Fungal Community Composition and Its Relationship with Volatile Compounds during Spontaneous Fermentation of Cabernet Sauvignon from Two Chinese Wine-Growing Regions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Winemaking and Sampling
2.2. Cabernet Sauvignon Inoculated Fermentation
2.3. DNA Extraction and Sequencing
2.4. Physicochemical Parameter Analysis
2.5. Analysis of Volatile Compounds
2.6. Statistical Analysis
3. Results
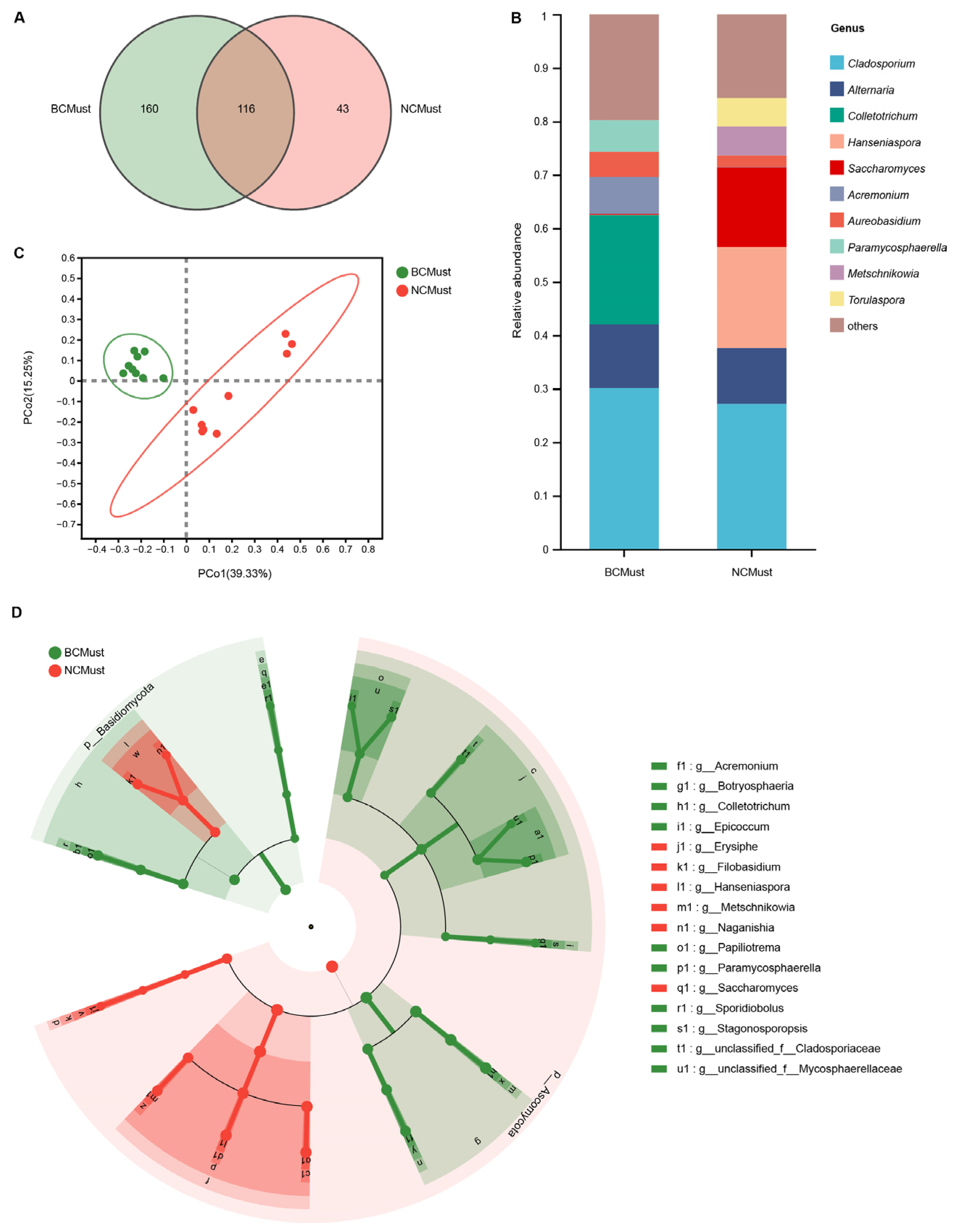
3.1. Fungal Communities Varied by Wine-Growing Region
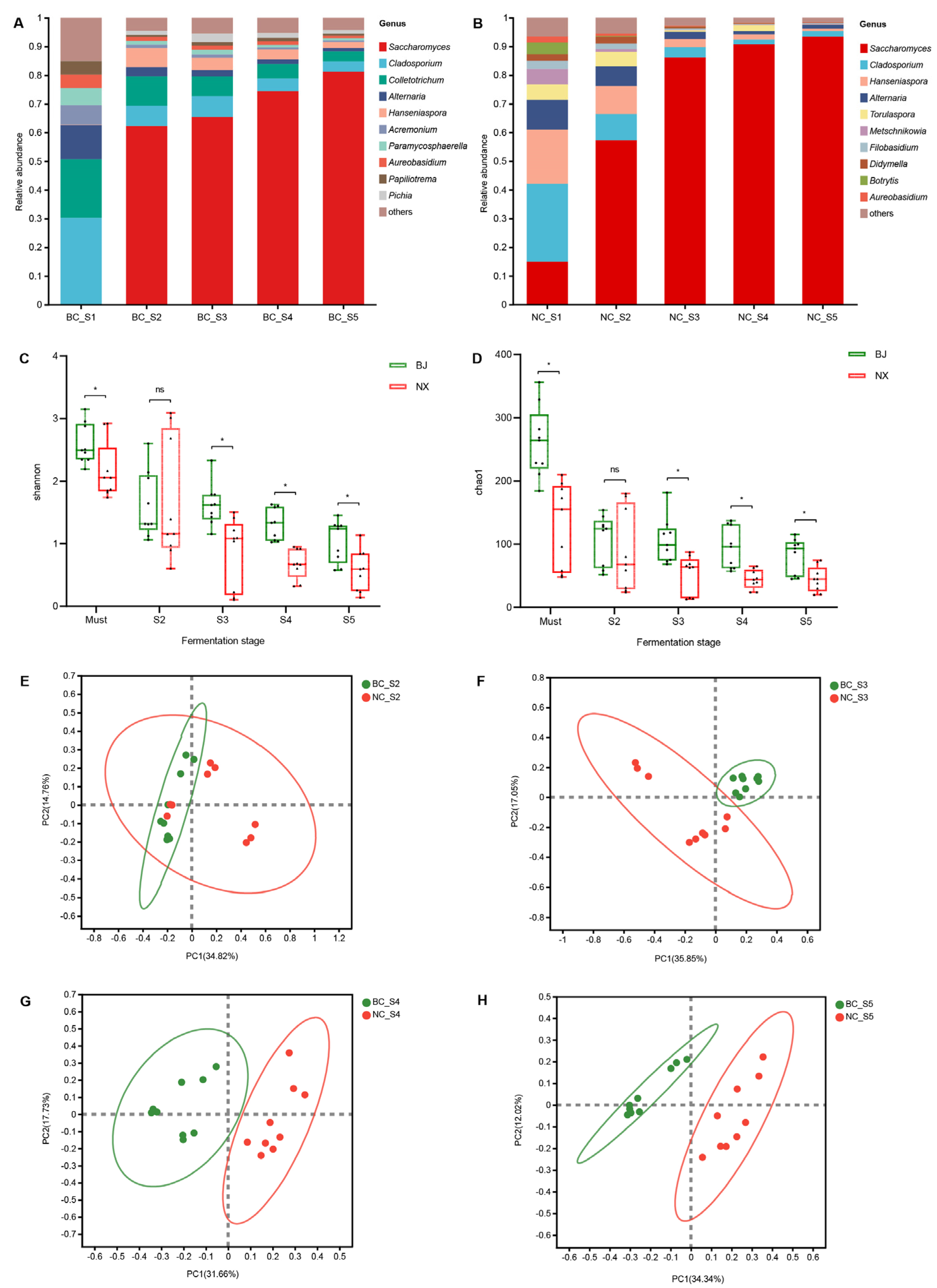
3.2. Dynamics of Fungal Communities during Spontaneous Fermentation
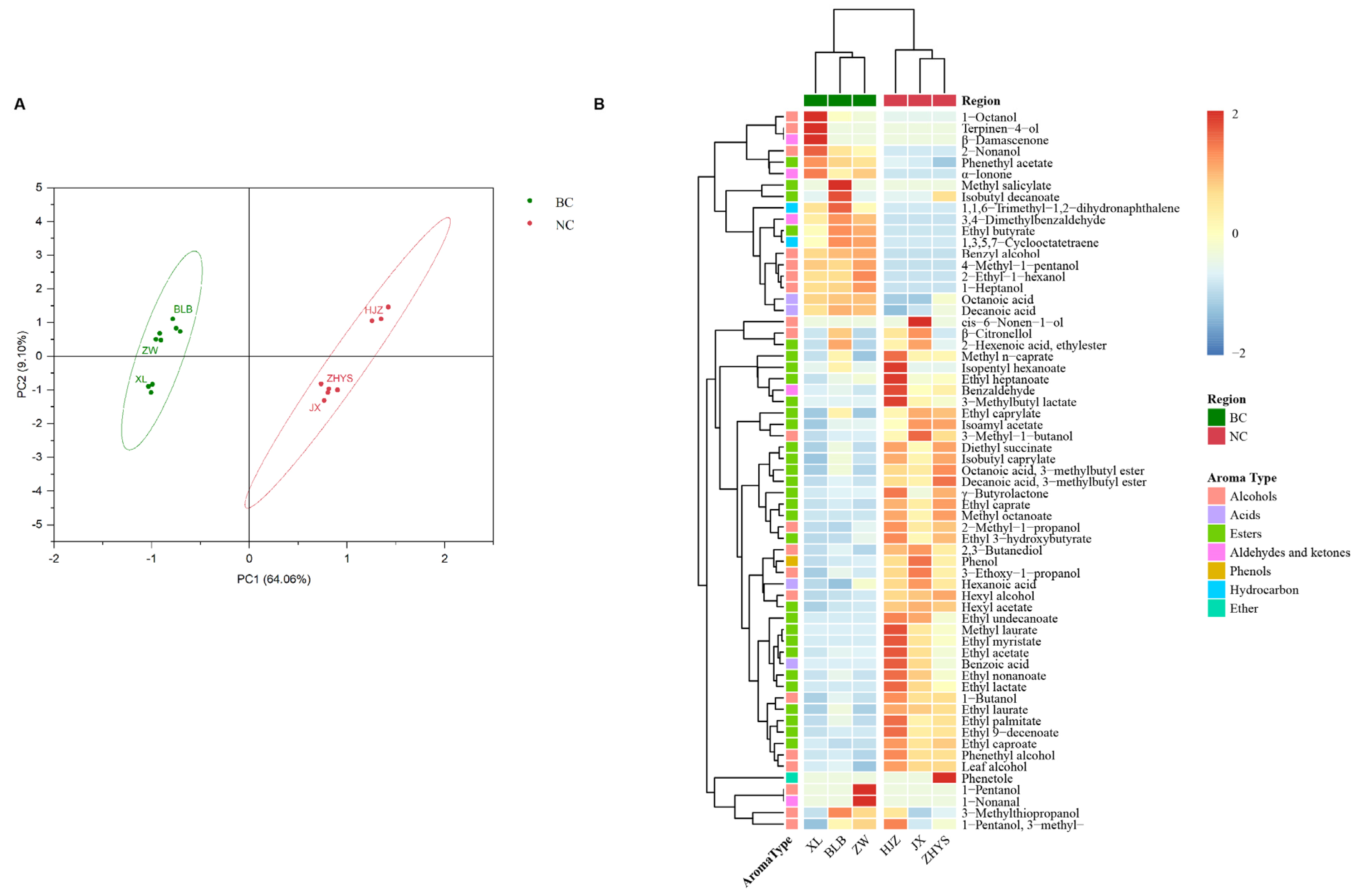
3.3. Aroma Characteristics of Wines Varied by Wine-Growing Region
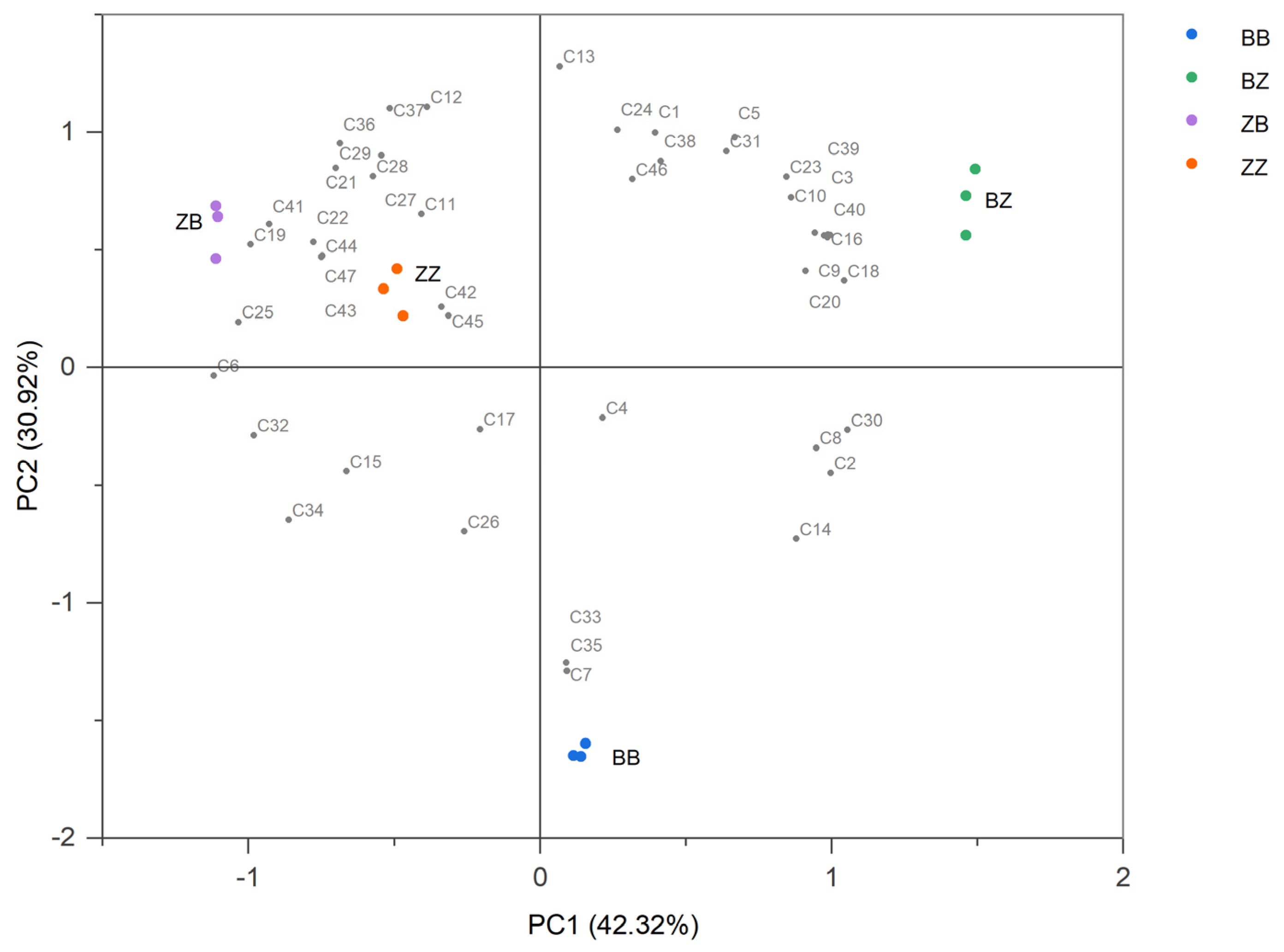
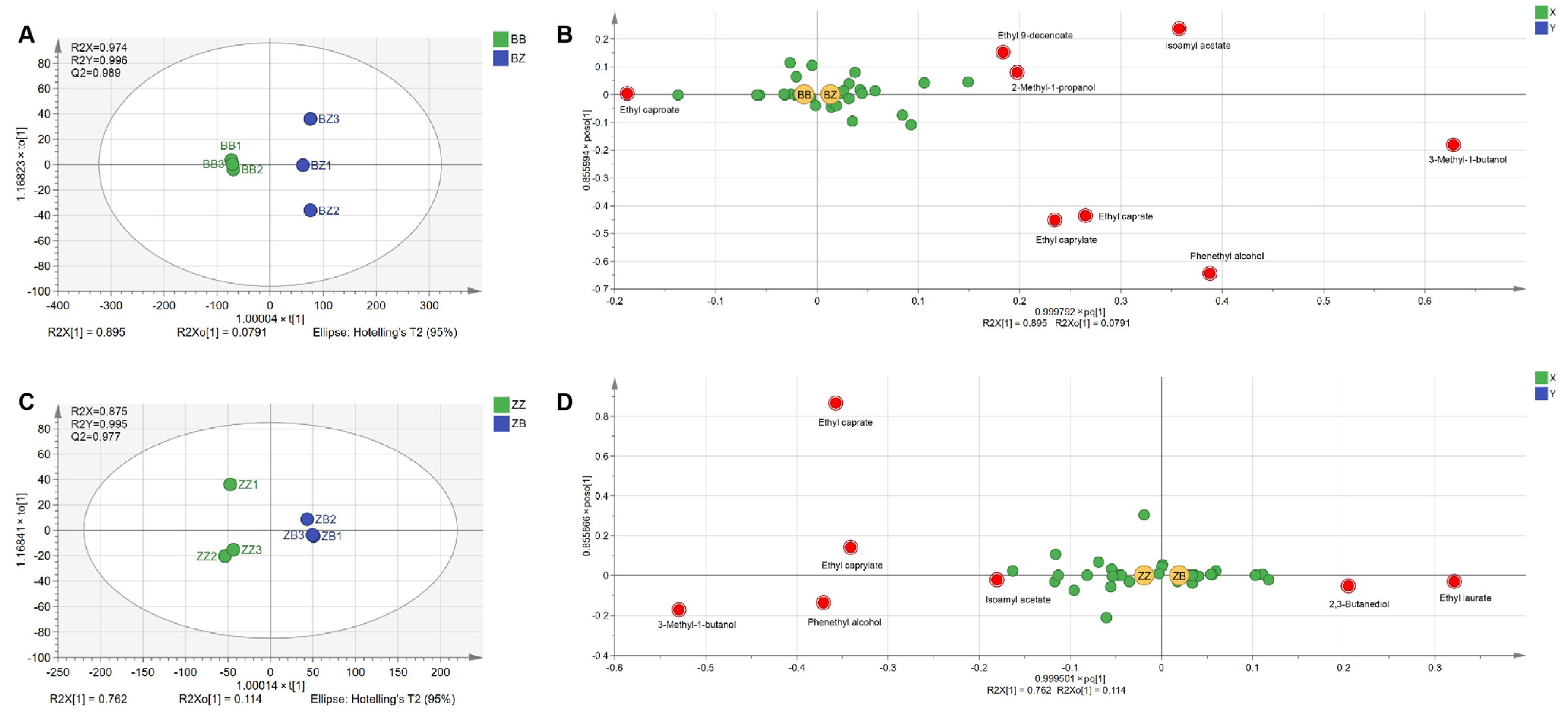
3.4. Wines Fermented by Microorganisms with Different Geographical Origins Had Distinctive Aroma Profiles
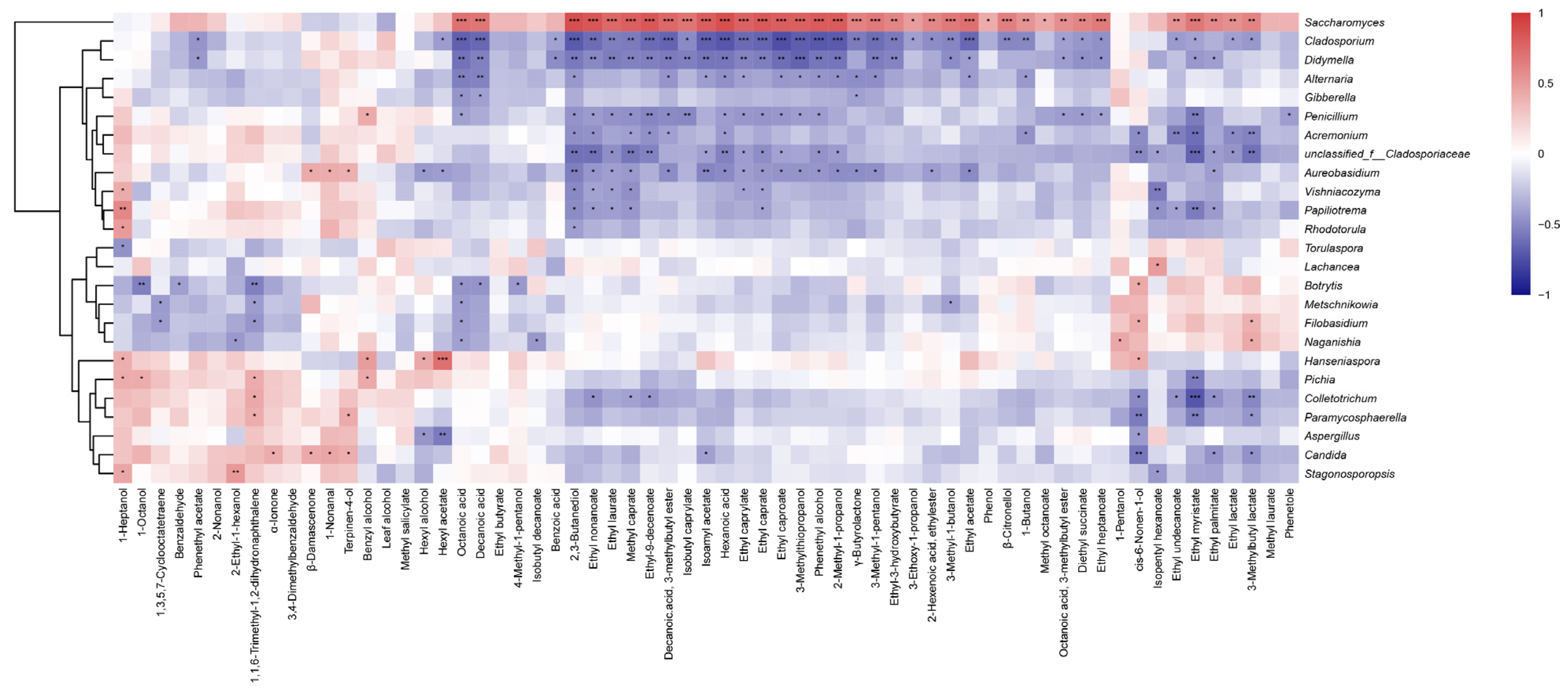
3.5. Fungal Communities Associated with Wine Aroma Profiles
4. Discussion
4.1. Biogeographical Distribution of Fungal Microbiota during Spontaneous Fermentation
4.2. Influence of Fungal Microbiota on Volatile Compounds in Wine
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Leeuwen, C.; Seguin, G. The Concept of Terroir in Viticulture. J. Wine Res. 2006, 17, 1–10. [Google Scholar] [CrossRef]
- López-Rituerto, E.; Savorani, F.; Avenoza, A.; Busto, J.H.; Peregrina, J.M.; Engelsen, S.B. Investigations of La Rioja Terroir for Wine Production using 1H NMR Metabolomics. J. Agric. Food Chem. 2012, 60, 3452–3461. [Google Scholar] [CrossRef] [PubMed]
