Construction of PCR-SERS Method for Detection of Vibrio parahaemolyticus
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Bacterial Strains and Culture Conditions
2.1.2. Main Reagents
2.2. Methods
2.2.1. PCR Method
2.2.2. Construction and Characterization of SERS Substrates
2.2.3. Establishment of Detection Methods
2.2.4. Screening of Signaling Molecules
2.2.5. Optimization of the PCR Amplification Time
2.2.6. Method Sensitivity Assessment
2.2.7. Method Specificity Assessment
2.2.8. Artificially Contaminated Sample Detection
2.2.9. Determination of the Actual Sample Recovery Rate
2.2.10. Data Processing
3. Results
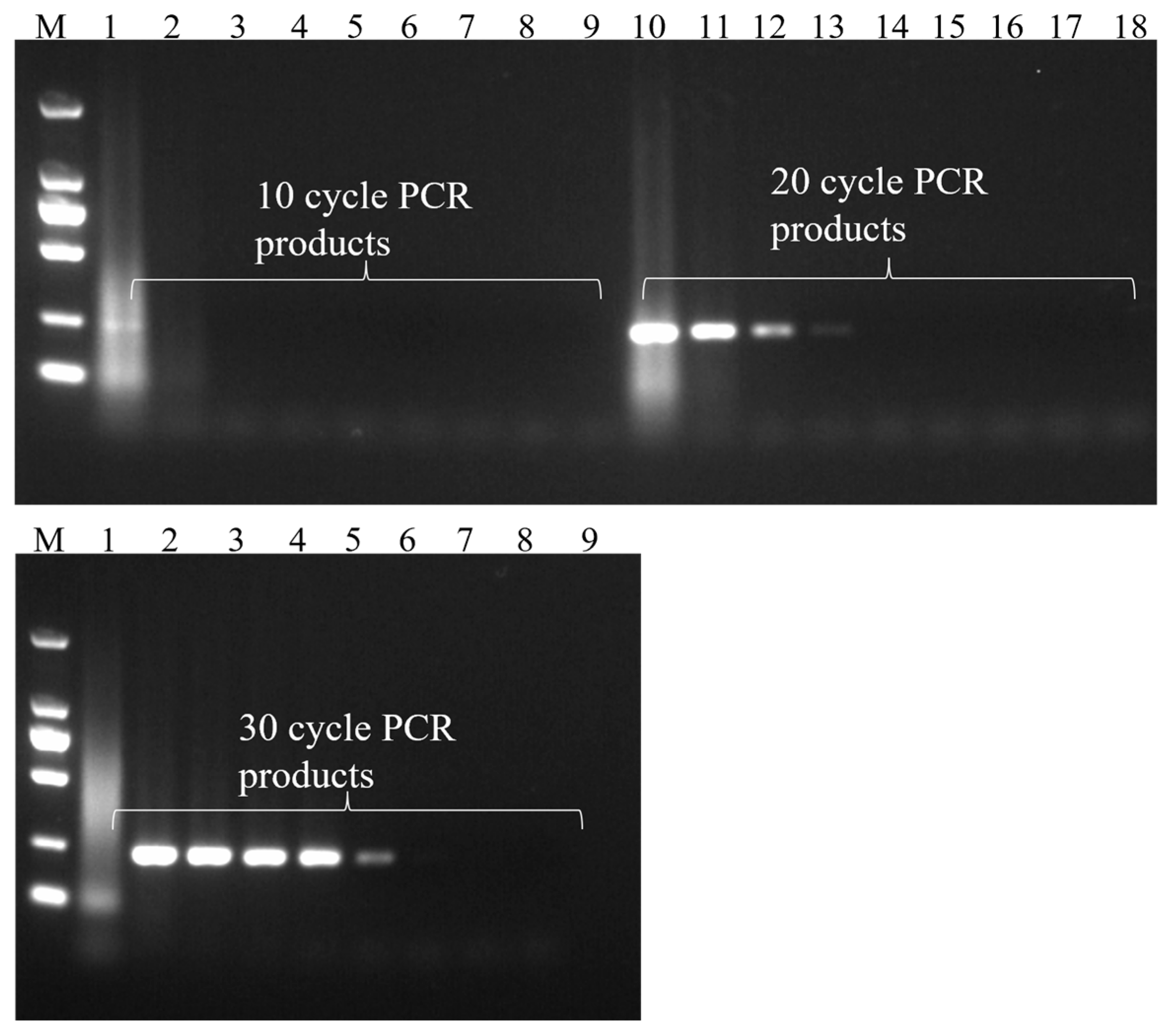
3.1. Establishment of Common PCR Methods
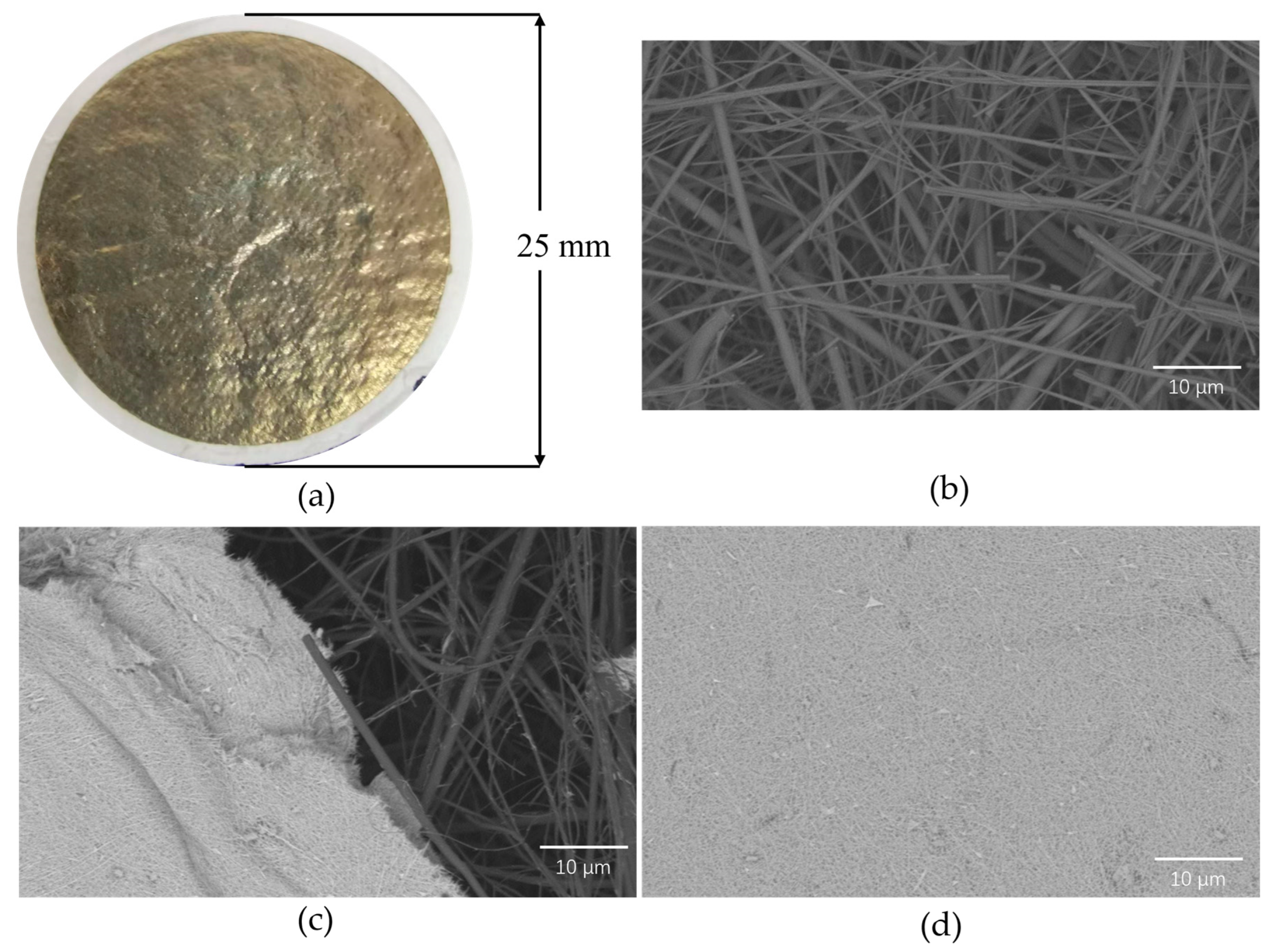
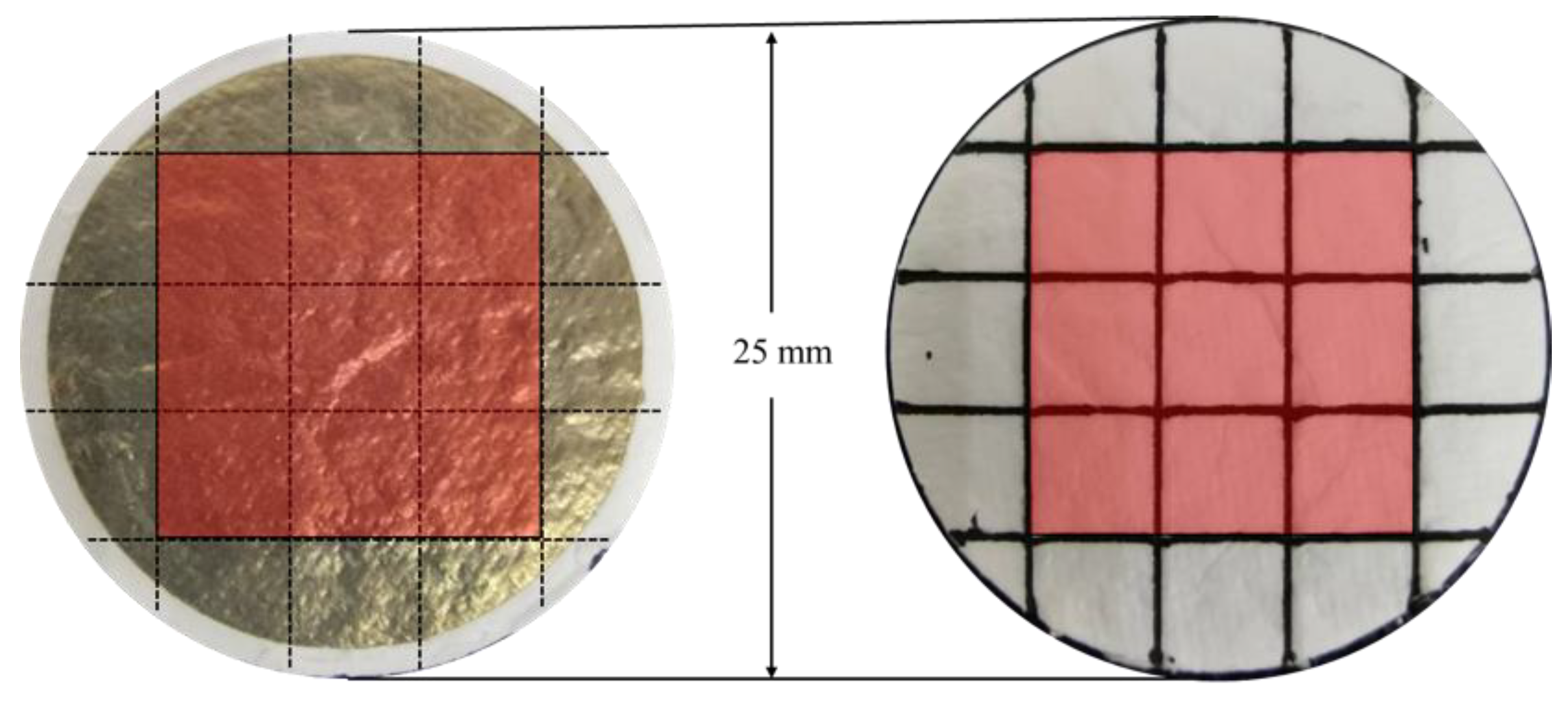
3.2. Construction of the Detection Substrate
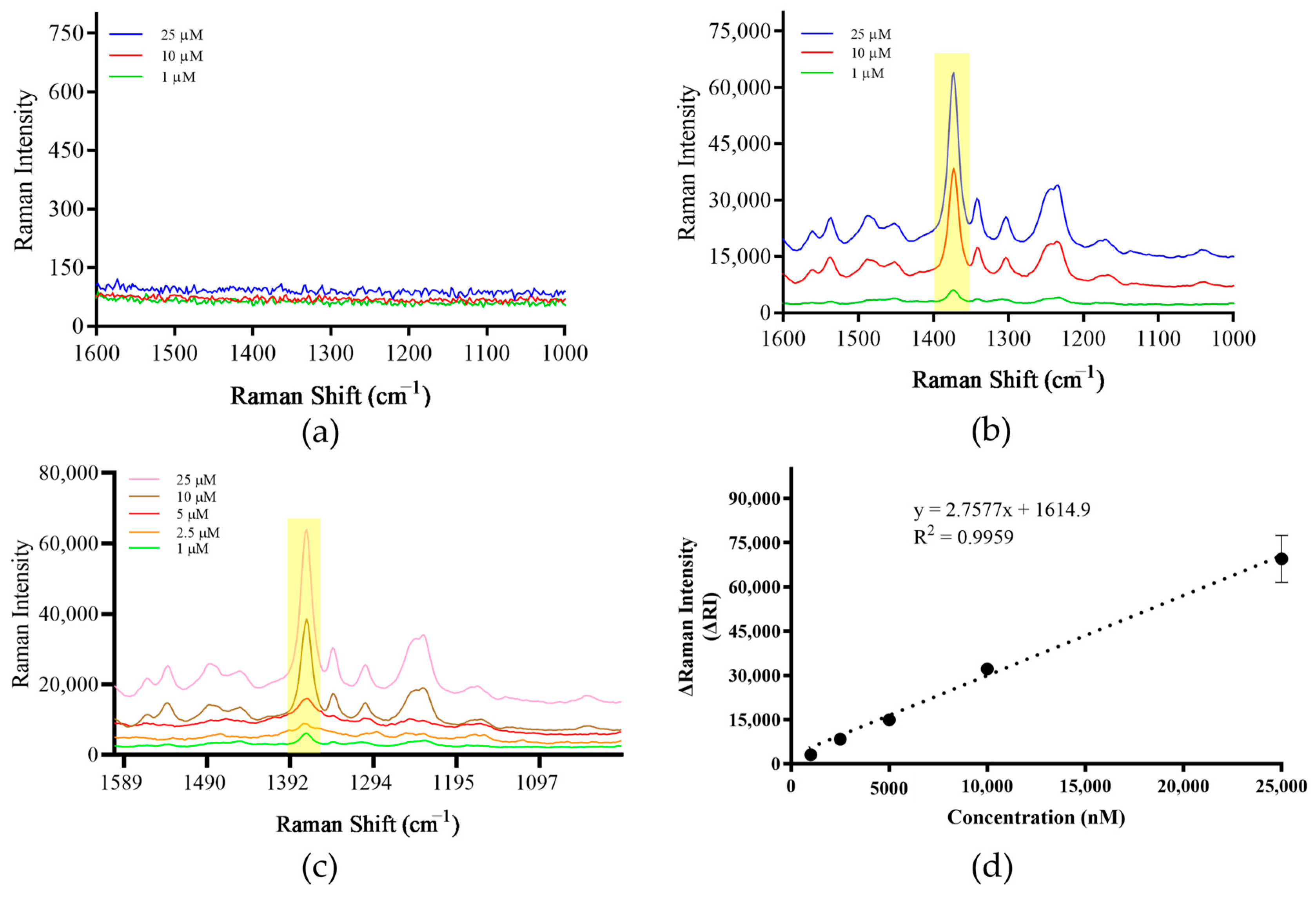
3.3. Establishment of Detection Methods
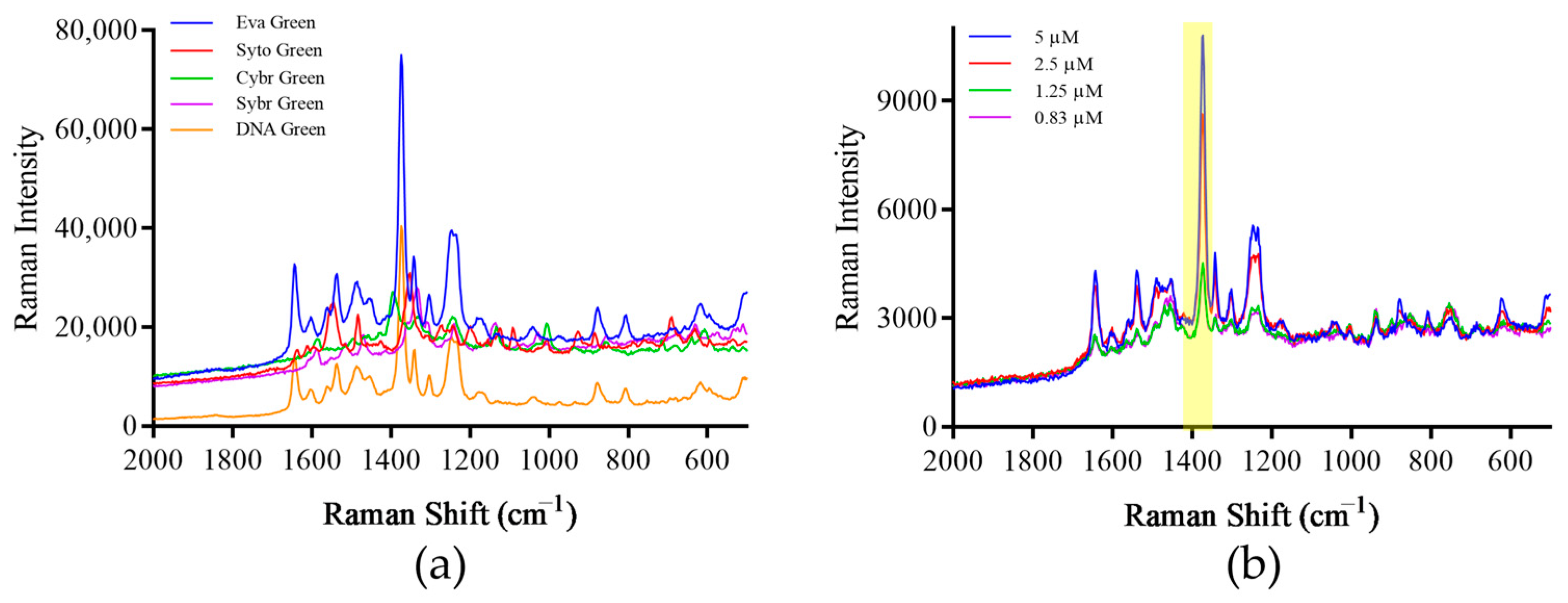
3.4. Screening of Signaling Molecules
3.5. PCR Amplification Time Optimization Results
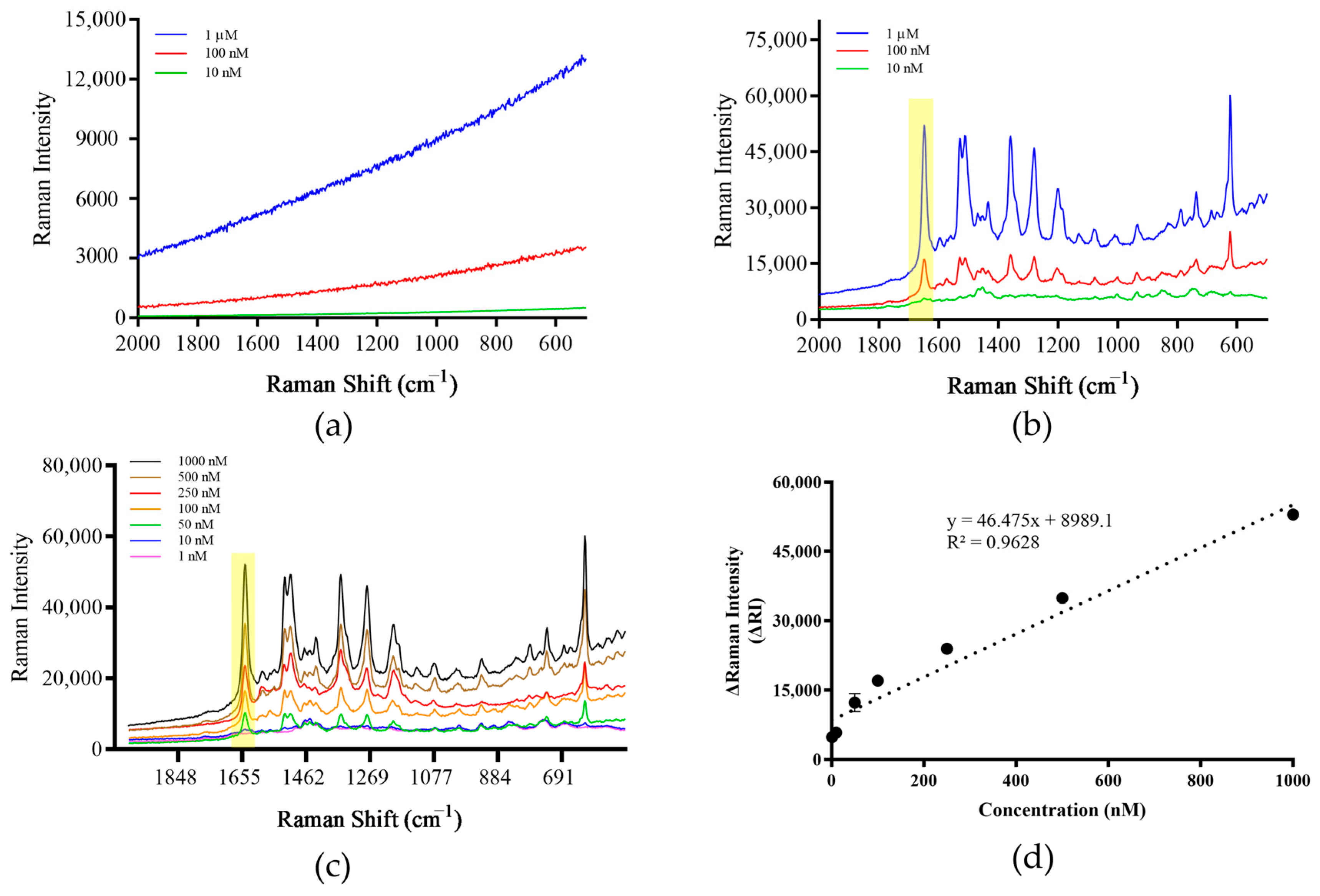
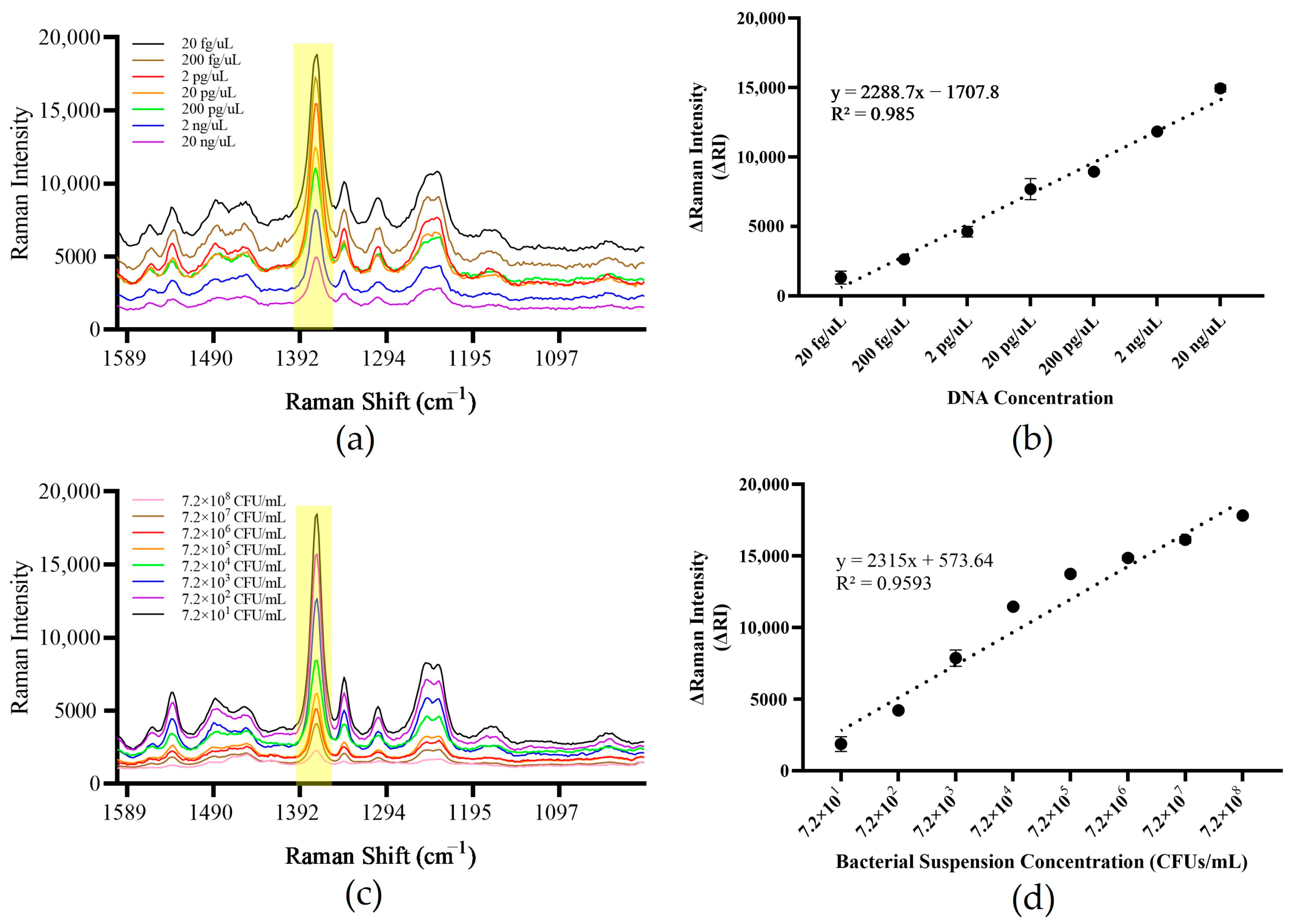
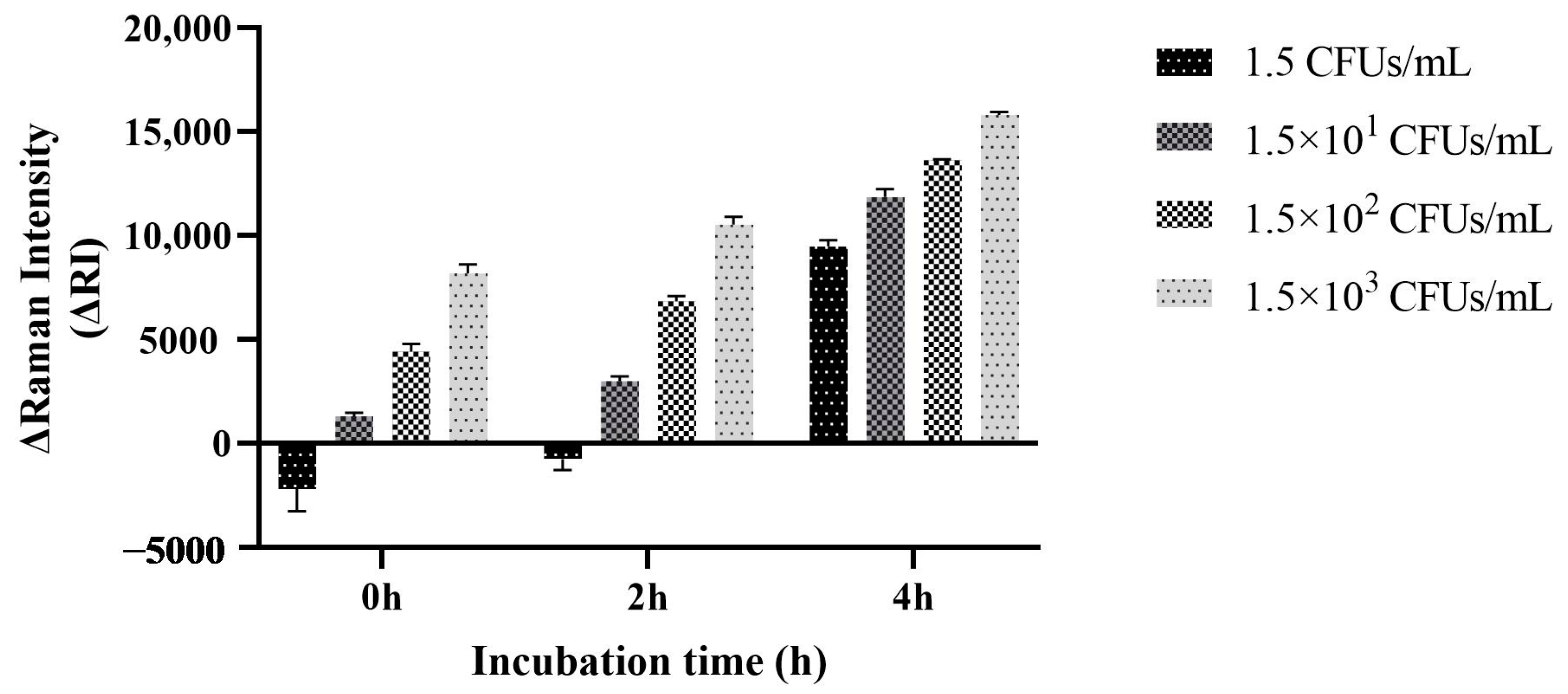
3.6. Method Sensitivity Assessment
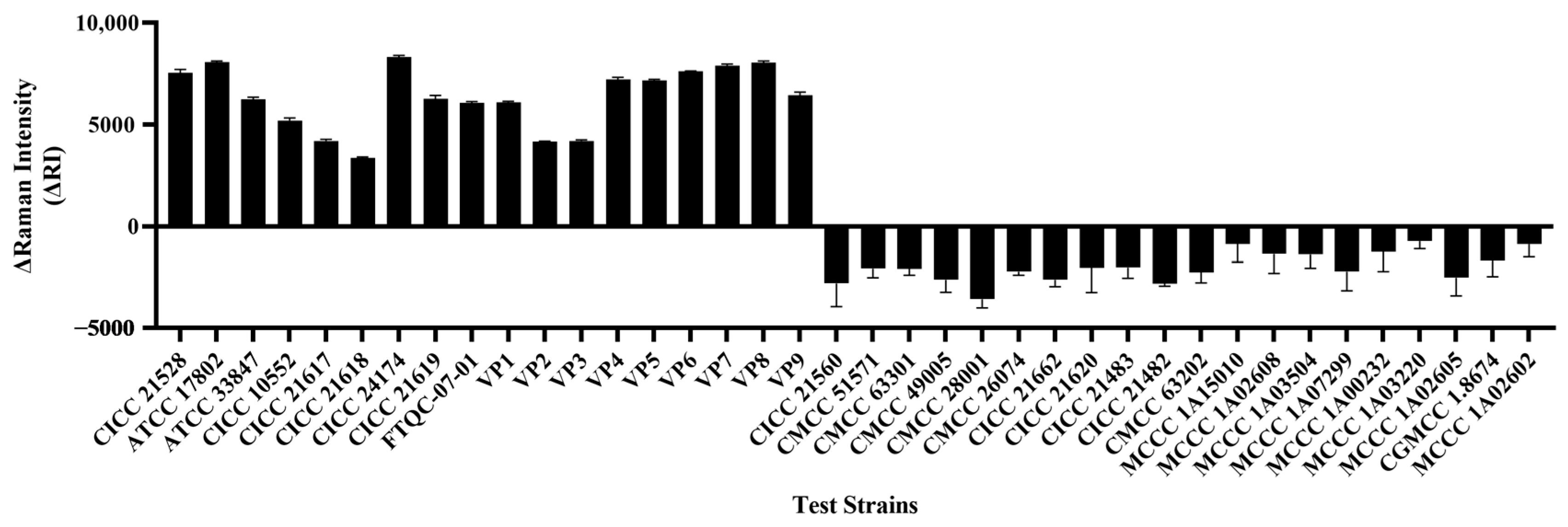
3.7. Method Specificity Assessment
3.8. Detection of Artificially Contaminated Samples
3.9. Determination of the Actual Sample Recovery Rate
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raman, C.V.; Krishnan, K.S. A New Type of Secondary Radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Raman, C.V. A Change of Wave-Length in Light Scattering. Nature 1928, 121, 619. [Google Scholar] [CrossRef]
- Liebel, M.; Calderon, I.; Pazos-Perez, N.; van Hulst, N.F.; Alvarez-Puebla, R.A. Widefield SERS for High-Throughput Nanoparticle Screening. Angew. Chem. Int. Ed. 2022, 61, e202200072. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ma, W.; Wang, X.; Li, S. Recent Progress of Surface-Enhanced Raman Spectroscopy for Bacteria Detection. Biosensors 2023, 13, 350. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, M.; Hill, I.R.; Pemble, M.E. Surface-Enhanced Raman Spectroscopy of 12CN− and 13CN− Adsorbed at Silver Electrodes. J. Electroanal. Chem. Interfacial Electrochem. 1982, 136, 361–370. [Google Scholar] [CrossRef]
- Ali, H.T.; Mateen, A.; Ashraf, F.; Javed, M.R.; Ali, A.; Mahmood, K.; Zohaib, A.; Amin, N.; Ikram, S.; Yusuf, M. A New SERS Substrate Based on Zn2GeO4 Nanostructures for the Rapid Identification of E. coli and Methylene Blue. Ceram. Int. 2021, 47, 27998–28003. [Google Scholar] [CrossRef]
- Hsu, K.-C.; Chen, D.-H. Microwave-Assisted Green Synthesis of Ag/Reduced Graphene Oxide Nanocomposite as a Surface-Enhanced Raman Scattering Substrate with High Uniformity. Nanoscale Res. Lett. 2014, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, G.; Zhang, C.; Wei, C.; Wang, X.; Gao, Y.; Li, H.; Huang, X.; Yuan, H.; Lu, G. Silver Nanowire-Templated Molecular Nanopatterning and Nanoparticle Assembly for Surface-Enhanced Raman Scattering. Chem.—A Eur. J. 2019, 25, 10561–10565. [Google Scholar] [CrossRef] [PubMed]
- Sohn, H.; Park, C.; Oh, J.-M.; Kang, S.W.; Kim, M.-J. Silver Nanowire Networks: Mechano-Electric Properties and Applications. Materials 2019, 12, 2526. [Google Scholar] [CrossRef]
- Kim, H.; Jang, H.; Moon, J.; Byun, J.; Jeong, J.; Jung, J.; Lim, E.-K.; Kang, T. Metal–Organic Frameworks: Metal–Organic Framework Coating for the Preservation of Silver Nanowire Surface-Enhanced Raman Scattering Platform. Adv. Mater. Interfaces 2019, 6, 1970088. [Google Scholar] [CrossRef]
- Lee, H.; Ye, W.; Lee, J.; Kim, H.; Byun, D. Silver Nanowire Micro-Ring Formation Using Immiscible Emulsion Droplets for Surface-Enhanced Raman Spectroscopy. Appl. Sci. 2020, 10, 8018. [Google Scholar] [CrossRef]
