Effects of Selenium Content on Growth, Antioxidant Activity, and Key Selenium-Enriched Gene Expression in Alfalfa Sprouts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Design
2.2. Determination of Total and Organic Selenium in Sprouts
2.3. Determination of Antioxidant Activity
2.3.1. Determination of Scavenging Ability of Alfalfa Sprout Extracts to Inhibit the Transfer of Hydrogen Atoms Using the 2,2-Diphenylpicrylhydrazine (DPPH) Assay
2.3.2. Determination of Scavenging Ability of Alfalfa Sprout Extracts on Hydroxyl Radicals Using the Salicylic Acid Method
2.3.3. Determination of Antioxidant Activity by Selenium-Related Enzymes
2.4. Transcriptomic Analysis
2.5. Construction of Gene Co-Expression Network Using WGCNA
2.6. Real-Time Quantitative Fluorescence PCR (qRT-PCR) Analysis
3. Results and Discussion
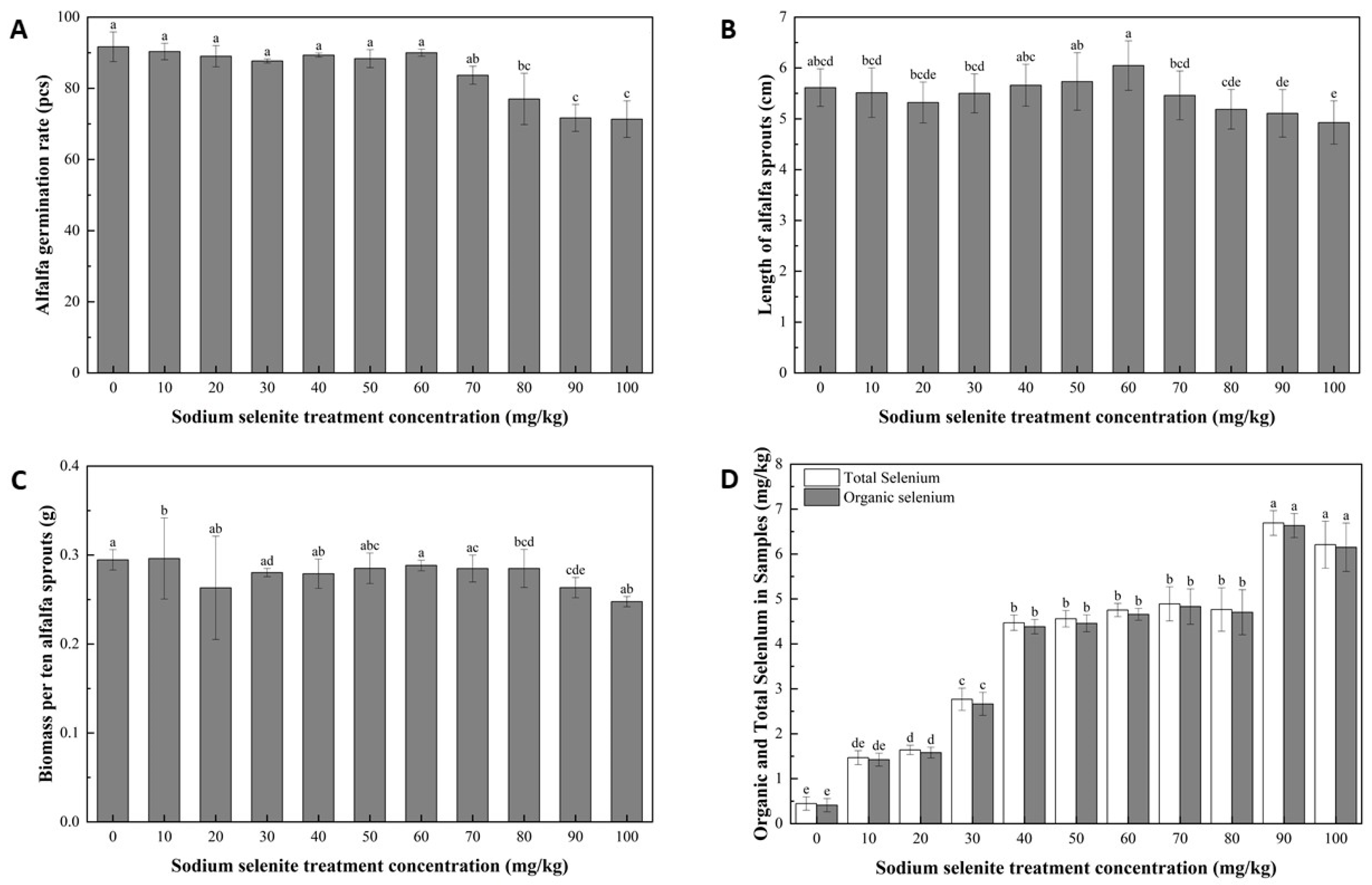
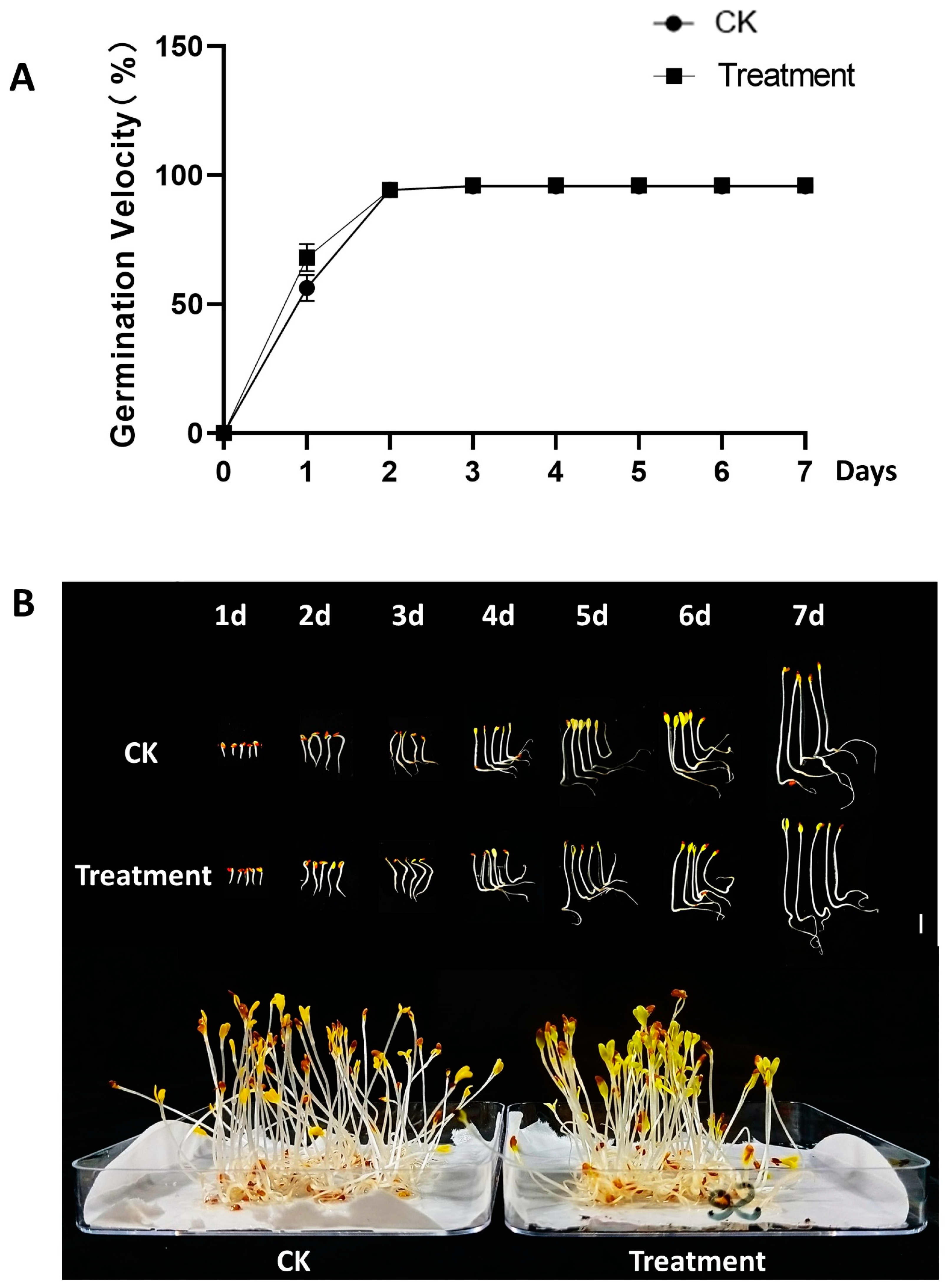
3.1. Growth and Physiological Characteristics of Alfalfa Sprouts
3.2. Analysis of Antioxidant Capacity
3.2.1. Hydroxyl Radical Scavenging Capacity
3.2.2. DPPH-Free Radical Scavenging Capacity
3.2.3. Antioxidant Capacity of Selenium-Related Enzymes
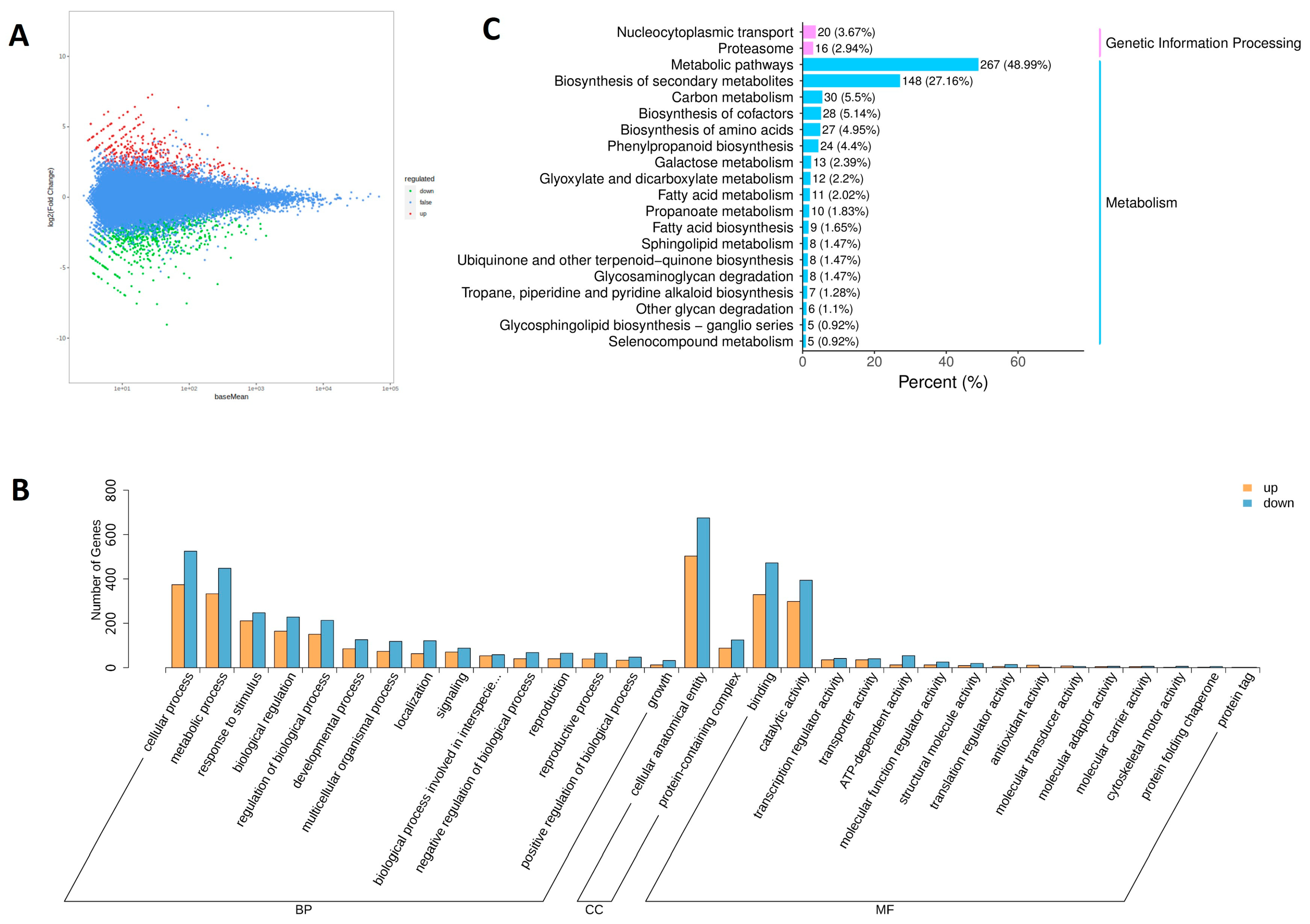
3.3. Analysis of Differentially Expressed Genes (DEGs)
3.4. Gene Ontology (GO) Enrichment Analysis of DEGs
3.5. KEGG Enrichment Analysis of DEGs
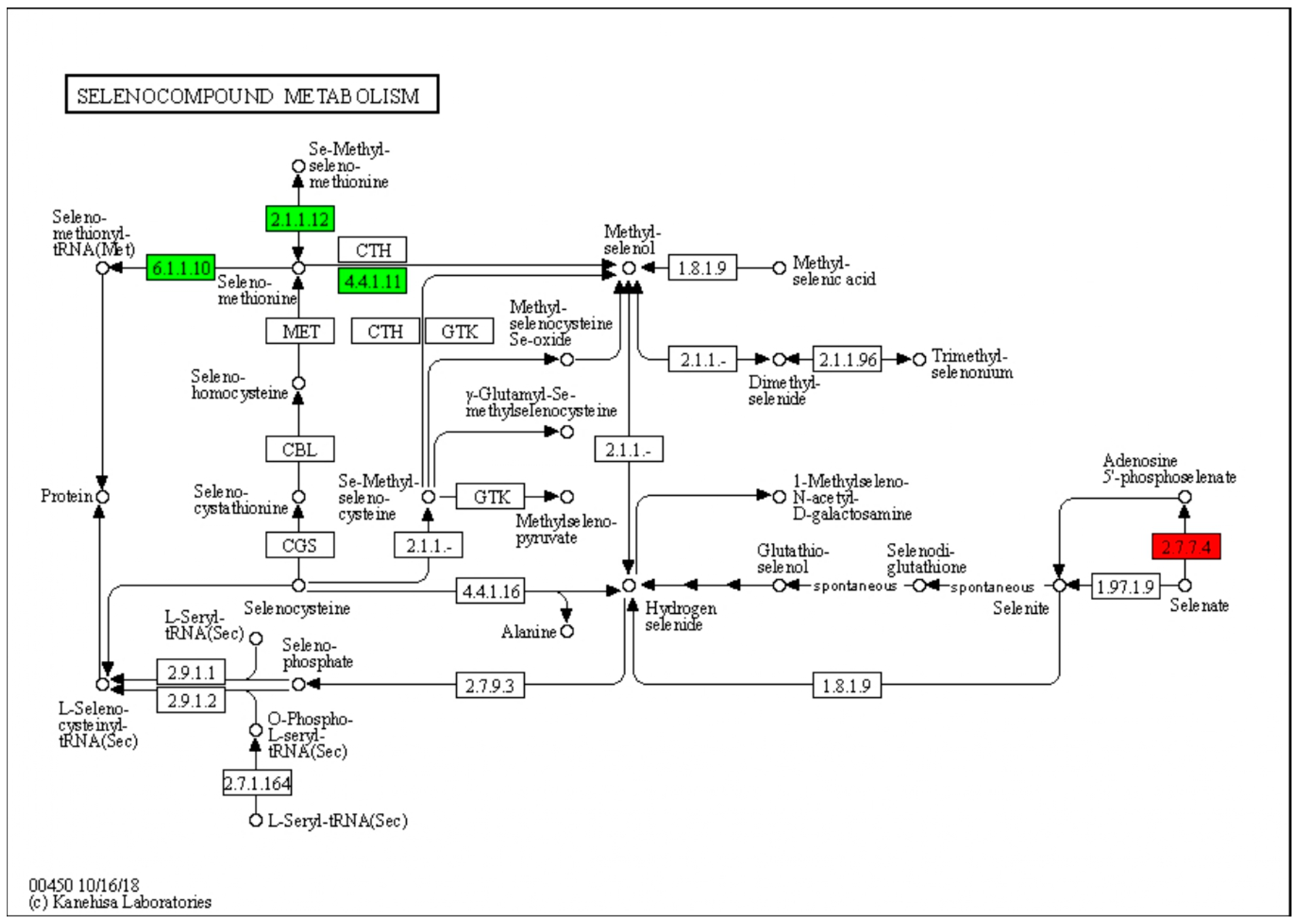
3.6. DEGs Involved in Selenium Metabolism
3.7. qRT-PCR Validation
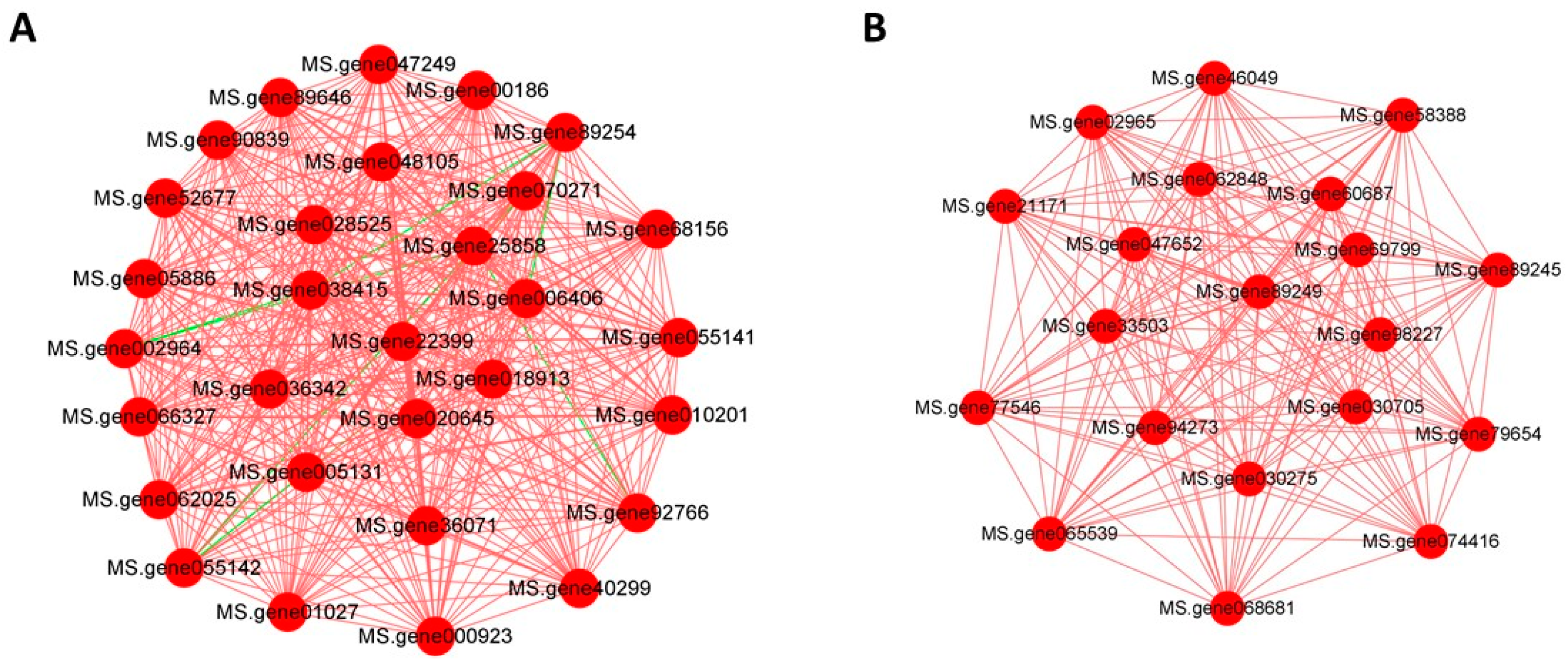
3.8. Gene Correlation Network Construction
3.9. Analysis of the Regulation of Key Gene Expression Modules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiavon, M.; Nardi, S.; dalla Vecchia, F.; Ertani, A. Selenium biofortification in the 21st century: Status and challenges for healthy human nutrition. Plant Soil 2020, 453, 245–270. [Google Scholar] [CrossRef]
- Peeler, J.C.; Weerapana, E. Chemical Biology Approaches to Interrogate the Selenoproteome. Acc. Chem. Res. 2019, 52, 2832–2840. [Google Scholar] [CrossRef]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef]
- Winther, K.H.; Rayman, M.P.; Bonnema, S.J.; Hegedüs, L. Selenium in thyroid disorders—Essential knowledge for clinicians. Nat. Rev. Endocrinol. 2020, 16, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jin, J.; Huang, B.; Ying, H.; He, J.; Jiang, L. Selenium Metabolism and Selenoproteins in Prokaryotes: A Bioinformatics Perspective. Biomolecules 2022, 12, 917. [Google Scholar] [CrossRef]