- Martiniuk, J.T.; Hamilton, J.; Dodsworth, T.; Measday, V. Grape-Associated Fungal Community Patterns Persist from Berry to Wine on a Fine Geographical Scale. FEMS Yeast Res. 2023, 23, foac067. [Google Scholar] [CrossRef] [PubMed]
- Kioroglou, D.; Kraeva-Deloire, E.; Schmidtke, L.M.; Mas, A.; Portillo, M.C. Geographical Origin Has a Greater Impact on Grape Berry Fungal Community than Grape Variety and Maturation State. Microorganisms 2019, 7, 669. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial Biogeography of Wine Grapes is Conditioned by Cultivar, Vintage, and Climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [PubMed]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.; Goddard, M.R. Quantifying Separation and Similarity in a Saccharomyces cerevisiae Metapopulation. ISME J. 2015, 9, 361–370. [Google Scholar] [CrossRef]
- Lam, S.S.; Howell, K.S. Drosophila-Associated Yeast Species in Vineyard Ecosystems. FEMS Microbiol. Lett. 2015, 362, fnv170. [Google Scholar] [CrossRef]
- Hamilton, W.D.; Lenton, T.M. Spora and Gaia: How Microbes Fly with Their Clouds. Ethol. Ecol. Evol. 1998, 10, 1–16. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.; Chen, D.; Howell, K. From the Vineyard to the Winery: How Microbial Ecology Drives Regional Distinctiveness of Wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef]
- Liu, D.; Legras, J.L.; Zhang, P.; Chen, D.; Howell, K. Diversity and Dynamics of Fungi During Spontaneous Fermentations and Association with Unique Aroma Profiles in Wine. Int. J. Food Microbiol. 2021, 338, 108983. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional Microbial Signatures Positively Correlate with Differential Wine Phenotypes: Evidence for a Microbial Aspect to Terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations Among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. mBio 2016, 7, e00631-16. [Google Scholar] [CrossRef] [PubMed]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and Bacterial Modulation of Wine Aroma and Flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Tufariello, M.; Chiriatti, M.A.; Grieco, F.; Perrotta, C.; Capone, S.; Rampino, P.; Tristezza, M.; Mita, G.; Grieco, F. Influence of Autochthonous Saccharomyces cerevisiae Strains on Volatile Profile of Negroamaro Wines. LWT Food Sci. Technol. 2014, 58, 35–48. [Google Scholar] [CrossRef]
- Saberi, S.; Cliff, M.A.; van Vuuren, H.J.J. Impact of Mixed S. cerevisiae Strains on the Production of Volatiles and Estimated Sensory Profiles of Chardonnay Wines. Food Res. Int. 2012, 48, 725–735. [Google Scholar] [CrossRef]
- Virdis, C.; Sumby, K.; Bartowsky, E.; Jiranek, V. Lactic Acid Bacteria in Wine: Technological Advances and Evaluation of Their Functional Role. Front. Microbiol. 2021, 11, 3192. [Google Scholar] [CrossRef]
- Liu, D.; Chen, Q.; Zhang, P.; Chen, D.; Howell, K.S. The Fungal Microbiome Is an Important Component of Vineyard Ecosystems and Correlates with Regional Distinctiveness of Wine. mSphere 2020, 5, e00534-20. [Google Scholar] [CrossRef]
- OIV. International Organization of Vine and Wine. Available online: https://www.oiv.int/what-we-do/statistics (accessed on 29 October 2023).
- Wang, X.; Xie, X.; Chen, N.; Wang, H.; Li, H. Study on Current Status and Climatic Characteristics of Wine Regions in China. Vitis 2018, 57, 9–16. [Google Scholar] [CrossRef]
- Li, R.; Yang, S.; Lin, M.; Guo, S.; Han, X.; Ren, M.; Du, L.; Song, Y.; You, Y.; Zhan, J.; et al. The Biogeography of Fungal Communities Across Different Chinese Wine-Producing Regions Associated with Environmental Factors and Spontaneous Fermentation Performance. Front. Microbiol. 2022, 12, 636639. [Google Scholar] [CrossRef]
