1. Introduction
Polygonati Rhizoma (PR), which is called Huangjing in China, is the rhizome of a liliaceous plant from the genus
Polygonatum Mill and has been used in traditional food and medicine in China for centuries [
1]. PR contains a range of essential compounds such as sugars, lipids, proteins, carotenoids, vitamins, amino acids, and trace elements, which can resist hidden hunger and makes it a potential high-quality crop [
2,
3]. Rich in compounds like polysaccharides and flavonoids, it offers numerous health benefits, including anti-aging, anti-diabetic, anti-fatigue, and anti-cancer effects [
4,
5,
6,
7]. PR has traditionally been used in clinical practices to treat age-related diseases, diabetes, lung diseases, fatigue, feebleness, and indigestion in China, India, Pakistan, Iran, and Japan [
4,
8]. The wide range of medicinal benefits and the increasing demand for PR in various therapeutic applications underscore the importance of ensuring its authenticity and quality. PR is cultivated in various geographical regions, with China being the main producer. However, the geographical origin of PR affects the quality, drug effect, and price [
9,
10]. Products certified as protected geographical indications (PGIs) are more popular with consumers and have higher prices. Consequently, unscrupulous traders often mislabel the origins or adulterate PGI products with inferior products or products from other regions to increase profits, causing both healthy and wealthy losses to consumers [
9]. In the market, consumers are concerned about whether the product is pure, adulterated, or pure counterfeit. Therefore, the accurate identification of adulterated PR from different geographical origins is essential to protect consumer health and maintain fair trade practices.
Current identification methods for the geographical origin of foods and medicinal materials primarily include manual identification, chromatography, mass spectrometry, and DNA molecular identification [
11,
12,
13,
14,
15]. However, manual identification requires extensive professional knowledge and is unsuitable for processing products (dry whole root, slice, powder). Chromatography, mass spectrometry, and DNA molecular identification are time-consuming, expensive, environmentally unfriendly, and complicated to operate [
15,
16,
17]. Also, the adulteration of powder samples from different geographical origins has created challenges in these technologies. Therefore, there is an urgent need for a real-time, rapid, direct, efficient, and high-precision method to identify adulterated foods or medicinal materials from different regions.
Currently, some researchers use laser-induced breakdown spectroscopy (LIBS) and near-infrared spectroscopy (NIR) LIBS to identify geographical origin and adulterated foods or medicinal materials products, due to their advantages such as fast and in situ analysis [
18,
19,
20,
21]. For instance, Nie et al. employed visible and near-infrared spectroscopy (VNIR) for the quantitative analysis of the adulteration of
Sophora flavescens powder or corn flour in Notoginseng powder, yielding a predictive R-squared value within the range of 0.86 to 0.94 [
22]. Zhao et al. demonstrated the utility of LIBS in analyzing Chinese yam adulterated with cassava and the rhizome of winged yam, with R-squared values reaching 0.9570 [
23]. Akin et al. employed LIBS in the analysis of corn and sorghum flour mixtures, achieving a good R-squared result of 0.965 [
24]. Some researchers have also fused LIBS and VNIR to achieve better identification results [
25]. For example, Zhao et al. used the fusion of LIBS and hyperspectral imaging (400–1000 nm) data to improve ginseng samples’ geographical origin identification accuracy from 96.9% and 94.75% to 98.8% [
26]. Collectively, these studies described above verified the potential of LIBS and NIR techniques in the identification of adulterated samples, especially the fusion of LIBS and VNIR, which has a better effect. Combining the elemental and molecular information obtained from these two techniques makes it possible to achieve a more comprehensive and accurate identification of adulterated materials. The subtle chemical and morphological changes between these materials of the same species but with different geographical origins pose a significant challenge for adulteration identification, and single-modal analysis makes it difficult to achieve a high level of identification accuracy. However, there is no report on the identification of adulterated PR from different geographical origins. For food quality identification, the research on the fusion method of LIBS and VNIR at the atomic and molecular levels is rarely studied.