- Reeves, M.A.; Hoffmann, P.R. The human selenoproteome: Recent insights into functions and regulation. Cell. Mol. Life Sci. 2009, 66, 2457–2478. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Cattaneo, D.M.I.R.; Abbate, R.; Manoni, M.; Ottoboni, M.; Luciano, A.; von Holst, C.; Pinotti, L. Advances in selenium supplementation: From selenium-enriched yeast to potential selenium-enriched insects, and selenium nanoparticles. Anim. Nutr. 2023, 14, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Pilon-Smits, E.A.; Zhao, F.J.; Williams, P.N.; Meharg, A.A. Selenium in higher plants: Understanding mechanisms for biofortification and phytoremediation. Trends Plant Sci. 2009, 14, 436–442. [Google Scholar] [CrossRef]
- Kawakami, Y.; Bhullar, N.K. Molecular processes in iron and zinc homeostasis and their modulation for biofortification in rice. J. Integr. Plant Biol. 2018, 60, 1181–1198. [Google Scholar] [CrossRef]
- Funes-Collado, V.; Morell-Garcia, A.; Rubio, R.; López-Sánchez, J.F. Study of selenocompounds from selenium-enriched culture of edible sprouts. Food Chem. 2013, 141, 3738–3743. [Google Scholar] [CrossRef]
- Liu, S.; Wang, W.; Lu, H.; Shu, Q.; Zhang, Y.; Chen, Q. New perspectives on physiological, biochemical and bioactive components during germination of edible seeds: A review. Trends Food Sci. Technol. 2022, 123, 187–197. [Google Scholar] [CrossRef]
- D’Amato, R.; Fontanella, M.C.; Falcinelli, B.; Beone, G.M.; Bravi, E.; Marconi, O.; Benincasa, P.; Businelli, D. Selenium Biofortification in Rice (Oryza sativa L.) Sprouting: Effects on Se Yield and Nutritional Traits with Focus on Phenolic Acid Profile. J. Agric. Food Chem. 2018, 66, 4082–4090. [Google Scholar] [CrossRef]
- Diao, M.; Ma, L.; Wang, J.; Cui, J.; Fu, A.; Liu, H.-Y. Selenium Promotes the Growth and Photosynthesis of Tomato Seedlings under Salt Stress by Enhancing Chloroplast Antioxidant Defense System. J. Plant Growth Regul. 2014, 33, 671–682. [Google Scholar] [CrossRef]
- Nordin, N.H.; Molan, A.L.; Chua, W.; Kruger, M.C. Total Phenolic Contents and Antioxidant Activities of Selenium-Rich Black Tea Versus Regular Black Tea. Am. J. Life Sci. Res. 2017, 5, 40–50. [Google Scholar] [CrossRef]
