Phenotypic Traits and Probiotic Functions of Lactiplantibacillus plantarum Y42 in Planktonic and Biofilm Forms
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and HT-29 Cell
2.2. Preparation of Planktonic and Biofilm Strains
2.3. Growth Curves and Biofilm Formation of L. plantarum Y42
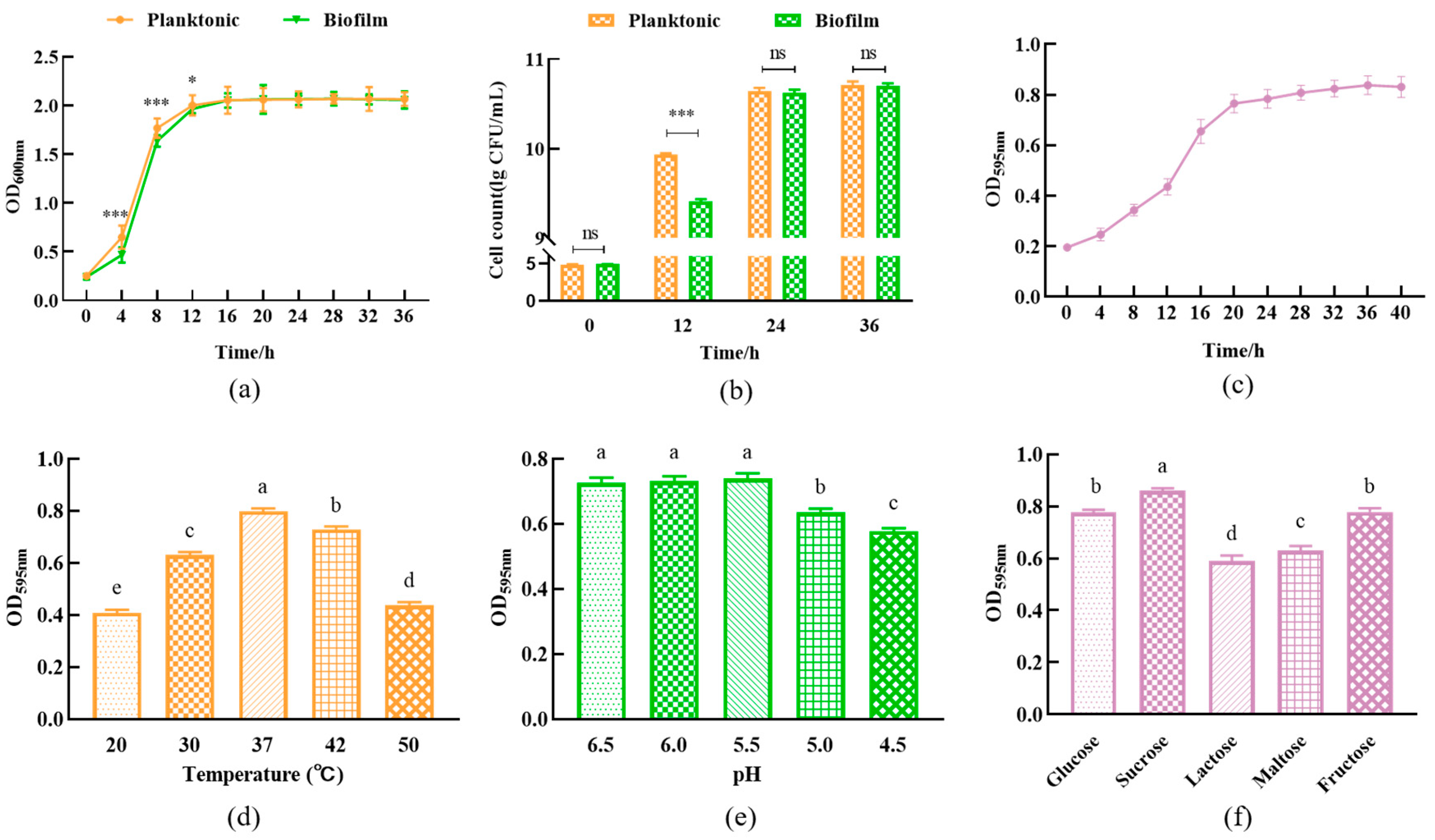
2.4. Factors Affecting the Formation of L. plantarum Y42 Biofilms
2.5. Morphological Observations of Planktonic and Biofilm L. plantarum Y42
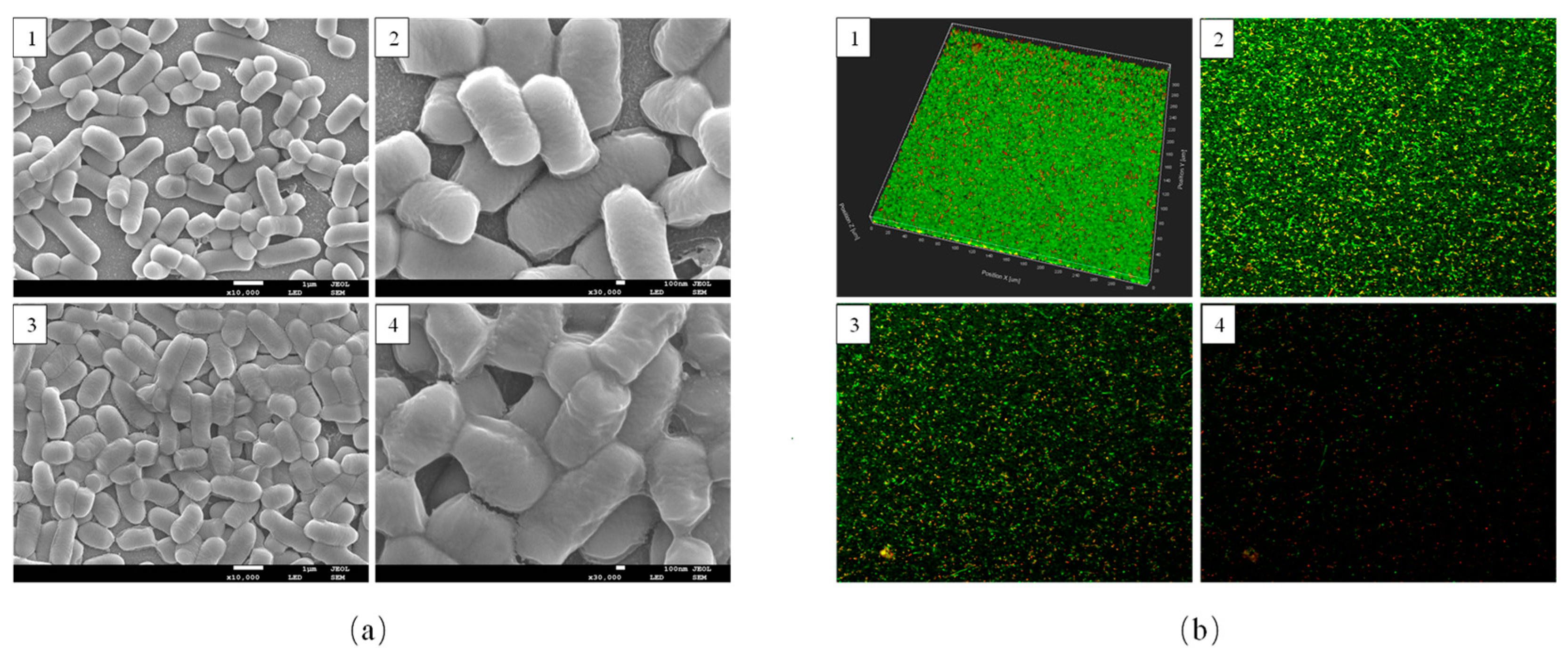
2.5.1. Field-Emission Scanning Electron Microscopy (SEM)
2.5.2. Confocal Laser Scanning Microscopy (CLSM)
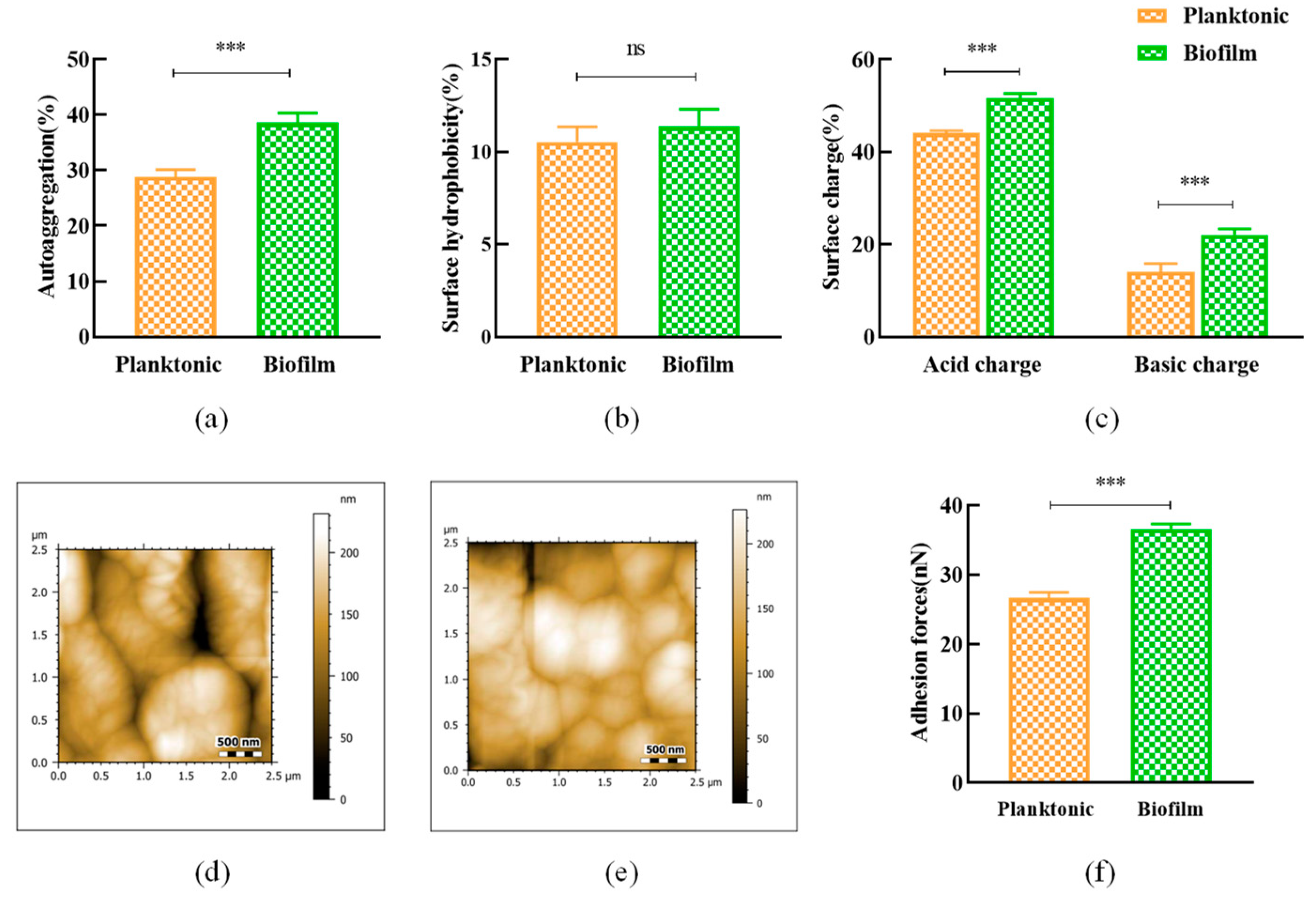
2.6. Surface Properties of Planktonic and Biofilm L. plantarum Y42
2.7. Measurement of Environmental Stress Tolerance
2.8. Tolerance of Gastro-Intestinal Fluid
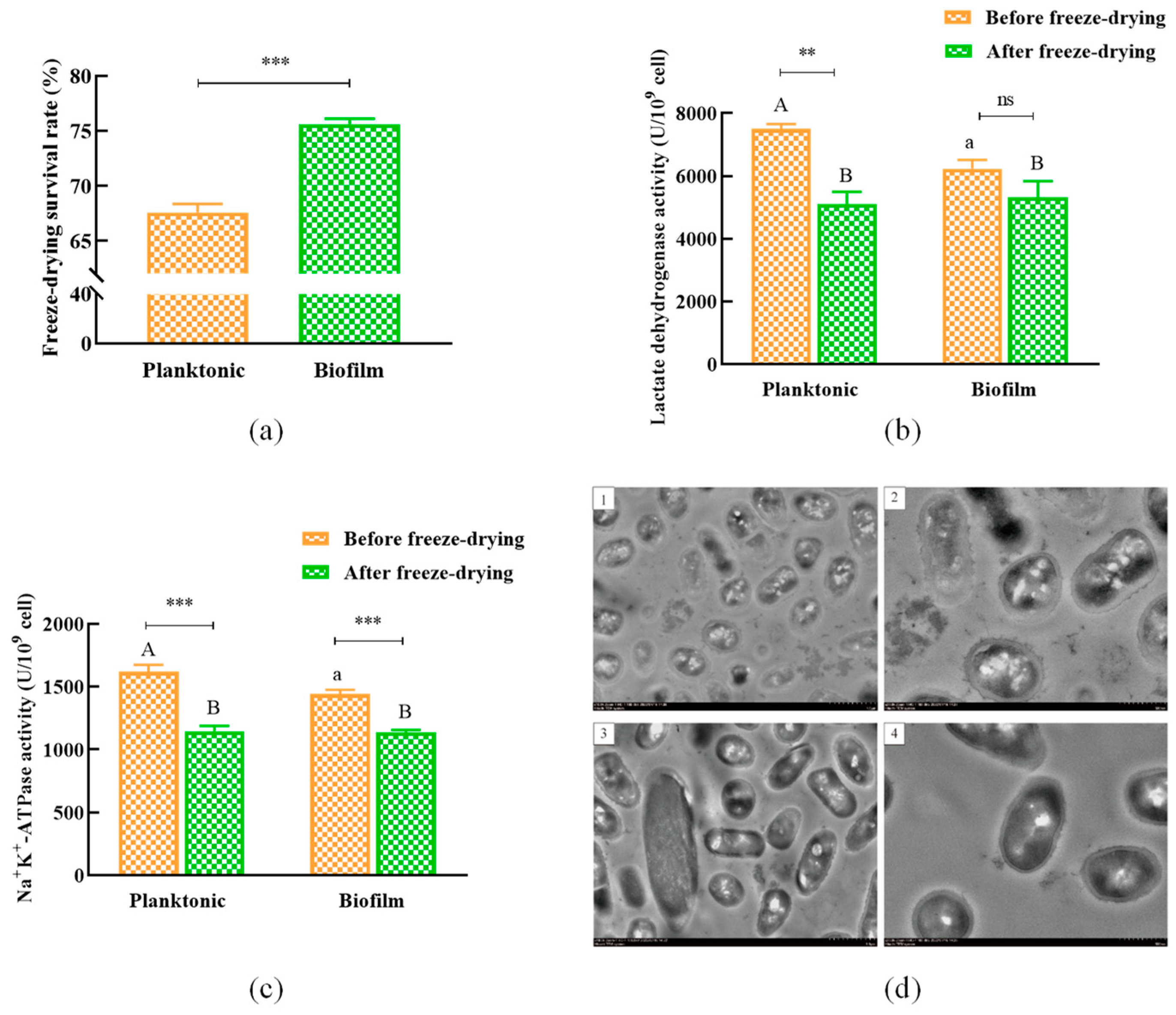
2.9. Effects of Vacuum Lyophilization on the Planktonic and Biofilm L. plantarum Y42
2.10. Adhesion of Planktonic and Biofilm L. plantarum Y42 on Human Enterocyte HT-29
2.10.1. Adhesion Ability
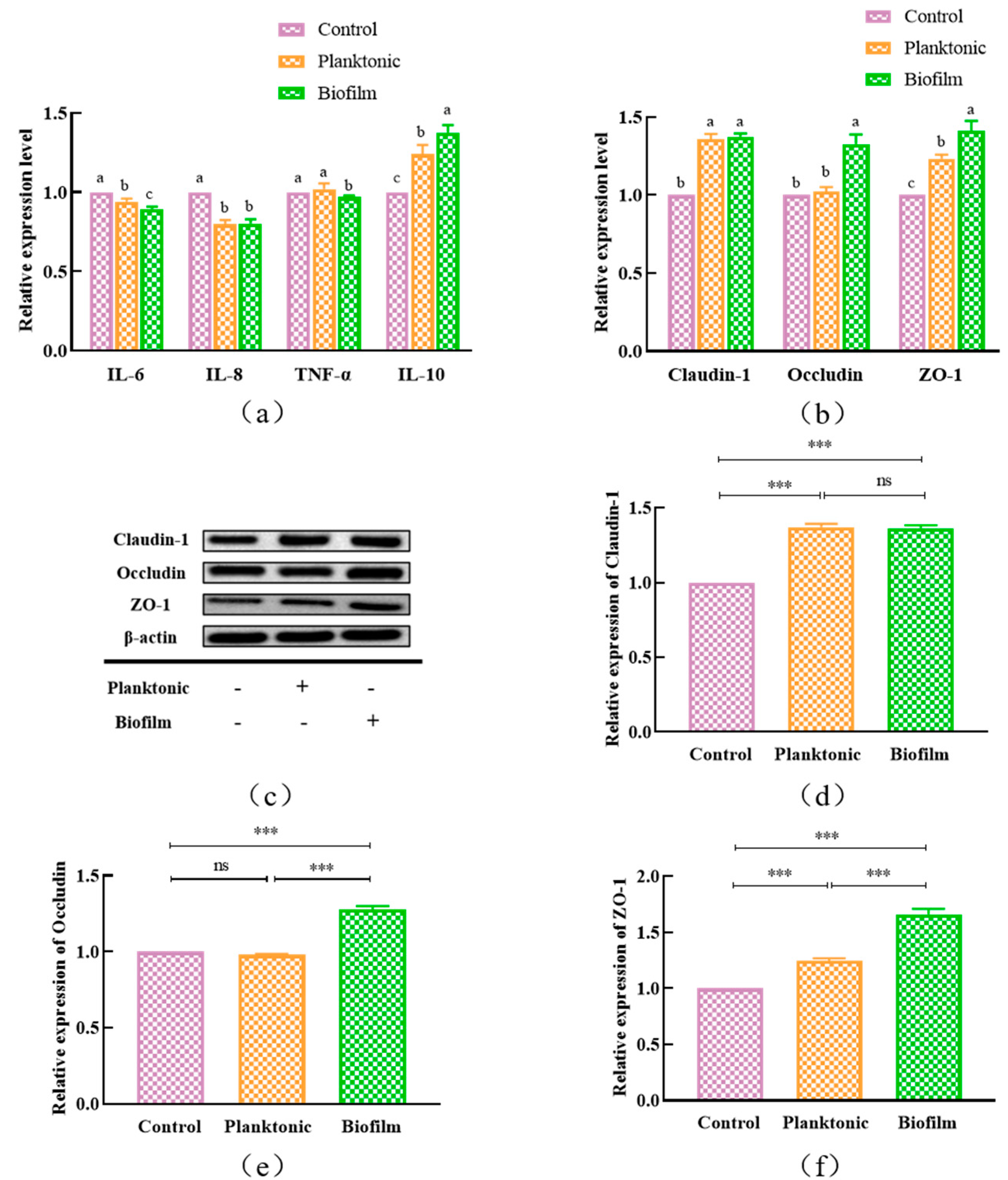
2.10.2. Secretion of Inflammatory Factors
2.10.3. Expressions of Tight Junction Proteins
2.11. Effect of Planktonic and Biofilm L. plantarum Y42 on L. monocytogenes ATCC 19115
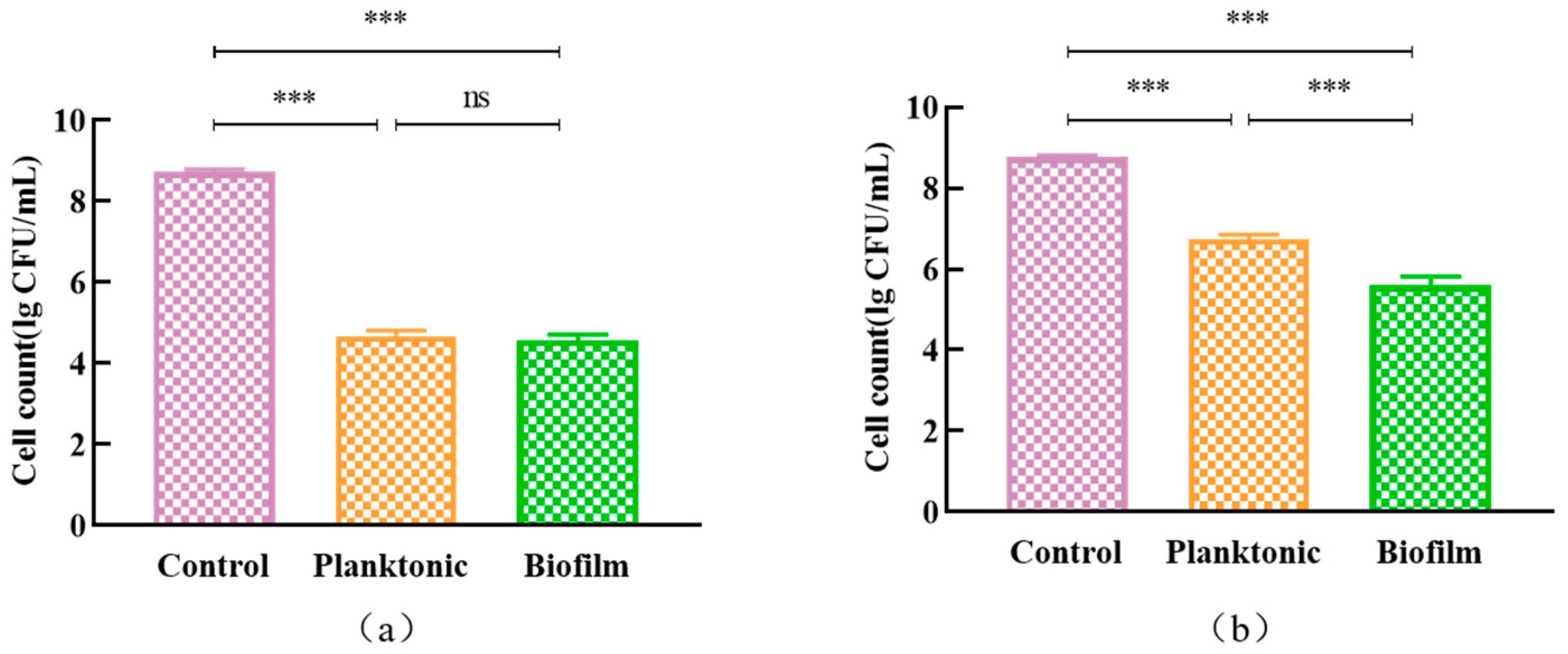
2.11.1. The Growth and Clearance of L. monocytogenes ATCC 19115 Biofilms
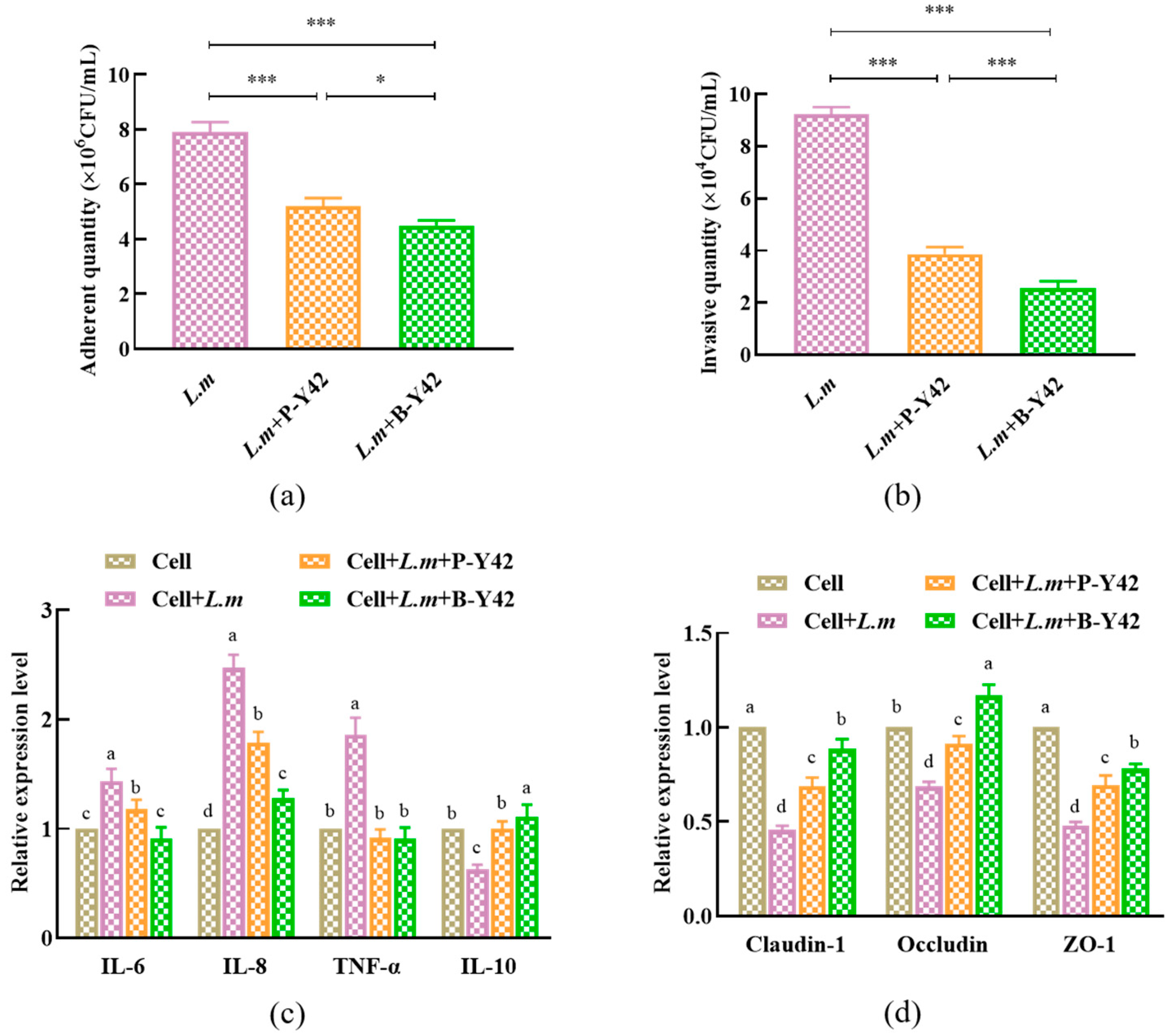
2.11.2. Adhesion of L. monocytogenes ATCC 19115 on HT-29 Cells
2.12. Statistical Analysis
3. Results
3.1. Influence Factors on the Biofilm Formation of L. plantarum Y42
3.2. Morphological Observations of L. plantarum Y42 in Biofilm and Planktonic Forms
3.3. Surface Properties of L. plantarum Y42 in Biofilm and Planktonic Forms
3.4. Tolerance of L. plantarum Y42 in Biofilm and Planktonic Forms to Harsh Environmental Conditions
3.5. Vacuum Lyophilization of L. plantarum Y42 in Biofilm or Planktonic Forms
3.6. Expressions of Inflammatory Factors and Tight Junction Proteins
3.7. Effect of Planktonic and Biofilm L. plantarum Y42 on L. monocytogenes ATCC 19115
3.7.1. The Growth and Clearance of L. monocytogenes ATCC 19115 Biofilms