- Wei, R.T.; Chen, N.; Ding, Y.T.; Wang, L.; Liu, Y.H.; Gao, F.F.; Zhang, L.; Li, H.; Wang, H. Correlations between microbiota with physicochemical properties and volatile compounds during the spontaneous fermentation of Cabernet Sauvignon (Vitis vinifera L.) wine. LWT-Food. Sci. Technol. 2022, 163, 113529. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, F.; Wang, W.; Liu, Y.; Wang, J.; Sun, J.; Mu, J.; Gao, Z. Effects of Spontaneous Fermentation on The Microorganisms Diversity and Volatile Compounds During ‘Marselan’ From Grape to Wine. LWT-Food. Sci. Technol. 2020, 134, 110193. [Google Scholar] [CrossRef]
- Ma, Y.; Li, T.; Xu, X.; Ji, Y.; Jiang, X.; Shi, X.; Wang, B. Investigation of Volatile Compounds, Microbial Succession, and Their Relation During Spontaneous Fermentation of Petit Manseng. Front. Microbiol. 2021, 12, 717387. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Jouanneau, S.; Nicolau, L.; Gardner, R.C.; Villas-Boas, S.G. Concentrations of the Volatile Thiol 3-Mercaptohexanol in Sauvignon blanc Wines: No Correlation with Juice Precursors. Am. J. Enol. Vitic. 2012, 63, 407–412. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, L.; Qiu, S.; Wu, X.; Li, J.; Ma, L. Freeze-Thaw Cycles Characterize Varietal Aroma of Vidal Blanc Grape During Late Harvest by Shaping Self-Assembled Microeukaryotic Communities. Food Chem. 2022, 384, 132553. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, M.; Huang, W.; You, Y.; Zhan, J. Indigenous Saccharomyces cerevisiae Could Better Adapt to the Physicochemical Conditions and Natural Microbial Ecology of Prince Grape Must Compared with Commercial Saccharomyces cerevisiae FX10. Molecules 2022, 27, 6892. [Google Scholar] [CrossRef] [PubMed]