- Jeon, Y.; Kim, D.; Kwon, G.; Lee, K.; Oh, C.-S.; Kim, U.-J.; You, J. Detection of Nanoplastics Based on Surface-Enhanced Raman Scattering with Silver Nanowire Arrays on Regenerated Cellulose Films. Carbohydr. Polym. 2021, 272, 118470. [Google Scholar] [CrossRef]
- Pandey, P.; Vongphachanh, S.; Yoon, J.; Kim, B.; Choi, C.-J.; Sohn, J.I.; Hong, W.-K. Silver Nanowire-Network-Film-Coated Soft Substrates with Wrinkled Surfaces for Use as Stretchable Surface Enhanced Raman Scattering Sensors. J. Alloys Compd. 2021, 859, 157862. [Google Scholar] [CrossRef]
- Ankudze, B.; Asare, B.; Goffart, S.; Koistinen, A.; Nuutinen, T.; Matikainen, A.; Andoh, S.S.; Roussey, M.; Pakkanen, T.T. Hydraulically Pressed Silver Nanowire-Cotton Fibers as an Active Platform for Filtering and Surface-Enhanced Raman Scattering Detection of Bacteria from Fluid. Appl. Surf. Sci. 2019, 479, 663–668. [Google Scholar] [CrossRef]
- Macdonald, D.; Smith, E.; Faulds, K.; Graham, D. DNA Detection by SERS: Hybridisation Parameters and the Potential for Asymmetric PCR. Analyst 2020, 145, 1871–1877. [Google Scholar] [CrossRef]
- Wu, Y.; Dang, H.; Park, S.-G.; Chen, L.; Choo, J. SERS-PCR Assays of SARS-CoV-2 Target Genes Using Au Nanoparticles-Internalized Au Nanodimple Substrates. Biosens. Bioelectron. 2022, 197, 113736. [Google Scholar] [CrossRef]
- Lee, H.G.; Choi, W.; Yang, S.Y.; Kim, D.-H.; Park, S.-G.; Lee, M.-Y.; Jung, H.S. PCR-Coupled Paper-Based Surface-Enhanced Raman Scattering (SERS) Sensor for Rapid and Sensitive Detection of Respiratory Bacterial DNA. Sens. Actuators B Chem. 2021, 326, 128802. [Google Scholar] [CrossRef]
- Hu, A.; Kong, L.; Lu, Z.; Qiao, J.; Lv, F.; Meng, F.; Bie, X. Research on Nanogold-Assisted HRM-qPCR Technology for Highly Sensitive and Accurate Detection of Vibrio parahaemolyticus. LWT 2022, 162, 113488. [Google Scholar] [CrossRef]