3.7.2. The Inhibitory Effects of Planktonic and Biofilm L. plantarum Y42 on the Adhesion and Invasion of L. monocytogenes ATCC 19115 of HT-29 Cell Monolayers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; Garcia, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.L.; Zhang, Y.; Guo, X.J.; Zhang, L.D.; Zhang, W.; Man, C.X.; Jiang, Y.J. Characterization and transcriptomic basis of biofilm formation by Lactobacillus plantarum J26 isolated from traditional fermented dairy products. LWT-Food Sci. Technol. 2020, 125, 9. [Google Scholar] [CrossRef]
- Nyabako, B.A.; Fang, H.; Cui, F.J.; Liu, K.Y.; Tao, T.L.; Zan, X.Y.; Sun, W.J. Enhanced Acid Tolerance in Lactobacillus acidophilus by Atmospheric and Room Temperature Plasma (ARTP) Coupled with Adaptive Laboratory Evolution (ALE). Appl. Biochem. Biotechnol. 2020, 191, 1499–1514. [Google Scholar] [CrossRef] [PubMed]
- Jingjing, E.; Ma, R.Z.; Chen, Z.C.; Yao, C.Q.; Wang, R.X.; Zhang, Q.L.; He, Z.B.; Sun, R.Y.; Wang, J.G. Improving the freeze-drying survival rate of Lactobacillus plantarum LIP-1 by increasing biofilm formation based on adjusting the composition of buffer salts in medium. Food Chem. 2021, 338, 10. [Google Scholar] [CrossRef]
- Chamignon, C.; Guéneau, V.; Medina, S.; Deschamps, J.; Gil-Izquierdo, A.; Briandet, R.; Mousset, P.Y.; Langella, P.; Lafay, S.; Microorganisms, L.B.-H.J. Evaluation of the Probiotic Properties and the Capacity to Form Biofilms of Various Lactobacillus Strains. Microorganisms 2020, 8, 1053. [Google Scholar] [CrossRef]
- Fusco, A.; Savio, V.; Stelitano, D.; Baroni, A.; Donnarumma, G. The Intestinal Biofilm of Pseudomonas aeruginosa and Staphylococcus aureus Is Inhibited by Antimicrobial Peptides HBD-2 and HBD-3. Appl. Sci. 2021, 11, 12. [Google Scholar] [CrossRef]
- Silva, D.R.; Sardi, J.D.O.; Pitangui, N.D.; Roque, S.M.; da Silva, A.C.B.; Rosalen, P.L. Probiotics as an alternative antimicrobial therapy: Current reality and future directions. J. Funct. Food 2020, 73, 12. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Giri, S.K. Probiotic functional foods: Survival of probiotics during processing and storage. J. Funct. Food 2014, 9, 225–241. [Google Scholar] [CrossRef]
- Hevia, A.; Delgado, S.; Sanchez, B.; Margolles, A. Molecular Players Involved in the Interaction between Beneficial Bacteria and the Immune System. Front. Microbiol. 2015, 6, 8. [Google Scholar] [CrossRef]