- Thukral, A.K. A review on measurement of Alpha diversity in biology. Agric. Res. J. 2017, 54, 1–10. [Google Scholar] [CrossRef]
- He, Y.; Wang, X.; Li, P.; Lv, Y.; Nan, H.; Wen, L.; Wang, Z. Research Progress of Wine Aroma Components: A Critical Review. Food Chem. 2023, 402, 134491. [Google Scholar] [CrossRef]
- Antonelli, A.; Castellari, L.; Zambonelli, C.; Carnacini, A. Yeast Influence on Volatile Composition of Wines. J. Agric. Food Chem. 1999, 47, 1139–1144. [Google Scholar] [CrossRef]
- Wang, H.L.; Hopfer, H.; Cockburn, D.W.; Wee, J. Characterization of Microbial Dynamics and Volatile Metabolome Changes During Fermentation of Chambourcin Hybrid Grapes from Two Pennsylvania Regions. Front. Microbiol. 2020, 11, 614278. [Google Scholar] [CrossRef]
- Gayevskiy, V.; Goddard, M.R. Geographic Delineations of Yeast Communities and Populations Associated with Vines and Wines in New Zealand. ISME J. 2012, 6, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Morrison-Whittle, P.; Goddard, M.R. From Vineyard to Winery: A Source Map of Microbial Diversity Driving Wine Fermentation. Environ. Microbiol. 2018, 20, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Andorrà, I.; Miró, G.; Espligares, N.; Mislata, A.M.; Puxeu, M.; Ferrer-Gallego, R. Wild Yeast and Lactic Acid Bacteria of Wine. In Yeasts in Biotechnology; Peixoto Basso, T., Ed.; IntechOpen: London, UK, 2019. [Google Scholar]
- Wang, X.; Schlatter, D.C.; Glawe, D.A.; Edwards, C.G.; Weller, D.M.; Paulitz, T.C.; Abatzoglou, J.T.; Okubara, P.A. Native Yeast and Non-Yeast Fungal Communities of Cabernet Sauvignon Berries from Two Washington State Vineyards, and Persistence in Spontaneous Fermentation. Int. J. Food Microbiol. 2021, 350, 109225. [Google Scholar] [CrossRef] [PubMed]