- Chao, W.; Rao, S.; Chen, Q.; Zhang, W.; Liao, Y.; Ye, J.; Cheng, S.; Yang, X.; Xu, F. Advances in Research on the Involvement of Selenium in Regulating Plant Ecosystems. Plants 2022, 11, 2712. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Zhang, L.; Peng, J.; Huang, T.; Yang, X.; Jeong, B.R.; Yang, Q. Seleno-Amino Acids in Vegetables: A Review of Their Forms and Metabolism. Front. Plant Sci. 2022, 13, 804368. [Google Scholar] [CrossRef]
- Ebert, A.W. Sprouts and Microgreens—Novel Food Sources for Healthy Diets. Plants 2022, 11, 571. [Google Scholar] [CrossRef]
- Almuhayawi, M.S.; Hassan, A.H.; Al Jaouni, S.K.; Alkhalifah, D.H.M.; Hozzein, W.N.; Selim, S.; AbdElgawad, H.; Khamis, G. Influence of elevated CO2 on nutritive value and health-promoting prospective of three genotypes of Alfalfa sprouts (Medicago sativa). Food Chem. 2021, 340, 128147. [Google Scholar] [CrossRef]
- Zhang, K.; Li, H.; Zhang, T.; Wang, S.; Liu, L.; Dong, X.; Cong, L.; Song, H.; Wang, A.; Yang, G.; et al. Comprehensive transcriptomic and metabolomic profiling reveals the differences between alfalfa sprouts germinated with or without light exposure. Front. Plant Sci. 2022, 13, 943740. [Google Scholar] [CrossRef]
- Cheng, H.; Li, L.; Dong, J.; Wang, S.; Wu, S.; Rao, S.; Li, L.; Cheng, S.; Li, L. Transcriptome and physiological determination reveal the effects of selenite on the growth and selenium metabolism in mung bean sprouts. Food Res. Int. 2023, 169, 112880. [Google Scholar] [CrossRef]
- Lintschinger, J.; Fuchs, N.; Moser, J.; Kuehnelt, D.; Goessler, W. Selenium-Enriched Sprouts. A Raw Material for Fortified Cereal-Based Diets. J. Agric. Food Chem. 2000, 48, 5362–5368. [Google Scholar] [CrossRef]
- Huang, Y.; Lei, N.; Xiong, Y.; Liu, Y.; Tong, L.; Wang, F.; Fan, B.; Maesen, P.; Blecker, C. Influence of Selenium Biofortification of Soybeans on Speciation and Transformation during Seed Germination and Sprouts Quality. Foods 2022, 11, 1200. [Google Scholar] [CrossRef]
- Bai, Y.-Z.; Zhang, S.-Q. Do selenium-enriched foods provide cognitive benefit? Metab. Brain Dis. 2023, 38, 1501–1502. [Google Scholar] [CrossRef]
