Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain, DNA Isolation and Genome Sequencing
2.2. Genome Features of L. salivarius P1CEA3
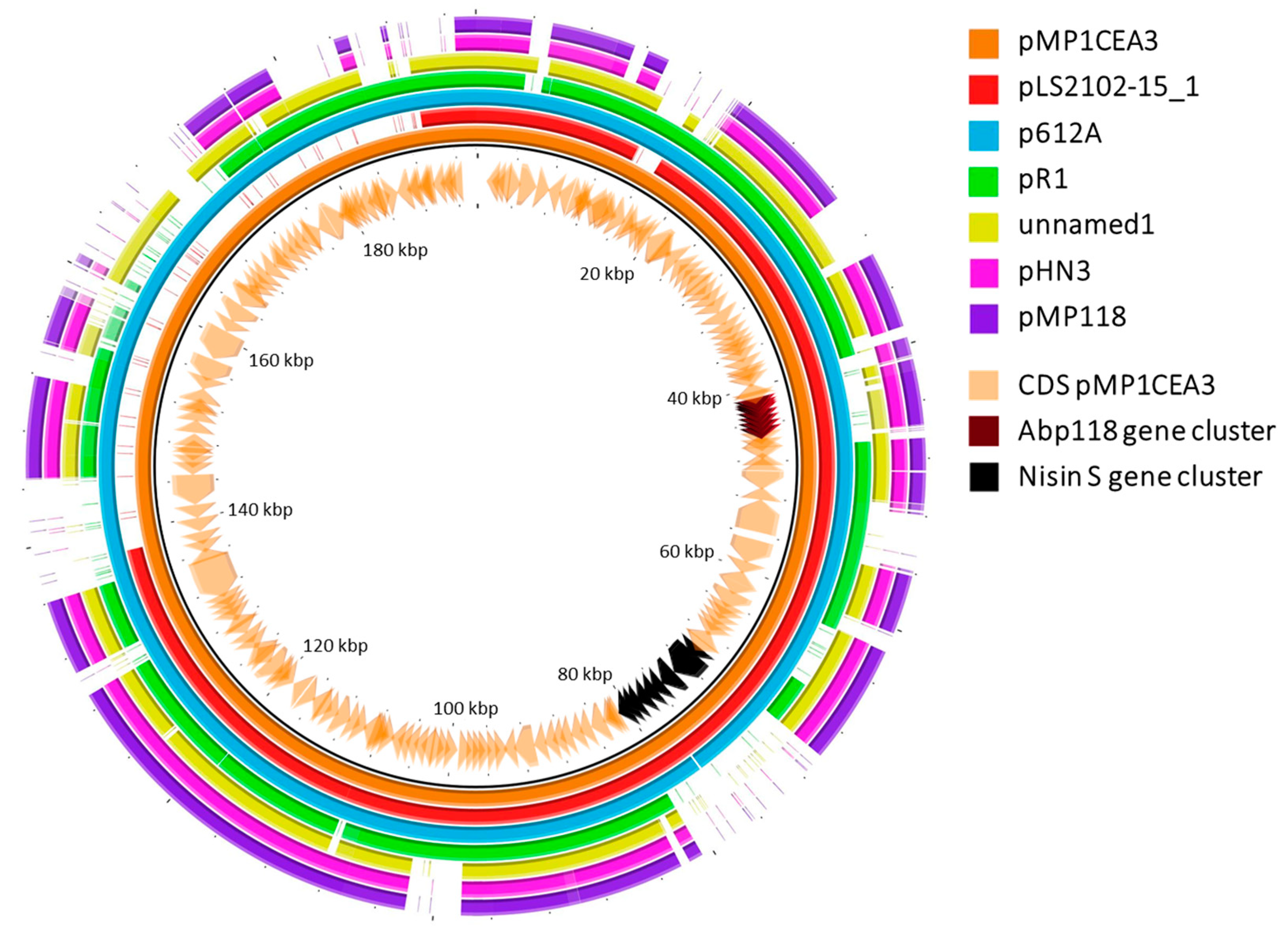
2.3. Comparison of Plasmids of L. salivarius P1CEA3 with Similar Plasmids
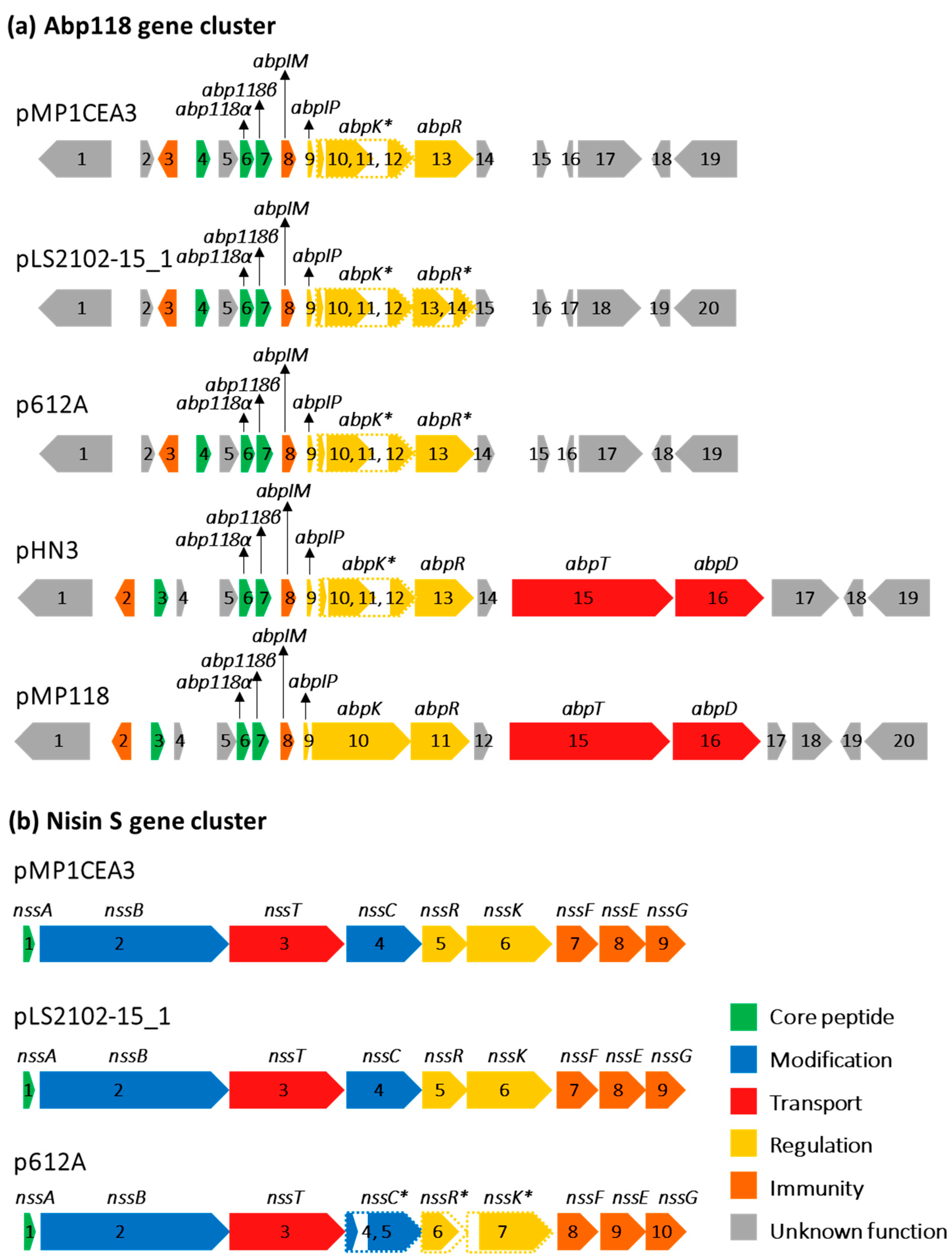
2.4. Bacteriocins and Secondary Metabolites
2.5. Transferable Antibiotic Resistances
2.6. Virulence and Pathogenicity
2.7. Mobile Genetic Elements (MGE): Insertion Sequences (IS), Genomic Islands (GI) and Prophages
2.8. CRISPR/CRISPR-Cas Systems
2.9. Production of Biogenic Amines (BA) and Hemolytic and Gelatinase Activities
2.10. Probiotic Associated Traits
3. Results and Discussion
3.1. Features Associated to the L. salivarius P1CEA3 Genome
3.2. Comparison of the L. salivarius P1CEA3 Plasmids with Other Plasmids
3.3. Bacteriocins and Secondary Metabolites
3.4. Transferable Antibiotic Resistances
3.5. Virulence and Pathogenicity
3.6. Mobile Genetic Elements (MGE)
3.7. CRISPR/CRISPR/Cas Systems
3.8. Biogenic Amines (BA), Hemolysin and Gelatinase Production
3.9. Probiotic-Related Genes
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial Resistance: One Health Approach. Vet. World 2022, 15, 743. [Google Scholar] [CrossRef]
- van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A Viable Alternative to Antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Ben Lagha, A.; Haas, B.; Gottschalk, M.; Grenier, D. Antimicrobial Potential of Bacteriocins in Poultry and Swine Production. Vet. Res. 2017, 48, 22. [Google Scholar] [CrossRef]
- Pircalabioru, G.G.; Popa, L.I.; Marutescu, L.; Gheorghe, I.; Popa, M.; Czobor Barbu, I.; Cristescu, R.; Chifiriuc, M.-C.; Millán, S.E. Pharmaceutics Bacteriocins in the Era of Antibiotic Resistance: Rising to the Challenge. Pharmaceutics 2021, 13, 196. [Google Scholar] [CrossRef]
- Rea, M.C.; Dobson, A.; O’Sullivan, O.; Crispie, F.; Fouhy, F.; Cotter, P.D.; Shanahan, F.; Kiely, B.; Hill, C.; Paul Ross, R. Effect of Broad- and Narrow-Spectrum Antimicrobials on Clostridium difficile and Microbial Diversity in a Model of the Distal Colon. Proc. Natl. Acad. Sci. USA 2011, 108, 4639–4644. [Google Scholar] [CrossRef]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and Emerging Applications of Bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Drider, D. Gut Microbiota Is an Important Source of Bacteriocins and Their In situ Expression Can Be Explored for Treatment of Bacterial Infections. Probiotics Antimicrob. Proteins 2021, 13, 1759–1765. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Stergiou, O.S.; Tegopoulos, K.; Kiousi, D.E.; Tsifintaris, M.; Papageorgiou, A.C.; Tassou, C.C.; Chorianopoulos, N.; Kolovos, P.; Galanis, A. Whole-Genome Sequencing, Phylogenetic and Genomic Analysis of Lactiplantibacillus pentosus L33, a Potential Probiotic Strain Isolated From Fermented Sausages. Front. Microbiol. 2021, 12, 746659. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, H.; Chen, Y.; Chen, H.; Wang, X.; Yuan, W. Lactobacillus paracasei S16 Alleviates Lumbar Disc Herniation by Modulating Inflammation Response and Gut Microbiota. Front. Nutr 2021, 8, 701644. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Lee, E.B.; Hsu, W.H.; Suk, K.; Sayem, S.A.J.; Ullah, H.M.A.; Lee, S.J.; Park, S.C. Probiotics and Postbiotics as an Alternative to Antibiotics: An Emphasis on Pigs. Pathogens 2023, 12, 874. [Google Scholar] [CrossRef] [PubMed]
- Indo, Y.; Kitahara, S.; Tomokiyo, M.; Araki, S.; Islam, M.A.; Zhou, B.; Albarracin, L.; Miyazaki, A.; Ikeda-Ohtsubo, W.; Nochi, T.; et al. Ligilactobacillus salivarius Strains Isolated from the Porcine Gut Modulate Innate Immune Responses in Epithelial Cells and Improve Protection Against Intestinal Viral-Bacterial Superinfection. Front. Immunol. 2021, 12, 652923. [Google Scholar] [CrossRef]
- O’Shea, E.F.; O’Connor, P.M.; Raftis, E.J.; O’Toole, P.W.; Stanton, C.; Cotter, P.D.; Paul Ross, R.; Hill, C. Subspecies Diversity in Bacteriocin Production by Intestinal Lactobacillus salivarius Strains. Gut Microbes 2012, 3, 468–473. [Google Scholar] [CrossRef]
- Draper, L.A.; Cotter, P.D.; Hill, C.; Ross, R.P. Lantibiotic Resistance. Microbiol. Mol. Biol. Rev. 2015, 79, 171–191. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.d.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the Characterisation of Microorganisms Used as Feed Additives or as Production Organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef]
- Castro-López, C.; García, H.S.; Cristian, G.; Martínez-Ávila, G.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Genomics-Based Approaches to Identify and Predict the Health-Promoting and Safety Activities of Promising Probiotic Strains—A Probiogenomics Review. Trends Food Sci. Technol. 2021, 108, 148–163. [Google Scholar] [CrossRef]