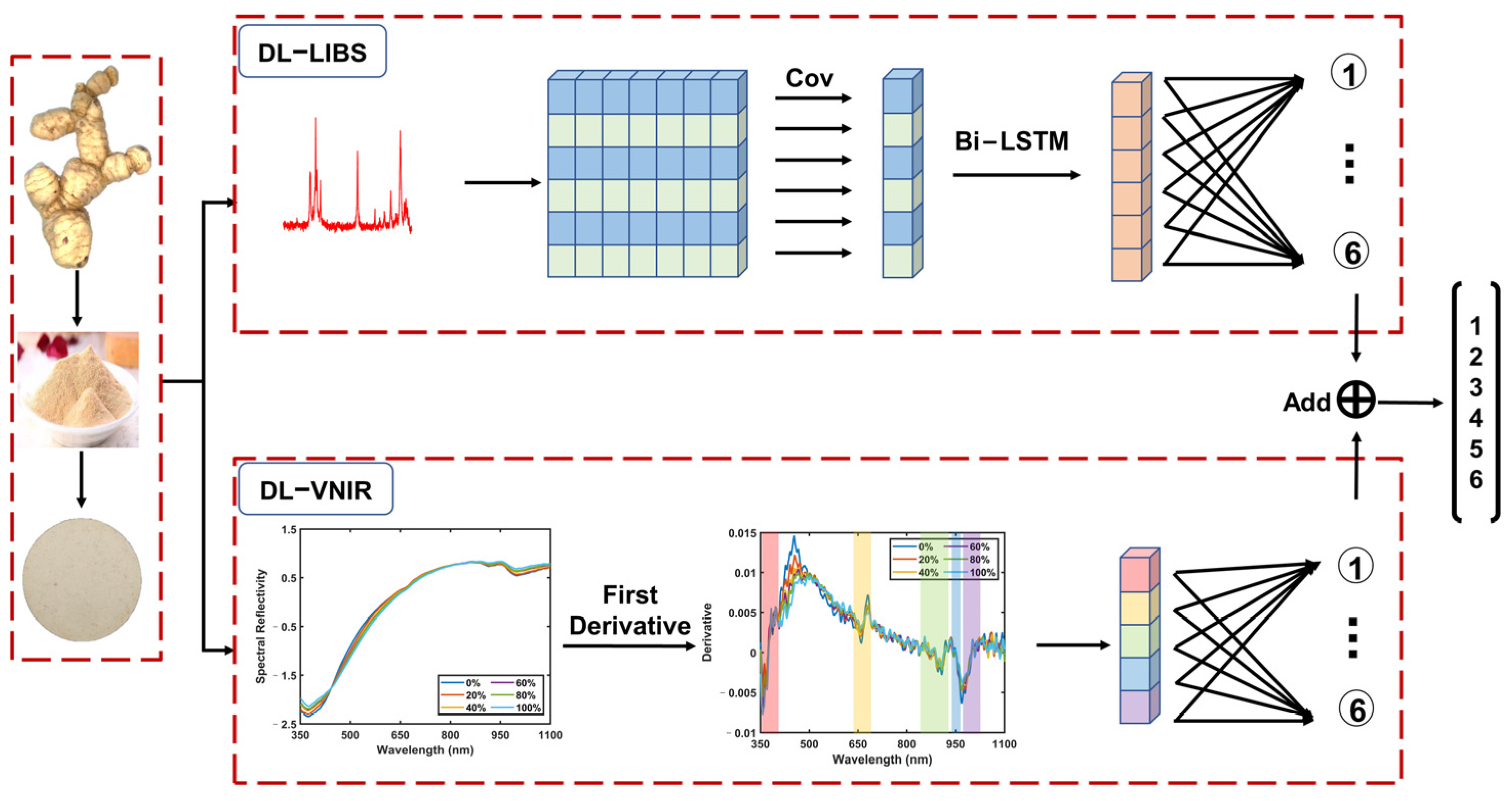
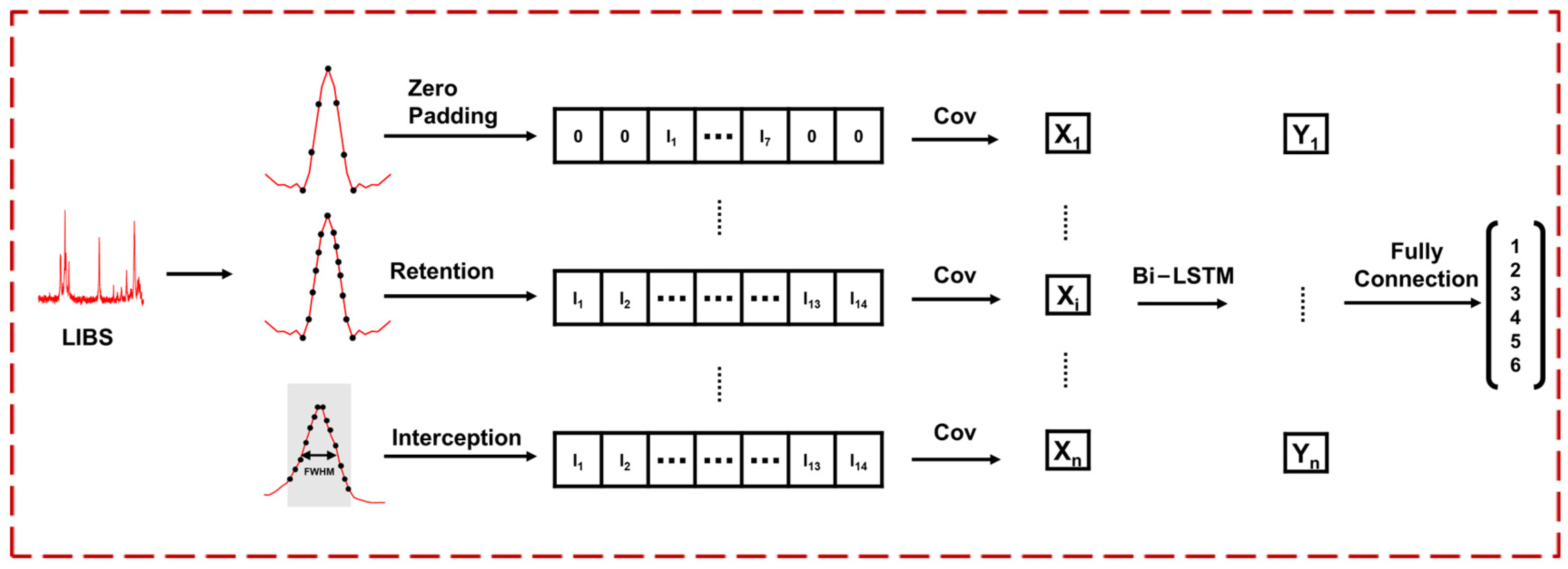
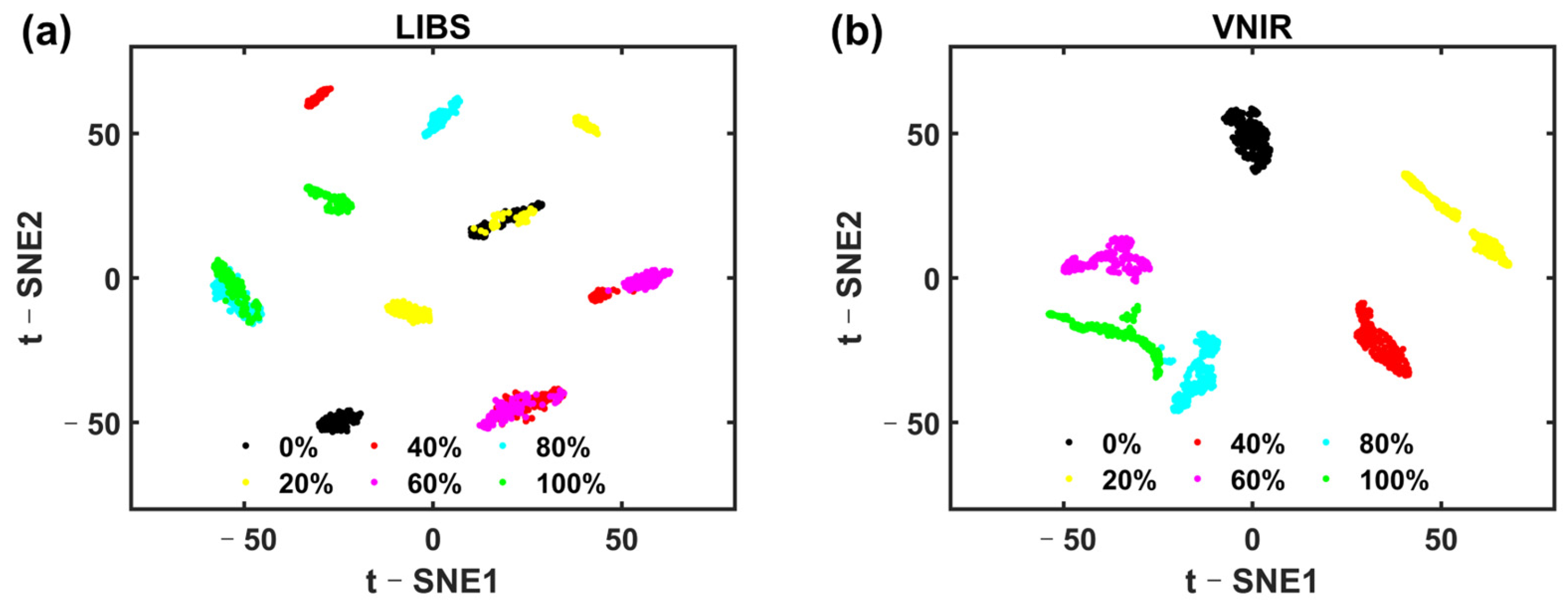
Since foods or herbal medicines are rich in elemental and molecular information, based on the complementary advantages of LIBS in elemental analysis and VNIR in molecular analysis, together, they can provide a comprehensive assessment of the authenticity of PR. Therefore, the purpose of this study is to propose a deep learning model that effectively combines LIBS and VNIR to improve the accuracy of adulteration identification. We proposed an LIBS-VNIR fusion based on a deep learning network (LVDLNet) to detect adulteration in PR sourced from different regions in this study. The model was explained and verified from different aspects. Finally, the study confirmed that the fusion of LIBS and VNIR was feasible and effective in identifying adulterated PR. This work provides a powerful solution for the efficient, accurate, precise, and robust detection of adulteration, which is expected to enhance the integrity and safety of the food supply chain.
2. Samples and Experimental System
2.1. Sample Preparation
The highest quality PR, produced in Jiuhua Mountain and its surrounding areas in Qingyang County, southern Anhui, is certified as a PGI in China [