- Ku, S.; Park, M.S.; Ji, G.E.; You, H.J. Review on Bifidobacterium bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism. Int. J. Mol. Sci. 2016, 17, 23. [Google Scholar] [CrossRef]
- Al-Hadidi, A.; Navarro, J.; Goodman, S.D.; Bailey, M.T.; Besner, G.E. Lactobacillus reuteri in Its Biofilm State Improves Protection from Experimental Necrotizing Enterocolitis. Nutrients 2021, 13, 12. [Google Scholar] [CrossRef]
- Liao, C.J.; Li, H.; Wang, S.Y.; Xiong, J.; Mei, C.; Liu, D.; He, Y.Z.; Peng, L.C.; Song, Z.H.; Chen, H.W. Bacterial biofilms: Novel strategies for intestinal colonization by probiotics. Chin. J. Biotechnol. 2022, 38, 2821–2839. (In Chinese) [Google Scholar]
- Song, X. Study on Inhibition of Lactobacillus plantarum Y42 Culture Supernatant aginst Listeria Monocytogenes Aiofilm Formation; Dalian Polytechnic University: Dalian, China, 2018. (In Chinese) [Google Scholar]
- Wang, J.Q. Study on Inhibition of the Protein Produced by Lactobacillus plantarum Y42 aginst Biofilm Formation of Listeria monocytogenes; Dalian Polytechnic University: Dalian, China, 2019. (In Chinese) [Google Scholar]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 11. [Google Scholar] [CrossRef]
- Yao, S.J.; Hao, L.Y.; Zhou, R.Q.; Jin, Y.; Huang, J.; Wu, C.D. Formation of Biofilm by Tetragenococcus halophilus Benefited Stress Tolerance and Anti-biofilm Activity against S. aureus and S. Typhimurium. Front. Microbiol. 2022, 13, 15. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Y.F.; Gu, Y.; Wang, Y.; Deng, Y.X. Stress resistance and antioxidant properties of lactic acid bacteria with high biofilm production. Trans. Chin. Soc. Agric. Eng. 2021, 37, 282–288. (In Chinese) [Google Scholar]
- Wang, R.X.; Sun, R.Y.; Yang, Y.; E, J.J.; Yao, C.Q.; Zhang, Q.L.; Chen, Z.C.; Ma, R.Z.; Li, J.; Zhang, J.Y.; et al. Effects of salt stress on the freeze-drying survival rate of Lactiplantibacillus plantarum LIP-1. Food Microbiol. 2022, 105, 10. [Google Scholar] [CrossRef]
- Zhang, X.N.; Chen, J.; Ma, L.L.; E, J.J.; Bao, Q.H.; Wang, J.G. The effect of optimized medium components on the activity of Lactobacillus plantarum LIP-1 after freeze-drying. Food Sci. Technol. 2019, 44, 4023–4032. (In Chinese) [Google Scholar]