- Pradhan, D.; Mallappa, R.H.; Grover, S. Comprehensive Approaches for Assessing the Safety of Probiotic Bacteria. Food Control 2020, 108, 106872. [Google Scholar] [CrossRef]
- Sevillano, E.; Peña, N.; Lafuente, I.; Cintas, L.M.; Muñoz-Atienza, E.; Hernández, P.E.; Borrero, J. Nisin S, a Novel Nisin Variant Produced by Ligilactobacillus salivarius P1CEA3. Int. J. Mol. Sci. 2023, 24, 6813. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comp. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.v.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An Integrated Platform for Visualization and Analysis of High-Throughput Sequence-Based Experimental Data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’Neill, K.R.; Haft, D.H.; Dicuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline Reach with Protein Family Model Curation. Nucleic Acids Res. 2021, 49, D1020. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid Identification, Annotation and Analysis of Secondary Metabolite Biosynthesis Gene Clusters in Bacterial and Fungal Genome Sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive Prediction of Secondary Metabolite Structure and Biological Activity from Microbial Genome Sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and Precise Alignment of Raw Reads against Redundant Databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Araújo, C.; Muñoz-Atienza, E.; Ramírez, M.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Safety Assessment, Genetic Relatedness and Bacteriocin Activity of Potential Probiotic Lactococcus lactis Strains from Rainbow Trout (Oncorhynchus mykiss, Walbaum) and Rearing Environment. Eur. Food Res. Technol. 2015, 241, 647–662. [Google Scholar] [CrossRef]
- Klare, I.; Konstabel, C.; Müller-Bertling, S.; Reissbrodt, R.; Huys, G.; Vancanneyt, M.; Swings, J.; Goossens, H.; Witte, W. Evaluation of New Broth Media for Microdilution Antibiotic Susceptibility Testing of Lactobacilli, Pediococci, Lactococci, and Bifidobacteria. Appl. Environ. Microbiol. 2005, 71, 8982–8986. [Google Scholar] [CrossRef]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. MobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e0099122. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of Mobile Genetic Elements Associated with Antibiotic Resistance in Salmonella Enterica Using a Newly Developed Web Tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2004, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Hsiao, W.W.L.; Brinkman, F.S.L. Evaluation of Genomic Island Predictors Using a Comparative Genomics Approach. BMC Bioinform. 2008, 9, 329. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, H.X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An Integrative Hunting Tool for Active Prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-Throughput Prophage Sequence Annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Atienza, E.; Gómez-Sala, B.; Araújo, C.; Campanero, C.; Del Campo, R.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Antimicrobial Activity, Antibiotic Susceptibility and Virulence Factors of Lactic Acid Bacteria of Aquatic Origin Intended for Use as Probiotics in Aquaculture. BMC Microbiol. 2013, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Raftis, E.J.; Forde, B.M.; Claesson, M.J.; O’Toole, P.W. Unusual Genome Complexity in Lactobacillus salivarius JCM1046. BMC Genom. 2014, 15, 771. [Google Scholar] [CrossRef] [PubMed]
- Leser, T.D.; Amenuvor, J.Z.; Jensen, T.K.; Lindecrona, R.H.; Boye, M.; Møller, K. Culture-Independent Analysis of Gut Bacteria: The Pig Gastrointestinal Tract Microbiota Revisited. Appl. Environ. Microbiol. 2002, 68, 673–690. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Flynn, S.; Li, Y.; Claesson, M.J.; van Pijkeren, J.P.; Collins, J.K.; van Sinderen, D.; O’Toole, P.W. Characterization of Endogenous Plasmids from Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 2008, 74, 3216–3228. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Canchaya, C.; Fang, F.; Raftis, E.; Ryan, K.A.; van Pijkeren, J.P.; van Sinderen, D.; O’Toole, P.W. Distribution of Megaplasmids in Lactobacillus salivarius and Other Lactobacilli. J. Bacteriol. 2007, 189, 6128. [Google Scholar] [CrossRef] [PubMed]
- Flynn, S.; van Sinderen, D.; Thornton, G.M.; Holo, H.; Nes, I.F.; Collins, J.K. Characterization of the Genetic Locus Responsible for the Production of ABP-118, a Novel Bacteriocin Produced by the Probiotic Bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 2002, 148, 973–984. [Google Scholar] [CrossRef]