6,
27]. In this experiment, PR from Qingyang County was adulterated with cheaper PR from Dandong City, Liaoning Province. To ensure the authenticity of the samples, our staff personally collected the PR from their respective regions of origin. After the collection, the samples were cleansed with deionized water to eliminate surface dust and debris. Subsequently, they were sliced to a thickness of approximately 2 mm. These slices were then dried to constant weight at a controlled temperature of 60 °C within an electric blast drying oven (101-0B, Shaoxing Shangcheng Instrument Manufacturing Co., Ltd., Shaoxing, China). The dried samples were subsequently crushed and ground to a fine powder, passing through an 80-mesh sieve to ensure uniformity.
In actual market conditions, there are instances where cheaper Polygonati Rhizoma (PR) is used to completely impersonate more expensive PR from famous origins. There are also situations where the cheaper PR is mixed into the more expensive PR for sale, and a small amount of adulteration is insignificant for counterfeiters. To simulate the market adulteration practices, PR from Dandong City was systematically blended with PR from Qingyang County in incremental proportions ranging from 0% to 100% in steps of 20% (ω/ω). The adulterated samples contained 0, 20, 40, 60, 80, and 100% (ω/ω) adulterated levels. Specifically, the adulteration percentages refer to the weight percentage of PR from Dandong City in the mixture. For instance, a 0% level indicates pure PR from Qingyang County, while a 100% level signifies a mixture composed entirely of PR from Dandong City. Intermediate levels at 20%, 40%, 60%, and 80% represent the respective proportions of the Dandong City PR in the blend. The resulting mixtures were then compacted into pellets, each weighing two grams, using an electric tablet press exerting a substantial pressure of 24 tons over one minute. The pressed pellets, characterized by a thickness of approximately 3 mm and a diameter of 20 mm, were employed for our subsequent analyses. Two replicate samples were made for each concentration gradient to eliminate individual differences in samples. A total of 12 pressed pellets were prepared for measurement without further treatment. To evaluate the robustness of our proposed model, we also prepared another batch of samples by blending PR from Baise City in Guangxi Province with authentic PR from Qingyang County, adhering to the stringent criteria outlined in our previous methodology.