- Shen, C.; Du, H.; Chen, Z.; Lu, H.; Zhu, F.; Chen, H.; Meng, X.; Liu, Q.; Liu, P.; Zheng, L.; et al. The Chromosome-Level Genome Sequence of the Autotetraploid Alfalfa and Resequencing of Core Germplasms Provide Genomic Resources for Alfalfa Research. Mol. Plant 2020, 13, 1250–1261. [Google Scholar] [CrossRef]
- Versini, A.; Di Tullo, P.; Aubry, E.; Bueno, M.; Thiry, Y.; Pannier, F.; Castrec-Rouelle, M. Influence of Se concentrations and species in hydroponic cultures on Se uptake, translocation and assimilation in non-accumulator ryegrass. Plant Physiol. Biochem. 2016, 108, 372–380. [Google Scholar] [CrossRef]
- Chantiratikul, A.; Borisuth, L.; Chinrasri, O.; Saenthaweesuk, N.; Chookhampaeng, S.; Thosaikham, W.; Sriart, N.; Chantiratikul, P. Evaluation of the toxicity of selenium from hydroponically produced selenium-enriched kale sprout in laying hens. J. Trace Elem. Med. Biol. 2016, 35, 116–121. [Google Scholar] [CrossRef]
- Bourgis, F.; Roje, S.; Nuccio, M.L.; Fisher, D.B.; Tarczynski, M.C.; Li, C.; Herschbach, C.; Rennenberg, H.; Pimenta, M.J.; Shen, T.L.; et al. S-Methylmethionine Plays a Major Role in Phloem Sulfur Transport and Is Synthesized by a Novel Type of Methyltransferase. Plant Cell 1999, 11, 1485. [Google Scholar] [CrossRef]
- Scherer, R.; Godoy, H.T. Antioxidant activity index (AAI) by the 2,2-diphenyl-1-picrylhydrazyl method. Food Chem. 2009, 112, 654–658. [Google Scholar] [CrossRef]
- Kurpios-Piec, D.; Majewska-Wierzbicka, M.; Czeczot, H.; Podsiad, M. Flavonoids as hydroxyl radical scavengers, iron reductants and chelators: In vitro antioxidant action. Acta Pol. Pharm. 2022, 78, 635. [Google Scholar] [CrossRef]
- Dervisi, I.; Valassakis, C.; Koletti, A.; Kouvelis, V.N.; Flemetakis, E.; Ouzounis, C.A.; Roussis, A. Evolutionary Aspects of Selenium Binding Protein (SBP). J. Mol. Evol. 2023, 91, 471–481. [Google Scholar] [CrossRef]
- Sekula, B.; Ruszkowski, M.; Dauter, Z. S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula. Int. J. Biol. Macromol. 2020, 151, 554–565. [Google Scholar] [CrossRef]
- Neuhierl, B.; Thanbichler, M.; Lottspeich, F.; Böck, A. A Family of S-Methylmethionine-dependent Thiol/Selenol Methyltransferases: Role in selenium tolerance and evolutionary relation. J. Biol. Chem. 1999, 274, 5407–5414. [Google Scholar] [CrossRef]
- Freeman, J.L.; Tamaoki, M.; Stushnoff, C.; Quinn, C.F.; Cappa, J.J.; Devonshire, J.; Fakra, S.C.; Marcus, M.A.; McGrath, S.P.; Van Hoewyk, D.; et al. Molecular Mechanisms of Selenium Tolerance and Hyperaccumulation in Stanleya pinnata. Plant Physiol. 2010, 153, 1630–1652. [Google Scholar] [CrossRef]