- Çataloluk, O. Molecular Characterization of the Gene Encoding for the Salivaricin B Activity and Its Flanking Sequences. Turk. J. Biol. 2001, 25, 379–386. [Google Scholar]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin Production: A Probiotic Trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef]
- Okoye, C.O.; Dong, K.; Wang, Y.; Gao, L.; Li, X.; Wu, Y.; Jiang, J. Comparative Genomics Reveals the Organic Acid Biosynthesis Metabolic Pathways among Five Lactic Acid Bacterial Species Isolated from Fermented Vegetables. New Biotechnol. 2022, 70, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Liu, L.; Fu, T.; Li, C.; Jin, N.; Zhang, H.; Li, C.; Liu, Y.; Zhao, C. Genome Sequence and Evaluation of Safety and Probiotic Potential of Lactiplantibacillus plantarum LPJZ-658. Microorganisms 2023, 11, 1620. [Google Scholar] [CrossRef]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Front. Microbiol. 2019, 10, 478460. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-Wide Assessment of Antibiotic Resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Lyu, L.; Wang, Y.; Zhang, Y.; Guo, X.; Chen, Q.; Liu, C. Safety Assessment and Probiotic Characteristics of Enterococcus lactis JDM1. Microb. Pathog. 2022, 163, 105380. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial Insertion Sequences: Their Genomic Impact and Diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; van der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic Islands: Tools of Bacterial Horizontal Gene Transfer and Evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef] [PubMed]
- Argov, T.; Azulay, G.; Pasechnek, A.; Stadnyuk, O.; Ran-Sapir, S.; Borovok, I.; Sigal, N.; Herskovits, A.A. Temperate Bacteriophages as Regulators of Host Behavior. Curr. Opin. Microbiol. 2017, 38, 81–87. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A Web Tool to Identify Clustered Regularly Interspaced Short Palindromic Repeats. Nucleic Acids Res. 2007, 35, W52. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Evolutionary Plasticity and Functional Versatility of CRISPR Systems. PLoS Biol. 2022, 20, e3001481. [Google Scholar] [CrossRef]