- Greppi, A.; Saubade, F.; Botta, C.; Humblot, C.; Guyot, J.P.; Cocolin, L. Potential probiotic Pichia kudriavzevii strains and their ability to enhance folate content of traditional cereal-based African fermented food. Food Microbiol. 2017, 62, 169–177. [Google Scholar] [CrossRef]
- Dai, J.H.; Zhang, Y.J.; Ou, W.H.; Yu, G.J.; Ai, Q.H.; Zhang, W.B.; Mai, K.S. Stachyose protects intestinal mucosal barrier via promotion of tight junction and Lactobacillus casei-drived inhibition of apoptosis in juvenile turbot, Scophthalmus maximus L. Aquaculture 2022, 556, 9. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, Y.; Ma, F.; Sun, M.; Song, Y.; Xu, D.; Mu, G.; Tuo, Y. Lactobacillus plantarum Y44 alleviates oxidative stress by regulating gut microbiota and colonic barrier function in Balb/C mice with subcutaneous d-galactose injection. Food Funct. 2021, 12, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yang, X.; Liu, L.C.; Chen, L.; Teng, J.; Zhu, X.P.; Zhao, J.M.; Wang, Q. Spatial and seasonal variations in biofilm formation on microplastics in coastal waters. Sci. Total Environ. 2021, 770, 13. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Chen, Z.Y.; Chen, P.T.; Ng, I.S. Production, characterization and antibacterial activity of exopolysaccharide from a newly isolated Weissella cibaria under sucrose effect. J. Biosci. Bioeng. 2018, 126, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Bellich, B.; Lagatolla, C.; Tossi, A.; Benincasa, M.; Cescutti, P.; Rizzo, R. Influence of Bacterial Biofilm Polysaccharide Structure on Interactions with Antimicrobial Peptides: A Study on Klebsiella pneumoniae. Int. J. Mol. Sci. 2018, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Lu, M.; Liu, R.M.; Tian, Y.Y.; Vu, V.H.; Li, Y.; Liu, B.; Kushmaro, A.; Li, Y.Q.; Sun, Q. Inhibition of Streptococcus mutans Biofilm Formation and Virulence by Lactobacillus plantarum K41 Isolated From Traditional Sichuan Pickles. Front. Microbiol. 2020, 11, 12. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Beganovic, J.; Frece, J.; Kos, B.; Pavunc, A.L.; Habjanic, K.; Suskovic, J. Functionality of the S-layer protein from the probiotic strain Lactobacillus helveticus M92. Antonie Van Leeuwenhoek 2011, 100, 43–53. [Google Scholar] [CrossRef]