- Marrs, K.A. The functions and regulation of glutathione s-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tian, Y.-S.; Xing, X.-J.; Xu, Z.-S.; Zhu, B.; Fu, X.-Y.; Peng, R.-H.; Yao, Q.-H. Enhancement of phenol stress tolerance in transgenic Arabidopsis plants overexpressing glutathione S-transferase. Plant Growth Regul. 2017, 82, 37–45. [Google Scholar] [CrossRef]
- Arteel, G.E.; Sies, H. The biochemistry of selenium and the glutathione system. Environ. Toxicol. Pharmacol. 2001, 10, 153–158. [Google Scholar] [CrossRef]
- Xue, T.; Hartikainen, H. Association of antioxidative enzymes with the synergistic effect of selenium and UV irradiation in enhancing plant growth. Agric. Food Sci. 2000, 9, 177–186. [Google Scholar] [CrossRef]
- Santosh, T.R.; Sreekala, M.; Lalitha, K. Oxidative stress during selenium deficiency in seedlings of Trigonella foenum-graecum and mitigation by mimosine part II. glutathione metabolism. Biol. Trace Elem. Res. 1999, 70, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-F.; McGrath, S.P.; Zhao, F.-J. Selenium uptake, translocation and speciation in wheat supplied with selenate or selenite. New Phytol. 2008, 178, 92–102. [Google Scholar] [CrossRef] [PubMed]
- White, P.J. Selenium metabolism in plants. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2333–2342. [Google Scholar] [CrossRef]
- Tangjaidee, P.; Swedlund, P.; Xiang, J.; Yin, H.; Quek, S.Y. Selenium-enriched plant foods: Selenium accumulation, speciation, and health functionality. Front. Nutr. 2022, 9, 962312. [Google Scholar] [CrossRef]

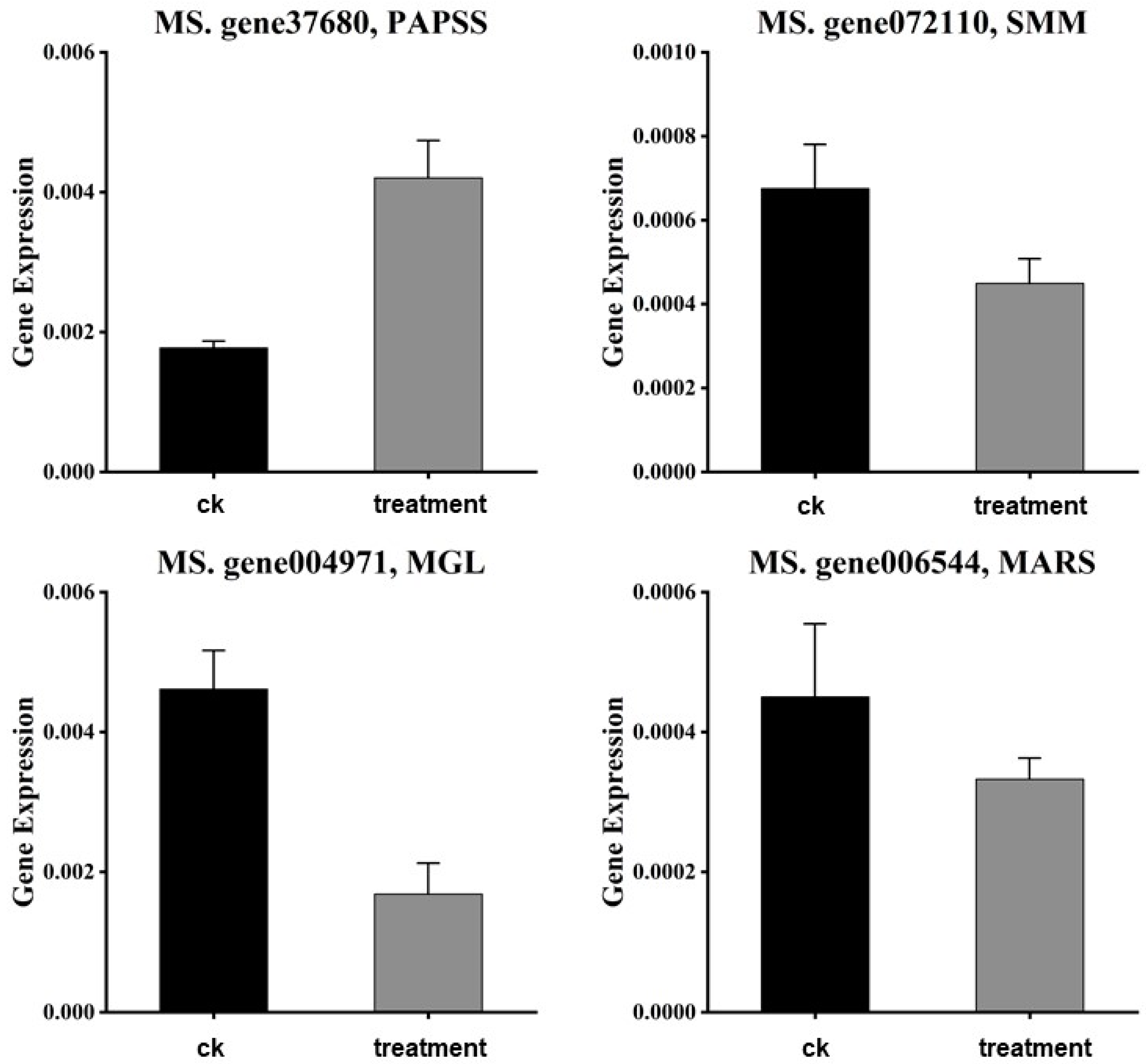
| Primer Name | Primer Sequence |
|---|---|
| PAPSS37680 F | TGATCCATCAAGGCCTCAGG |
| PAPSS37680 R | CAACTAGCACCTTCCAGCCA |
| SMM072110 F | TCTCACCTCGGCTTGCAATT |
| SMM072110 R | TCACCACCACTTGAGGCTTC |
| MGL004971 F | CCTAGAAGACCATCCACATCATGA |
| MGL004971 R | GTGCTACTTCCAGAACAAGACATAA |
| MARS006544 F | GTTGAGTTGGTTGAACCGCC |
| MARS006544 R | AATGCCAGCTGATGTGGTCA |
| Module Colors | Gene ID | KEGG |
|---|---|---|
| yellow | MS.gene000923 | K22366 thioredoxin reductase-like selenoprotein T|(GenBank) selT/selW/selH selenoprotein domain protein (A) |