- Teklemariam, A.D.; Al-Hindi, R.R.; Qadri, I.; Alharbi, M.G.; Ramadan, W.S.; Ayubu, J.; Al-Hejin, A.M.; Hakim, R.F.; Hakim, F.F.; Hakim, R.F.; et al. The Battle between Bacteria and Bacteriophages: A Conundrum to Their Immune System. Antibiotics 2023, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- Crawley, A.B.; Henriksen, E.D.; Stout, E.; Brandt, K.; Barrangou, R. Characterizing the Activity of Abundant, Diverse and Active CRISPR-Cas Systems in Lactobacilli. Sci. Rep. 2018, 8, 11544. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.E.; Manca de Nadra, M.C. Biogenic Amine Production by Lactobacillus. J. Appl. Microbiol. 2001, 90, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Ladero, V.; Alvarez, M.A.; Lucas, P.M. Putrescine Production via the Ornithine Decarboxylation Pathway Improves the Acid Stress Survival of Lactobacillus brevis and Is Part of a Horizontally Transferred Acid Resistance Locus. Int. J. Food Microbiol. 2014, 175, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Jiménez, E.; Olivares, M.; Marín, M.L.; Fernández, L.; Xaus, J.; Rodríguez, J.M. Lactobacillus salivarius CECT 5713, a Potential Probiotic Strain Isolated from Infant Feces and Breast Milk of a Mother–Child Pair. Int. J. Food Microbiol. 2006, 112, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.; van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Umanets, A.; Surono, I.S.; Venema, K. I Am Better than I Look: Genome Based Safety Assessment of the Probiotic Lactiplantibacillus plantarum IS-10506. BMC Genom. 2023, 24, 518. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Li, Y.; Leahy, S.; Canchaya, C.; van Pijkeren, J.P.; Cerdeño-Tárraga, A.M.; Parkhill, J.; Flynn, S.; O’Sullivan, G.C.; Collins, J.K.; et al. Multireplicon Genome Architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 2006, 103, 6718–6723. [Google Scholar] [CrossRef]
- Yang, C.; Huang, W.; Sun, Y.; You, L.; Jin, H.; Sun, Z. Effect of Probiotics on Diversity and Function of Gut Microbiota in Moschus Berezovskii. Arch. Microbiol. 2021, 203, 3305–3315. [Google Scholar] [CrossRef]
- Gokey, T.; Halavaty, A.S.; Minasov, G.; Anderson, W.F.; Kuhn, M.L. Structure of the Bacillus anthracis DTDP-l-Rhamnose Biosynthetic Pathway Enzyme: DTDP-α-d-Glucose 4,6-Dehydratase, RfbB. J. Struct. Biol. 2018, 202, 175–181. [Google Scholar] [CrossRef]
- Boels, I.C.; Beerthuyzen, M.M.; Kosters, M.H.W.; van Kaauwen, M.P.W.; Kleerebezem, M.; de Vos, W.M. Identification and Functional Characterization of the Lactococcus lactis Rfb Operon, Required for DTDP-Rhamnose Biosynthesis. J. Bacteriol. 