- Wang, H.-X.; Zhao, Y.-W.; Li, Z.; Liu, B.-S.; Zhang, D. Development and Application of Aptamer-Based Surface-Enhanced Raman Spectroscopy Sensors in Quantitative Analysis and Biotherapy. Sensors 2019, 19, 3806. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.; Wang, J.; Li, H.; Jiao, T.; Chen, Q. Trends in the Bacterial Recognition Patterns Used in Surface Enhanced Raman Spectroscopy. TrAC Trends Anal. Chem. 2021, 142, 116310. [Google Scholar] [CrossRef]
- Rippa, M.; Castagna, R.; Pannico, M.; Musto, P.; Borriello, G.; Paradiso, R.; Galiero, G.; Bolletti Censi, S.; Zhou, J.; Zyss, J.; et al. Octupolar Metastructures for a Highly Sensitive, Rapid, and Reproducible Phage-Based Detection of Bacterial Pathogens by Surface-Enhanced Raman Scattering. ACS Sens. 2017, 2, 947–954. [Google Scholar] [CrossRef]
- Ho, C.-S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid Identification of Pathogenic Bacteria Using Raman Spectroscopy and Deep Learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef]
- Xu, M.-L.; Gao, Y.; Han, X.-X.; Zhao, B. Innovative Application of SERS in Food Quality and Safety: A Brief Review of Recent Trends. Foods 2022, 11, 2097. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, C.; Huang, Z.; Jiang, L. Simultaneous Quantitative Determination of Low-Concentration Preservatives and Heavy Metals in Tricholoma Matsutakes Based on SERS and FLU Spectral Data Fusion. Foods 2023, 12, 4267. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, C.; Li, W.; Li, L.; Zhang, P.; Zhu, C.; Ding, Y.; Sun, H. Rapid Indentification of Auramine O Dyeing Adulteration in Dendrobium Officinale, Saffron and Curcuma by SERS Raman Spectroscopy Combined with SSA-BP Neural Networks Model. Foods 2023, 12, 4124. [Google Scholar] [CrossRef]
- Lee, H.K.; Lee, Y.H.; Koh, C.S.L.; Phan-Quang, G.C.; Han, X.; Lay, C.L.; Sim, H.Y.F.; Kao, Y.-C.; An, Q.; Ling, X.Y. Designing Surface-Enhanced Raman Scattering (SERS) Platforms beyond Hotspot Engineering: Emerging Opportunities in Analyte Manipulations and Hybrid Materials. Chem. Soc. Rev. 2019, 48, 731–756. [Google Scholar] [CrossRef]