- Hu, M.X.; Li, J.N.; Guo, Q.; Zhu, Y.Q.; Niu, H.M. Probiotics Biofilm-Integrated Electrospun Nanofiber Membranes: A New Starter Culture for Fermented Milk Production. J. Agric. Food Chem. 2019, 67, 3198–3208. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, Y.; Wu, R.; Zheng, Y.; Wang, Y.; Nie, L.; Qiao, R.; He, Y. Exploring the relationship between the signal molecule AI-2 and the biofilm formation of Lactobacillus sanfranciscensis. LWT 2022, 154, 112704. [Google Scholar] [CrossRef]
- Dufrene, Y.F.; Persat, A. Mechanomicrobiology: How bacteria sense and respond to forces. Nat. Rev. Microbiol. 2020, 18, 227–240. [Google Scholar] [CrossRef]
- Tytgat, H.L.P.; Nobrega, F.L.; van der Oost, J.; de Vos, W.M. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol. 2019, 27, 17–25. [Google Scholar] [CrossRef]
- Liu, L.; Wu, R.Y.; Li, J.; Li, P.L. Adhesion ability of Lactobacillus paraplantarum L-ZS9 and influencing factors of its biofilm formation. Food Sci. 2016, 37, 136–143. (In Chinese) [Google Scholar]
- Nhu, N.T.K.; Phan, M.D.; Peters, K.M.; Lo, A.W.; Forde, B.M.; Chong, T.M.; Yin, W.F.; Chan, K.G.; Chromek, M.; Brauner, A.; et al. Discovery of New Genes Involved in Curli Production by a Uropathogenic Escherichia coli Strain from the Highly Virulent O45:K1:H7 Lineage. mBio 2018, 9, 16. [Google Scholar] [CrossRef]
- Desai, S.; Sanghrajka, K.; Gajjar, D. High Adhesion and Increased Cell Death Contribute to Strong Biofilm Formation in Klebsiella pneumoniae. Pathogens 2019, 8, 16. [Google Scholar] [CrossRef]
- Wang, H.H.; Wu, N.; Jiang, Y.; Ye, K.P.; Xu, X.L.; Zhou, G.H. Response of long-term acid stress to biofilm formation of meat-related Salmonella Enteritidis. Food Control 2016, 69, 214–220. [Google Scholar] [CrossRef]
- di Biase, A.; Kowalski, M.S.; Devlin, T.R.; Oleszkiewicz, J.A. Physicochemical methods for biofilm removal allow for control of biofilm retention time in a high rate MBBR. Environ. Technol. 2022, 43, 1593–1602. [Google Scholar] [CrossRef]
- Welman, A.D.; Maddox, I.S. Exopolysaccharides from lactic acid bacteria: Perspectives and challenges. Trends Biotechnol. 2003, 21, 269–274. [Google Scholar] [CrossRef]