- Louw, N.L.; Lele, K.; Ye, R.; Edwards, C.B.; Wolfe, B.E. Microbiome Assembly in Fermented Foods. Annu. Rev. Microbiol. 2023, 77, 381–402. [Google Scholar] [CrossRef] [PubMed]
- Steensels, J.; Verstrepen, K.J. Taming Wild Yeast: Potential of Conventional and Nonconventional Yeasts in Industrial Fermentations. Annu. Rev. Microbiol. 2014, 68, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.R. Quantifying the Complexities of Saccharomyces Cerevisiae’s Ecosystem Engineering Via Fermentation. Ecology 2008, 89, 2077–2082. [Google Scholar] [CrossRef]
- Holm Hansen, E.; Nissen, P.; Sommer, P.; Nielsen, J.C.; Arneborg, N. The Effect of Oxygen on the Survival of Non-Saccharomyces Yeasts During Mixed Culture Fermentations of Grape Juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 2001, 91, 541–547. [Google Scholar] [CrossRef]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not Your Ordinary Yeast: Non-Saccharomyces Yeasts in Wine Production Uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef]
- Sumby, K.M.; Grbin, P.R.; Jiranek, V. Microbial Modulation of Aromatic Esters in Wine: Current Knowledge and Future Prospects. Food Chem. 2010, 121, 1–16. [Google Scholar] [CrossRef]
- De-la-Fuente-Blanco, A.; Saenz-Navajas, M.P.; Ferreira, V. On the Effects of Higher Alcohols on Red Wine Aroma. Food Chem. 2016, 210, 107–114. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeast Interactions and Wine Flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Esteve-Zarzoso, B.; Arroyo, T. Non-Saccharomyces Yeasts: Biotechnological Role for Wine Production. In Grape and Wine Biotechnology; IntechOpen: London, UK, 2016. [Google Scholar]