- Tang, X.; Chen, X.; Wang, H.; Yang, J.; Li, L.; Zhu, J.; Liu, Y. Virtual Screening Technology for Two Novel Peptides in Soybean as Inhibitors of α-Amylase and α-Glucosidase. Foods 2023, 12, 4387. [Google Scholar] [CrossRef] [PubMed]
- Cialla, D.; März, A.; Böhme, R.; Theil, F.; Weber, K.; Schmitt, M.; Popp, J. Surface-Enhanced Raman Spectroscopy (SERS): Progress and Trends. Anal. Bioanal. Chem. 2012, 403, 27–54. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Cao, L.; Zhang, X.; Jiao, R.; Ou, D.; Wang, Y.; Zhang, D.; Shen, Y.; Ling, N.; Ye, Y. A Novel Fluorescence Resonance Energy Transfer (FRET)-Based Paper Sensor with Smartphone for Quantitative Detection of Vibrio parahaemolyticus. Food Control 2023, 145, 109412. [Google Scholar] [CrossRef]
- Wang, S.; Hu, J.; Xiao, S.; Wang, M.; Yu, J.; Jia, Z.; Yu, Z.; Gan, N. Fluorescent/Electrochemical Dual-Signal Response Biosensing Strategy Mediated by DNAzyme-Ferrocene-Triggered Click Chemistry for Simultaneous Rapid Screening and Quantitative Detection of Vibrio parahaemolyticus. Sens. Actuators B Chem. 2023, 380, 133393. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, W.; Fath, T.; Li, F.; Fang, L.; Zhou, Z.; Zhang, D. Rapid and Simultaneous Detection of Viable Vibrio parahaemolyticus, Vibrio alginolyticus, and Vibrio cholerae by PMA-mPCR Assay in Aquatic Products. LWT 2023, 180, 114663. [Google Scholar] [CrossRef]
- Du, Q.; Ding, Y.; Wang, Y.; Wang, J.; Shao, Y.; Wang, X. A Novel Phagomagnetic Separation-ATP Bioluminescence (PhMS-BL) for Rapid and Sensitive Detection of Viable Vibrio parahaemolyticus in Aquatic Product. Food Chem. 2024, 439, 138113. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xiao, S.; Zeng, M.; Xie, H.; Gan, N. Dual-Mode Colorimetric-Electrochemical Biosensor for Vibrio parahaemolyticus Detection Based on CuO2 Nanodot-Encapsulated Metal-Organic Framework Nanozymes. Sens. Actuators B Chem. 2023, 387, 133835. [Google Scholar] [CrossRef]