- Savijoki, K.; Nyman, T.A.; Kainulainen, V.; Miettinen, I.; Siljamaki, P.; Fallarero, A.; Sandholm, J.; Satokari, R.; Varmanen, P. Growth Mode and Carbon Source Impact the Surfaceome Dynamics of Lactobacillus rhamnosus GG. Front. Microbiol. 2019, 10, 17. [Google Scholar] [CrossRef]
- Liu, L. Regulation Mechanism of LuxS/AI-2 Quorum Sensing System on Biofilm Formation of Lactobacillus paraplantarum L-ZS9; China Agricultural University: Beijing, China, 2017. (In Chinese) [Google Scholar]
- Shenkutie, A.M.; Zhang, J.Y.; Yao, M.Z.; Asrat, D.; Chow, F.W.N.; Leung, P.H.M. Effects of Sub-Minimum Inhibitory Concentrations of Imipenem and Colistin on Expression of Biofilm-Specific Antibiotic Resistance and Virulence Genes in Acinetobacter baumannii Sequence Type 1894. Int. J. Mol. Sci. 2022, 23, 19. [Google Scholar] [CrossRef]
- Feldman-Salit, A.; Hering, S.; Messiha, H.L.; Veith, N.; Cojocaru, V.; Sieg, A.; Westerhoff, H.V.; Kreikemeyer, B.; Wade, R.C.; Fiedler, T. Regulation of the activity of lactate dehydrogenases from four lactic acid bacteria. J. Biol. Chem. 2013, 288, 21295–21306. [Google Scholar] [CrossRef]
- Gong, P.M. Research on Damage Mechanism and Protection Methods of Lactobacillus bulgaricus during Spray Drying Process; Harbin Institute of Technology: Harbin, China, 2019. (In Chinese) [Google Scholar]
- Kalaiarasan, E.; Kottha, T.; Harish, B.N.; Gnanasambandam, V.; Sali, V.K.; John, J. Inhibition of quorum sensing-controlled biofilm formation in Pseudomonas aeruginosa by quorum-sensing inhibitors. Microb. Pathog. 2017, 111, 99–107. [Google Scholar] [CrossRef]
- Derakhshandeh, S.; Shahrokhi, N.; Khalaj, V.; Habibi, M.; Moazzezy, N.; Karam, M.R.A.; Bouzari, S. Surface display of uropathogenic Escherichia coli FimH in Lactococcus lactis: In vitro characterization of recombinant bacteria and its protectivity in animal model. Microb. Pathog. 2020, 141, 9. [Google Scholar] [CrossRef]
- Liu, L.; Gui, M.; Wu, R.Y.; Li, P.L. Progress in research on biofilm formation regulated by LuxS/AI-2 quorum sensing. Food Sci. 2016, 37, 254–262. (In Chinese) [Google Scholar]
- Lin, L.; Zhang, J.Q. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017, 18, 25. [Google Scholar] [CrossRef]