2004, 186, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Balzaretti, S.; Taverniti, V.; Guglielmetti, S.; Fiore, W.; Minuzzo, M.; Ngo, H.N.; Ngere, J.B.; Sadiq, S.; Humphreys, P.N.; Laws, A.P. A Novel Rhamnose-Rich Hetero-Exopolysaccharide Isolated from Lactobacillus paracasei DG Activates THP-1 Human Monocytic Cells. Appl. Environ. Microbiol. 2017, 83, 2702–2718. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.S.; Kumar, S.; Mohanty, A.K.; Grover, S.; Kaushik, J.K. Mechanistic Insights into the Host-Microbe Interaction and Pathogen Exclusion Mediated by the Mucus-Binding Protein of Lactobacillus plantarum. Sci. Rep. 2018, 8, 14198. [Google Scholar] [CrossRef] [PubMed]
- Edelman, S.M.; Lehti, T.A.; Kainulainen, V.; Antikainen, J.; Kylväjä, R.; Baumann, M.; Westerlund-Wikström, B.; Korhonen, T.K. Identification of a High-Molecular-Mass Lactobacillus Epithelium Adhesin (LEA) of Lactobacillus crispatus ST1 That Binds to Stratified Squamous Epithelium. Microbiology 2012, 158, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Zuo, F.; Appaswamy, A.; Gebremariam, H.G.; Jonsson, A.B. Role of Sortase A in Lactobacillus gasseri Kx110A1 Adhesion to Gastric Epithelial Cells and Competitive Exclusion of Helicobacter pylori. Front. Microbiol. 2019, 10, 480331. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Amaretti, A.; Raimondi, S. Folate Production by Probiotic Bacteria. Nutrients 2011, 3, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, R.; Oh, J.K.; Song, J.H.; Kang, D.K. Gut Microbiome-Produced Metabolites in Pigs: A Review on Their Biological Functions and the Influence of Probiotics. J. Anim. Sci. Technol. 2022, 64, 671–695. [Google Scholar] [CrossRef]
- Pessione, E. Lactic Acid Bacteria Contribution to Gut Microbiota Complexity: Lights and Shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef]
- Yang, F.; Hou, C.; Zeng, X.; Qiao, S. The Use of Lactic Acid Bacteria as a Probiotic in Swine Diets. Pathogens 2015, 4, 34–45. [Google Scholar] [CrossRef]
- Vitetta, L.; Coulson, S.; Thomsen, M.; Nguyen, T.; Hall, S. Probiotics, D–Lactic Acidosis, Oxidative Stress and Strain Specificity. Gut Microbes 2017, 8, 311–322. [Google Scholar] [CrossRef]