- Wang, K.; Zeng, L.; Gong, L.; Ouyang, Y.; Yang, T.; Sun, T.; Zeng, H. Rapid and Accurate Sensing Platform Based on Lanthanide Metal-Organic Framework and Aptamer for Vibrio parahaemolyticus Diagnosis. Sens. Actuators B Chem. 2023, 389, 133881. [Google Scholar] [CrossRef]
- Li, J.; Lin, X.; Wu, J.; Ying, D.; Duan, N.; Wang, Z.; Wu, S. Multifunctional Magnetic Composite Nanomaterial for Colorimetric-SERS Dual-Mode Detection and Photothermal Sterilization of Vibrio parahaemolyticus. Chem. Eng. J. 2023, 477, 147113. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Chen, S.; Gong, T.; Zhu, Y. Surface-Enhanced Raman Scattering Properties of Multi-Walled Carbon Nanotubes Arrays-Ag Nanoparticles. Carbon 2016, 100, 395–407. [Google Scholar] [CrossRef]
- Pyrak, E.; Krajczewski, J.; Kowalik, A.; Kudelski, A.; Jaworska, A. Surface Enhanced Raman Spectroscopy for DNA Biosensors-How Far Are We? Molecules 2019, 24, 4423. [Google Scholar] [CrossRef] [PubMed]
- Jakhmola, A.; Krishnan, S.; Onesto, V.; Gentile, F.; Profeta, M.; Manikas, A.; Battista, E.; Vecchione, R.; Netti, P.A. Sustainable Synthesis and Theoretical Studies of Polyhedral Gold Nanoparticles Displaying High SERS Activity, NIR Absorption, and Cellular Uptake. Mater. Today Chem. 2022, 26, 101016. [Google Scholar] [CrossRef]
- Qiu, M.; Tang, L.; Wang, J.; Xu, Q.; Zheng, S.; Weng, S. SERS with Flexible β-CD@AuNP/PTFE Substrates for In Situ Detection and Identification of PAH Residues on Fruit and Vegetable Surfaces Combined with Lightweight Network. Foods 2023, 12, 3096. [Google Scholar] [CrossRef]
- Ha, M.; Kim, J.-H.; You, M.; Li, Q.; Fan, C.; Nam, J.-M. Multicomponent Plasmonic Nanoparticles: From Heterostructured Nanoparticles to Colloidal Composite Nanostructures. Chem. Rev. 2019, 119, 12208–12278. [Google Scholar] [CrossRef]
- Li, J.-F.; Zhang, Y.-J.; Ding, S.-Y.; Panneerselvam, R.; Tian, Z.-Q. Core–Shell Nanoparticle-Enhanced Raman Spectroscopy. Chem. Rev. 2017, 117, 5002–5069. [Google Scholar] [CrossRef]