- Vazquez-Armenta, F.J.; Hernandez-Onate, M.A.; Martinez-Tellez, M.A.; Lopez-Zavala, A.A.; Gonzalez-Aguilar, G.A.; Gutierrez-Pacheco, M.M.; Ayala-Zavala, J.F. Quercetin repressed the stress response factor (sigB) and virulence genes (prfA, actA, inlA, and inlC), lower the adhesion, and biofilm development of L. monocytogenes. Food Microbiol. 2020, 87, 103377. [Google Scholar] [CrossRef]
- Wells, J.M.; Rossi, O.; Meijerink, M.; van Baarlen, P. Epithelial crosstalk at the microbiota-mucosal interface. Proc. Natl. Acad. Sci. USA 2011, 108, 4607–4614. [Google Scholar] [CrossRef]
- Siciliano, R.A.; Mazzeo, M.F. Molecular mechanisms of probiotic action: A proteomic perspective. Curr. Opin. Microbiol. 2012, 15, 390–396. [Google Scholar] [CrossRef]
- Sharma, R.; Young, C.; Neu, J. Molecular Modulation of Intestinal Epithelial Barrier: Contribution of Microbiota. J. Biomed. Biotechnol. 2010, 15, 305879. [Google Scholar] [CrossRef]
- Burger, E.; Araujo, A.; Lopez-Yglesias, A.; Rajala, M.W.; Geng, L.D.; Levine, B.; Hooper, L.V.; Burstein, E.; Yarovinsky, F. Loss of Paneth Cell Autophagy Causes Acute Susceptibility to Toxoplasma gondii-Mediated Inflammation. Cell Host Microbe 2018, 23, 177. [Google Scholar] [CrossRef]
- Hossain, M.I.; Mizan, M.F.R.; Roy, P.K.; Nahar, S.; Toushik, S.H.; Ashrafudoulla, M.; Jahid, I.K.; Lee, J.; Ha, S.D. Listeria monocytogenes biofilm inhibition on food contact surfaces by application of postbiotics from Lactobacillus curvatus B.67 and Lactobacillus plantarum M.2. Food Res. Int. 2021, 148, 13. [Google Scholar] [CrossRef]
- Bujnakova, D.; Strakova, E. Safety, probiotic and technological properties of Lactobacilli isolated from unpasteurised ovine and caprine cheeses. Ann. Microbiol. 2017, 67, 813–826. [Google Scholar] [CrossRef]
- Chervinets, Y.; Chervinets, V.; Shenderov, B.; Belyaeva, E.; Troshin, A.; Lebedev, S.; Danilenko, V. Adaptation and Probiotic Potential of Lactobacilli, Isolated from the Oral Cavity and Intestines of Healthy People. Probiotics Antimicrob. Proteins 2018, 10, 22–33. [Google Scholar] [CrossRef]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vazquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017, 2, 7. [Google Scholar] [CrossRef]
