidemiological Investigations in Trichinella Outbreaks on Pig Farms. Foods 2023, 12, 1320. https://doi.org/10.3390/foods12061320
Bilska-Zając E, Korpysa-Dzirba W, Bełcik A, Karamon J, Sroka J, Cencek T. Scheme of Effective Epidemiological Investigations in Trichinella Outbreaks on Pig Farms. Foods. 2023; 12(6):1320. https://doi.org/10.3390/foods12061320
Chicago/Turabian StyleBilska-Zając, Ewa, Weronika Korpysa-Dzirba, Aneta Bełcik, Jacek Karamon, Jacek Sroka, and Tomasz Cencek. 2023. "Scheme of Effective Epidemiological Investigations in Trichinella Outbreaks on Pig Farms" Foods 12, no. 6: 1320. https://doi.org/10.3390/foods12061320
APA StyleBilska-Zając, E., Korpysa-Dzirba, W., Bełcik, A., Karamon, J., Sroka, J., & Cencek, T. (2023). Scheme of Effective Epidemiological Investigations in Trichinella Outbreaks on Pig Farms. Foods, 12(6), 1320. https://doi.org/10.3390/foods12061320