| yellow | MS.gene00186 | K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]|(GenBank) dihydrolipoamide dehydrogenase (A) |
| yellow | MS.gene002964 | K05907 adenylyl-sulfate reductase (glutathione) [EC:1.8.4.9]|(GenBank) 5′-adenylylsulfate reductase (A) |
| yellow | MS.gene005131 | K12859 U5 snRNP protein, DIM1 family|(GenBank) mRNA splicing factor, thioredoxin-like U5 snRNP protein (A) |
| yellow | MS.gene006406 | K08967 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase [EC:1.13.11.53 1.13.11.54]|(GenBank) 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase (A) |
| yellow | MS.gene010201 | K23870 putative pectin methyltransferase [EC:2.1.1.-]|(GenBank) S-adenosylmethionine-dependent methyltransferase, putative (A) |
| yellow | MS.gene01027 | K05907 adenylyl-sulfate reductase (glutathione) [EC:1.8.4.9]|(GenBank) 5′-adenylylsulfate reductase (A) |
| yellow | MS.gene018913 | K03671 thioredoxin 1|(GenBank) thioredoxin H-type 1 protein (A) |
| yellow | MS.gene020645 | K03671 thioredoxin 1|(GenBank) thioredoxin (A) |
| yellow | MS.gene028525 | K07305 peptide-methionine (R)-S-oxide reductase [EC:1.8.4.12]|(GenBank) methionine sulfoxide reductase B 2 (A) |
| yellow | MS.gene036342 | K00559 sterol 24-C-methyltransferase [EC:2.1.1.41]|(GenBank) S-adenosyl-methionine-sterol-C- methyltransferase (A) |
| yellow | MS.gene038415 | K07304 peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11]|(GenBank) peptide methionine sulfoxide reductase family protein (A) |
| yellow | MS.gene047249 | K08247 methionine S-methyltransferase [EC:2.1.1.12]|(GenBank) methionine S-methyltransferase (A) |
| yellow | MS.gene048105 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| yellow | MS.gene055141 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| yellow | MS.gene055142 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| yellow | MS.gene05886 | K01759 lactoylglutathione lyase [EC:4.4.1.5]|(GenBank) lactoylglutathione lyase-like protein (A) |
| yellow | MS.gene062025 | K00789 S-adenosylmethionine synthetase [EC:2.5.1.6]|(GenBank) S-adenosylmethionine synthase-like protein (A) |
| yellow | MS.gene066327 | K23870 putative pectin methyltransferase [EC:2.1.1.-]|(GenBank) S-adenosylmethionine-dependent methyltransferase, putative (A) |