- Rodríguez, M.E.; Lopes, C.A.; Barbagelata, R.J.; Barda, N.B.; Caballero, A.C. Influence of Candida pulcherrima Patagonian Strain on Alcoholic Fermentation Behaviour and Wine Aroma. Int. J. Food Microbiol. 2010, 138, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Li, A.H.; Dizy, M.; Ullah, N.; Sun, W.X.; Tao, Y.S. Evaluation of aroma enhancement for “Ecolly” dry white wines by mixed inoculation of selected Rhodotorula mucilaginosa and Saccharomyces cerevisiae. Food Chem. 2017, 228, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Q.; Shen, J.Y.; Duan, C.Q.; Yan, G.L. Use of Indigenous Hanseniaspora vineae and Metschnikowia pulcherrima Co-fermentation with Saccharomyces cerevisiae to Improve the Aroma Diversity of Vidal Blanc Icewine. Front. Microbiol. 2018, 9, 2303. [Google Scholar] [CrossRef]
- Shi, W.K.; Wang, J.; Chen, F.S.; Zhang, X.Y. Effect of Issatchenkia terricola and Pichia kudriavzevii on Wine Flavor and Quality Through Simultaneous and Sequential Co-fermentation with Saccharomyces cerevisiae. LWT Food Sci. Technol. 2019, 116, 108477. [Google Scholar] [CrossRef]
| Region | Winery | Cabernet Sauvignon Musts (Must) | Cabernet Sauvignon Wines (Stage 5) | ||||
|---|---|---|---|---|---|---|---|
| Glucose (g/L) | Fructose (g/L) | Glucose (g/L) | Fructose (g/L) | Glycerol (g/L) | Ethanol (% v/v) | ||
| BC | BLB | 127.13 ± 0.21 | 169.89 ± 0.25 | 1.58 ± 0.26 | 0.50 ± 0.09 | 11.03 ± 0.12 | 16.56 ± 0.37 |
| XL | 112.58 ± 0.40 | 149.50 ± 0.91 | 1.27 ± 0.13 | 1.18 ± 0.06 | 9.88 ± 0.13 | 14.54 ± 0.07 | |
| ZW | 116.07 ± 1.04 | 151.80 ± 1.47 | 0.23 ± 0.06 | 3.00 ± 0.33 | 8.34 ± 0.10 | 14.33 ± 0.07 | |