- Terraf, M.C.L.; Tomas, M.S.J.; Nader-Macias, M.E.F.; Silva, C. Screening of biofilm formation by beneficial vaginal lactobacilli and influence of culture media components. J. Appl. Microbiol. 2012, 113, 1517–1529. [Google Scholar] [CrossRef]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C.J. Host interactions of probiotic bacterial surface molecules: Comparison with commensals and pathogens. Nat. Rev. Microbiol. 2010, 8, 171–184. [Google Scholar] [CrossRef]
- Li, P.N.; Herrmann, J.; Tolar, B.B.; Poitevin, F.; Ramdasi, R.; Bargar, J.R.; Stahl, D.A.; Jensen, G.J.; Francis, C.A.; Wakatsuki, S.; et al. Nutrient transport suggests an evolutionary basis for charged archaeal surface layer proteins. Isme J. 2018, 12, 2389–2402. [Google Scholar] [CrossRef]

| Gene | Primer Sequence (5′-3′) |
|---|---|
| GAPDH | F: GGAAGGTGAAGGTCGGAGTC |
| R: TCAGCCTTGACGGTGCCATG | |
| IL-6 | F: AAGCCAGAGCTGTGCAGATGAGTA |
| R: TGTCCTGCAGCCACTGGTTC | |
| IL-8 | F: ACACAGAGCTGCAGAAATCAGG |
| R: GGCACAAACTTTCAGAGACAG | |
| TNF-α | F: CTGCCTGCTGCACTTTGGAG |
| R: ACATGGGCTACAGGCTTGTCACT | |
| IL-10 | F: GAGATGCCTTCAGCAGAGTGAAGA |
| R: AGTTCACATGCGCCTTGATGTC | |
| β-actin | F: ACTCTGGTGATGGTGTTAC |
| R: GGCTGTGATCTCCTTCTG | |
| Claudin-1 | F: CATACTCCTGGGTCTGGTTGGT |
| R: GACAGCCATCCGCATCTTCT | |
| Occludin | F: ACGGCAGCACCTACCTCAA |
| R: GGGCGAAGAAGCAGATGAG | |
| ZO-1 | F: CTTCAGGTGTTTCTCTTCCTCCTC |
| R: CTGTGGTTTCATGGCTGGATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Mu, G.; Tuo, Y. Phenotypic Traits and Probiotic Functions of Lactiplantibacillus plantarum Y42 in Planktonic and Biofilm Forms. Foods 2023, 12, 1516. https://doi.org/10.3390/foods12071516
Li J, Mu G, Tuo Y. Phenotypic Traits and Probiotic Functions of Lactiplantibacillus plantarum Y42 in Planktonic and Biofilm Forms. Foods. 2023; 12(7):1516. https://doi.org/10.3390/foods12071516
Chicago/Turabian StyleLi, Jiayi, Guangqing Mu, and Yanfeng Tuo. 2023. "Phenotypic Traits and Probiotic Functions of Lactiplantibacillus plantarum Y42 in Planktonic and Biofilm Forms" Foods 12, no. 7: 1516. https://doi.org/10.3390/foods12071516
APA StyleLi, J., Mu, G., & Tuo, Y. (2023). Phenotypic Traits and Probiotic Functions of Lactiplantibacillus plantarum Y42 in Planktonic and Biofilm Forms. Foods, 12(7), 1516. https://doi.org/10.3390/foods12071516