| yellow | MS.gene070271 | K19306 18S rRNA (guanine1575-N7)-methyltransferase [EC:2.1.1.309]|(GenBank) S-adenosyl-L-methionine-dependent methyltransferase (A) |
| yellow | MS.gene22399 | K00789 S-adenosylmethionine synthetase [EC:2.5.1.6]|(GenBank) S-adenosylmethionine synthase-like protein (A) |
| yellow | MS.gene25858 | K11820 N-hydroxythioamide S-beta-glucosyltransferase [EC:2.4.1.195]|(GenBank) UDP-glucosyltransferase family protein (A) |
| yellow | MS.gene36071 | K03671 thioredoxin 1|(GenBank) thioredoxin M-type protein (A) |
| yellow | MS.gene40299 | K01759 lactoylglutathione lyase [EC:4.4.1.5]|(GenBank) lactoylglutathione lyase-like protein (A) |
| yellow | MS.gene52677 | K03671 thioredoxin 1|(GenBank) thioredoxin (A) |
| yellow | MS.gene68156 | K01244 5′-methylthioadenosine nucleosidase [EC:3.2.2.16]|(GenBank) 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (A) |
| yellow | MS.gene89254 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| yellow | MS.gene89646 | K07304 peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11]|(GenBank) peptide methionine sulfoxide reductase family protein (A) |
| yellow | MS.gene90839 | K21888 glutathione dehydrogenase/transferase [EC:1.8.5.1 2.5.1.18]|(GenBank) glutathione S-transferase (A) |
| yellow | MS.gene92766 | K03671 thioredoxin 1|(GenBank) thioredoxin (A) |
| magenta | MS.gene02965 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| magenta | MS.gene030275 | K03671 thioredoxin 1|(GenBank) thioredoxin H-type 1 protein (A) |
| magenta | MS.gene030705 | K15111 solute carrier family 25 (mitochondrial S-adenosylmethionine transporter), member 26|(GenBank) S-adenosylmethionine carrier protein (A) |
| magenta | MS.gene047652 | K01265 methionyl aminopeptidase [EC:3.4.11.18]|(GenBank) methionine aminopeptidase 1B (A) |
| magenta | MS.gene062848 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| magenta | MS.gene065539 | K00658 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide succinyltransferase) [EC:2.3.1.61]|(GenBank) 2-oxoacid dehydrogenase acyltransferase family protein (A) |
| magenta | MS.gene068681 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| magenta | MS.gene074416 | K00121 S-(hydroxymethyl)glutathione dehydrogenase/alcohol dehydrogenase [EC:1.1.1.284 1.1.1.1]|(GenBank) zinc-binding alcohol dehydrogenase family protein (A) |