- Guo, Q.; Xu, M.; Yuan, Y.; Gu, R.; Yao, J. Self-Assembled Large-Scale Monolayer of Au Nanoparticles at the Air/Water Interface Used as a SERS Substrate. Langmuir 2016, 32, 4530–4537. [Google Scholar] [CrossRef]
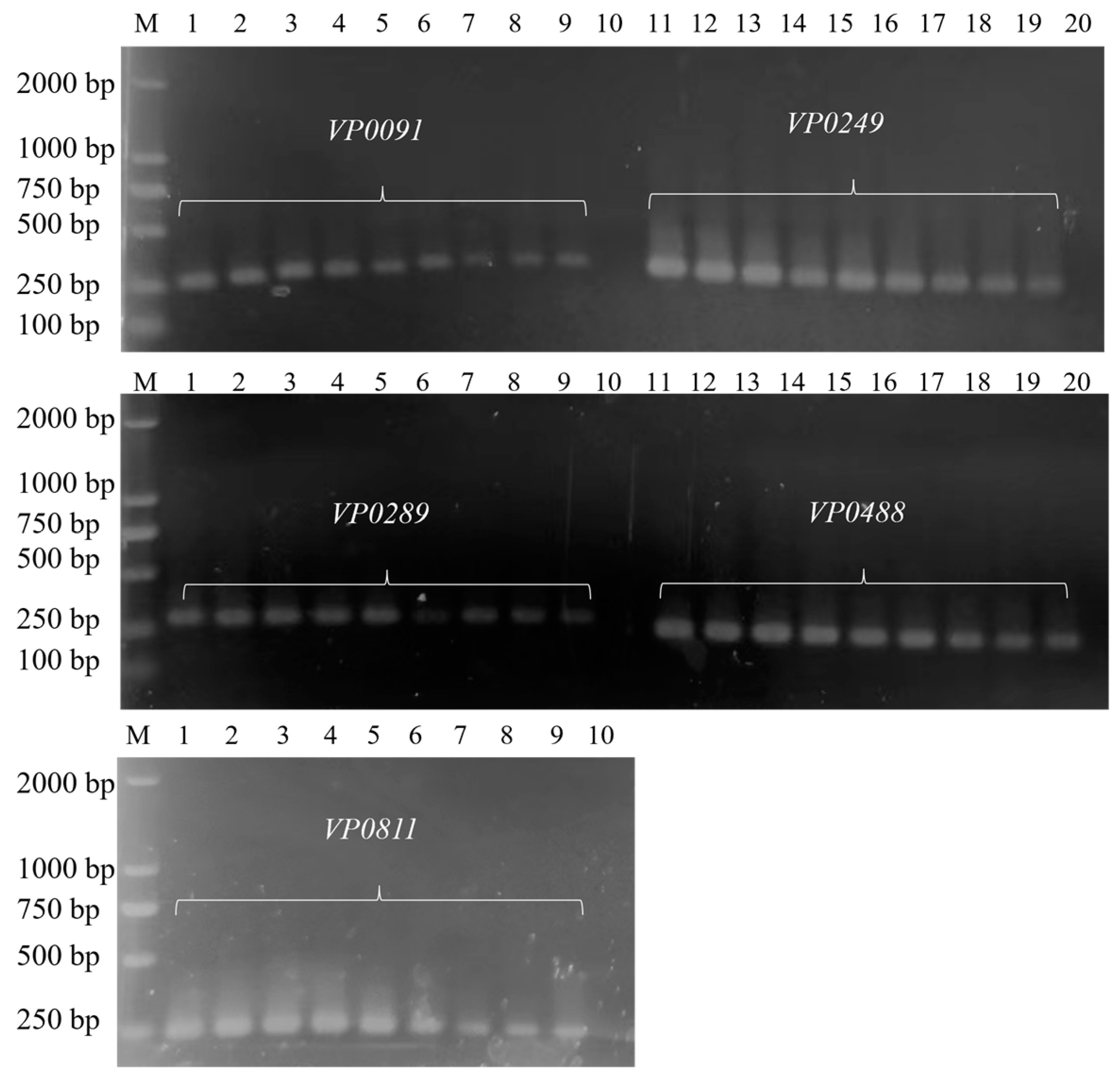

| Gene | Sequences (5′-3′) | Product Length (bp) |
|---|---|---|
| VP0091 | F: TCAGGCGATGATTGTGCTGT | 270 |
| R: GCGAAATTCATTACCCCCGC | ||
| VP0249 | F: GGCCGATTTTCATCGTCAGC | 258 |
| R: GATTCATGGTATCCGCCCGT | ||
| VP0289 | F: GACTGGGTGATCGGCTTTGA | 289 |
| R: CGAGGTAAGCCGCAATTTGG | ||
| VP0488 | F: GTGAACTGGCAACAAGGCAG | 252 |
| R: AGTGCGAGTTTCCAAGTCCC | ||
| VP0811 | F: AACAAGCCGAGTTCTAGCCC | 244 |
| R: GTCTCGCCTTCAGAGCCAAT |
| Strain Category | Strain No. | Detection Result |
|---|---|---|
| Vibrio parahaemolyticus | CICC 21528 | + |
| Vibrio parahaemolyticus | ATCC 17802 | + |
| Vibrio parahaemolyticus | ATCC 33847 | + |
| Vibrio parahaemolyticus | CICC 10552 | + |
| Vibrio parahaemolyticus | CICC 21617 | + |
| Vibrio parahaemolyticus | CICC 21618 | + |
| Vibrio parahaemolyticus | CICC 24174 | + |
| Vibrio parahaemolyticus | CICC 21619 | + |
| Vibrio parahaemolyticus | FTQC-07-01 | + |
| Vibrio parahaemolyticus | VP1-9 | + |
| Enterobacter sakazakii | CICC 21560 | - |
| Shigella flexneri | CMCC 51571 | - |
| Bacillus cereus | CMCC 63301 | - |
| Proteus mirabilis | CMCC 49005 | - |
| Micrococcus luteus | CMCC 28001 | - |
| Staphylococcus aureus | CMCC 26074 | - |
| Listeria monocytogenes | CICC 21662 | - |
| Pseudomonas fluorescens | CICC 21620 | - |
| Salmonella Typhimurium | CICC 21483 | - |
| Salmonella enteritidis | CICC 21482 | - |
| Bacillus pumilus | CMCC 63202 | - |
| Vibrio jasicida | MCCC 1A15010 | - |
| Vibrio cholerae | MCCC 1A02608 | - |
| Vibrio diabolicus - | MCCC 1A03504 | - |
| Vibrio anguillarum - | MCCC 1A07299 | - |
| Vibrio harveyi | MCCC 1A00232 | - |
| Vibrio alginolyticus - | MCCC 1A03220 | - |