| NC | HJZ | 140.42 ± 1.12 | 193.19 ± 1.50 | 1.04 ± 0.56 | 2.29 ± 0.16 | 12.00 ± 0.15 | 16.87 ± 0.06 |
| JX | 140.51 ± 0.16 | 177.16 ± 0.35 | 2.05 ± 0.22 | 1.17 ± 0.24 | 9.07 ± 0.07 | 17.13 ± 0.33 | |
| ZHYS | 146.93 ± 1.83 | 186.05 ± 2.37 | 0.25 ± 0.08 | 1.53 ± 0.28 | 9.84 ± 0.18 | 18.87 ± 0.02 | |
| Sample | Origin of Cabernet Sauvignon Juice | Origin of Microorganisms |
|---|---|---|
| BB | B | B |
| BZ | B | Z |
| ZZ | Z | Z |
| ZB | Z | B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Geng, H.; Chai, R.; Wu, T.; Huang, W.; You, Y.; Zhan, J. Fungal Community Composition and Its Relationship with Volatile Compounds during Spontaneous Fermentation of Cabernet Sauvignon from Two Chinese Wine-Growing Regions. Foods 2024, 13, 106. https://doi.org/10.3390/foods13010106
Gao J, Geng H, Chai R, Wu T, Huang W, You Y, Zhan J. Fungal Community Composition and Its Relationship with Volatile Compounds during Spontaneous Fermentation of Cabernet Sauvignon from Two Chinese Wine-Growing Regions. Foods. 2024; 13(1):106. https://doi.org/10.3390/foods13010106
Chicago/Turabian StyleGao, Jie, Huiying Geng, Ruru Chai, Tianyang Wu, Weidong Huang, Yilin You, and Jicheng Zhan. 2024. "Fungal Community Composition and Its Relationship with Volatile Compounds during Spontaneous Fermentation of Cabernet Sauvignon from Two Chinese Wine-Growing Regions" Foods 13, no. 1: 106. https://doi.org/10.3390/foods13010106
APA StyleGao, J., Geng, H., Chai, R., Wu, T., Huang, W., You, Y., & Zhan, J. (2024). Fungal Community Composition and Its Relationship with Volatile Compounds during Spontaneous Fermentation of Cabernet Sauvignon from Two Chinese Wine-Growing Regions. Foods, 13(1), 106. https://doi.org/10.3390/foods13010106