| magenta | MS.gene21171 | K22966 sulfate adenylyltransferase (ADP)/adenylylsulfatase [EC:2.7.7.5 3.6.2.1]|(GenBank) scavenger mRNA decapping enzyme carboxy-term-binding protein (A) |
| magenta | MS.gene33503 | K22366 thioredoxin reductase-like selenoprotein T|(GenBank) selT/selW/selH selenoprotein domain protein (A) |
| magenta | MS.gene46049 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| magenta | MS.gene58388 | K01265 methionyl aminopeptidase [EC:3.4.11.18]|(GenBank) methionine aminopeptidase 2B-like protein (A) |
| magenta | MS.gene60687 | K03671 thioredoxin 1|(GenBank) thioredoxin M-type protein (A) |
| magenta | MS.gene69799 | K03671 thioredoxin 1|(GenBank) thioredoxin (A) |
| magenta | MS.gene77546 | K11820 N-hydroxythioamide S-beta-glucosyltransferase [EC:2.4.1.195]|(GenBank) UDP-glucosyltransferase family protein (A) |
| magenta | MS.gene79654 | K03671 thioredoxin 1|(GenBank) thioredoxin H2-like protein (A) |
| magenta | MS.gene89245 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase (A) |
| magenta | MS.gene89249 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| magenta | MS.gene94273 | K00799 glutathione S-transferase [EC:2.5.1.18]|(GenBank) glutathione S-transferase, amino-terminal domain protein (A) |
| magenta | MS.gene98227 | K00235 succinate dehydrogenase (ubiquinone) iron-sulfur subunit [EC:1.3.5.1]|(GenBank) succinate dehydrogenase [ubiquinone] iron-sulfur subunit (A) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Zhang, Q.; Li, X.; Zhang, T.; Tian, D.; Liu, L.; Dong, X.; Wang, Z.-Y.; Chai, M. Effects of Selenium Content on Growth, Antioxidant Activity, and Key Selenium-Enriched Gene Expression in Alfalfa Sprouts. Foods 2024, 13, 2261. https://doi.org/10.3390/foods13142261
Ren Y, Zhang Q, Li X, Zhang T, Tian D, Liu L, Dong X, Wang Z-Y, Chai M. Effects of Selenium Content on Growth, Antioxidant Activity, and Key Selenium-Enriched Gene Expression in Alfalfa Sprouts. Foods. 2024; 13(14):2261. https://doi.org/10.3390/foods13142261
Chicago/Turabian StyleRen, Yaru, Qian Zhang, Xiang Li, Tianyi Zhang, Daicai Tian, Liang Liu, Xuyan Dong, Zeng-Yu Wang, and Maofeng Chai. 2024. "Effects of Selenium Content on Growth, Antioxidant Activity, and Key Selenium-Enriched Gene Expression in Alfalfa Sprouts" Foods 13, no. 14: 2261. https://doi.org/10.3390/foods13142261