| Vibrio campbellii | MCCC 1A02605 | - |
| Vibrio vulnificus - | CGMCC 1.8674 | - |
| Vibrio mimicus | MCCC 1A02602 | - |
| Samples | Added (CFUs/mL) | Recovered (CFUs/mL) | Recovery % | RSD % |
|---|---|---|---|---|
| oysters | 5000 | 5701.416 | 114.03 | 4.49 |
| clams | 5000 | 5628.17 | 112.56 | 2.75 |
| cod | 5000 | 6771.934 | 135.48 | 2.90 |
| river shrimp | 5000 | 9755.377 | 195.10 | 1.05 |
| Detection Method | Detection Limit (CFUs/mL) | Ref. |
|---|---|---|
| FRET-based paper sensor with a smartphone | 67 | [30] |
| Biosensing strategy mediated by DNAzyme–ferrocene-triggered click chemistry | 6 | [31] |
| PMA-mPCR | 1.05 × 103 | [32] |
| Phagomagnetic separation–ATP bioluminescence (PhMS-BL) | 78 | [33] |
| Dual-mode colorimetric-electrochemical biosensor | 5 | [34] |
| Visual analysis strategy based on lanthanide metal–organic frameworks and a FAM-aptamer | 100 | [35] |
| Colorimetric and SERS dual-mode detection using composite magnetic materials combined with magnetophoretic chromatography technology | 7 | [36] |
| PCR-SERS | 72 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, A.; Song, X.; Sun, X.; Lu, Z.; Liu, X.; Bie, X.; Yang, J. Construction of PCR-SERS Method for Detection of Vibrio parahaemolyticus. Foods 2024, 13, 1743. https://doi.org/10.3390/foods13111743
Hu A, Song X, Sun X, Lu Z, Liu X, Bie X, Yang J. Construction of PCR-SERS Method for Detection of Vibrio parahaemolyticus. Foods. 2024; 13(11):1743. https://doi.org/10.3390/foods13111743
Chicago/Turabian StyleHu, Antuo, Xiaoting Song, Xiaojie Sun, Zhaoxin Lu, Xinmei Liu, Xiaomei Bie, and Jun Yang. 2024. "Construction of PCR-SERS Method for Detection of Vibrio parahaemolyticus" Foods 13, no. 11: 1743. https://doi.org/10.3390/foods13111743
APA StyleHu, A., Song, X., Sun, X., Lu, Z., Liu, X., Bie, X., & Yang, J. (2024). Construction of PCR-SERS Method for Detection of Vibrio parahaemolyticus. Foods, 13(11), 1743. https://doi.org/10.3390/foods13111743