- Parente, E.; Ciocia, F.; Ricciardi, A.; Zotta, T.; Felis, G.E.; Torriani, S. Diversity of Stress Tolerance in Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus paraplantarum: A Multivariate Screening Study. Int. J. Food Microbiol. 2010, 144, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Sánchez, M.; Carrasco-Navarro, U.; Juárez-Castelán, C.; Lozano-Aguirre Beltrán, L.; Pérez-Chabela, M.L.; Ponce-Alquicira, E. Probiotic Properties and Proteomic Analysis of Pediococcus Pentosaceus 1101. Foods 2023, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Feito, J.; Contente, D.; Ponce-Alonso, M.; Díaz-Formoso, L.; Araújo, C.; Peña, N.; Borrero, J.; Gómez-Sala, B.; del Campo, R.; Muñoz-Atienza, E.; et al. Draft Genome Sequence of Lactococcus lactis subsp. cremoris WA2-67: A Promising Nisin-Producing Probiotic Strain Isolated from the Rearing Environment of a Spanish Rainbow Trout (Oncorhynchus mykiss, Walbaum) Farm. Microorganisms 2022, 10, 521. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Han, G.G.; Kim, E.B.; Choi, Y.J. Comparative Genomics of Lactobacillus salivarius Strains Focusing on Their Host Adaptation. Microbiol. Res. 2017, 205, 48–58. [Google Scholar] [CrossRef]
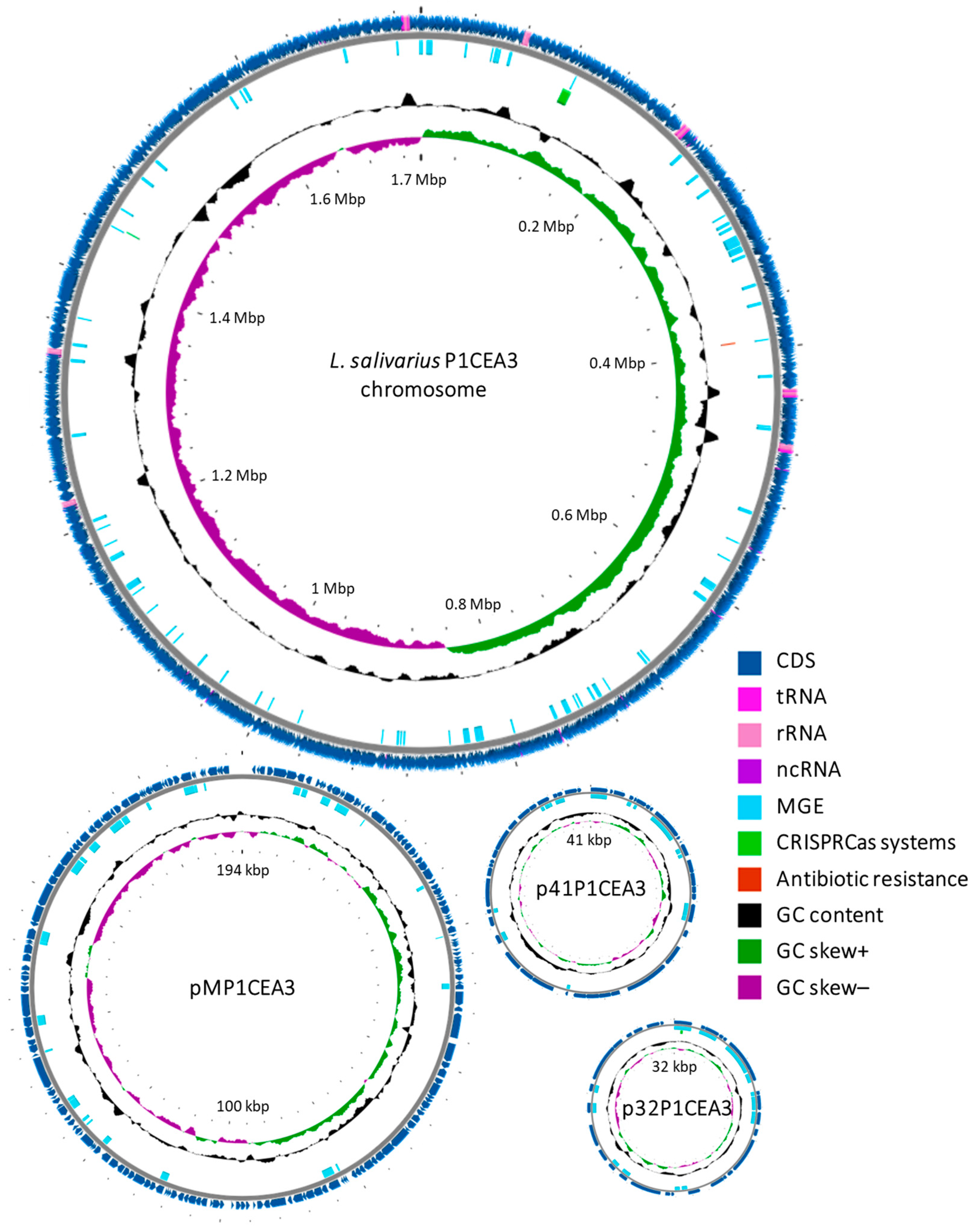
| Feature | Chromosome | pMP1CEA3 | p41P1CEA3 | p32P1CEA3 |
|---|---|---|---|---|
| Replicon size (bp) | 1,739,667 | 194,140 | 41,764 | 32,196 |
| GC content (%) | 32.5 | 31.5 | 39.5 | 38 |
| Topology | Circular | Circular | Circular | Circular |
| % of genome size | 86.6 | 9.7 | 2.1 | 1.6 |
| Coding genes | 1624 | 191 | 40 | 27 |
| Pseudogenes | 15 | 7 | 5 | 6 |
| rRNA | 22 | 0 | 0 | 0 |
| tRNAs | 77 | 0 | 0 | 0 |
| ncRNAs | 4 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sevillano, E.; Lafuente, I.; Peña, N.; Cintas, L.M.; Muñoz-Atienza, E.; Hernández, P.E.; Borrero, J. Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S. Foods 2024, 13, 107. https://doi.org/10.3390/foods13010107
Sevillano E, Lafuente I, Peña N, Cintas LM, Muñoz-Atienza E, Hernández PE, Borrero J. Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S. Foods. 2024; 13(1):107. https://doi.org/10.3390/foods13010107
Chicago/Turabian StyleSevillano, Ester, Irene Lafuente, Nuria Peña, Luis M. Cintas, Estefanía Muñoz-Atienza, Pablo E. Hernández, and Juan Borrero. 2024. "Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S" Foods 13, no. 1: 107. https://doi.org/10.3390/foods13010107
APA StyleSevillano, E., Lafuente, I., Peña, N., Cintas, L. M., Muñoz-Atienza, E., Hernández, P. E., & Borrero, J. (2024). Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S. Foods, 13(1), 107. https://doi.org/10.3390/foods13010107