2.2. Setup and Measurement
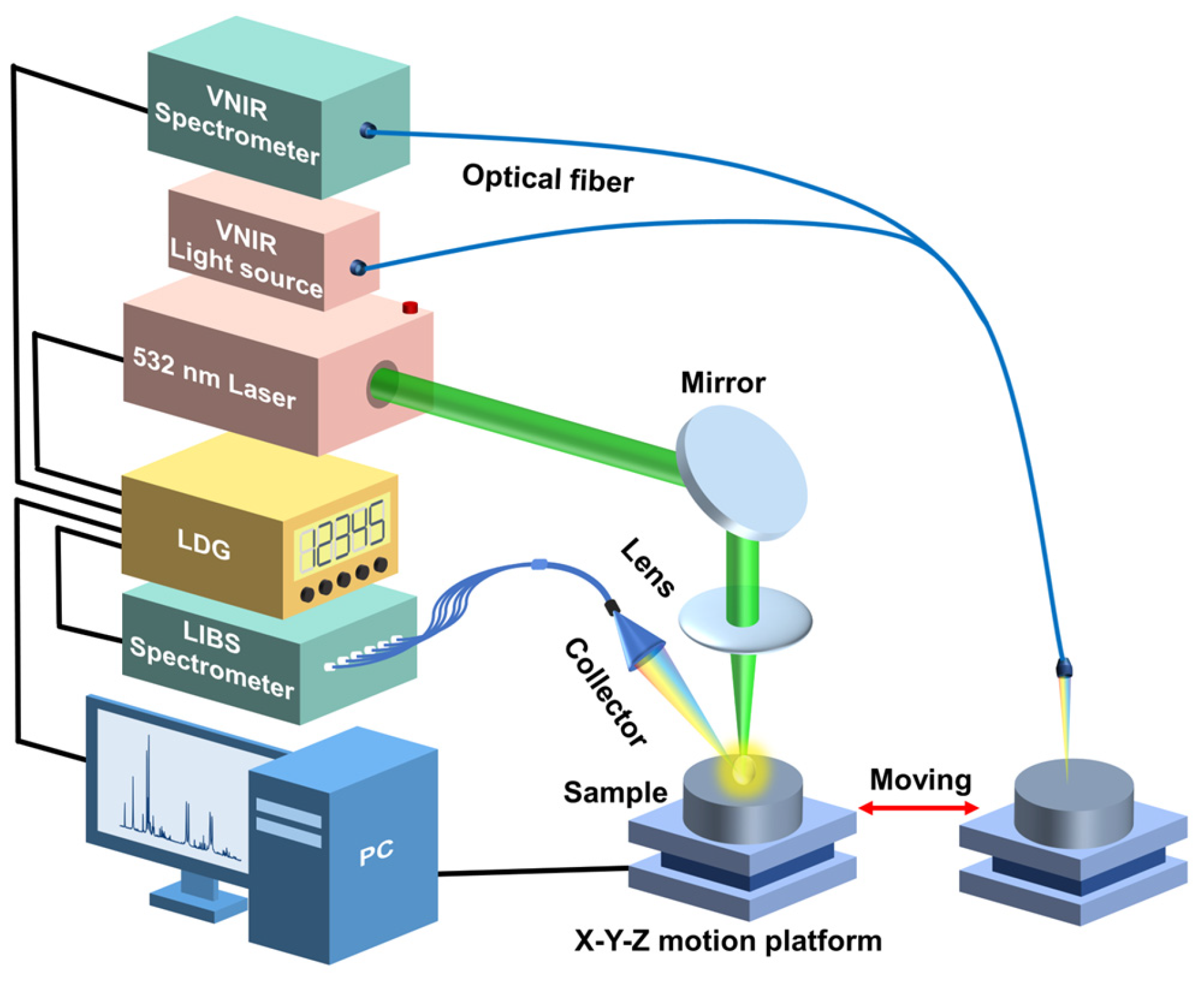
The schematic diagram of the experimental setup used in this work is shown in
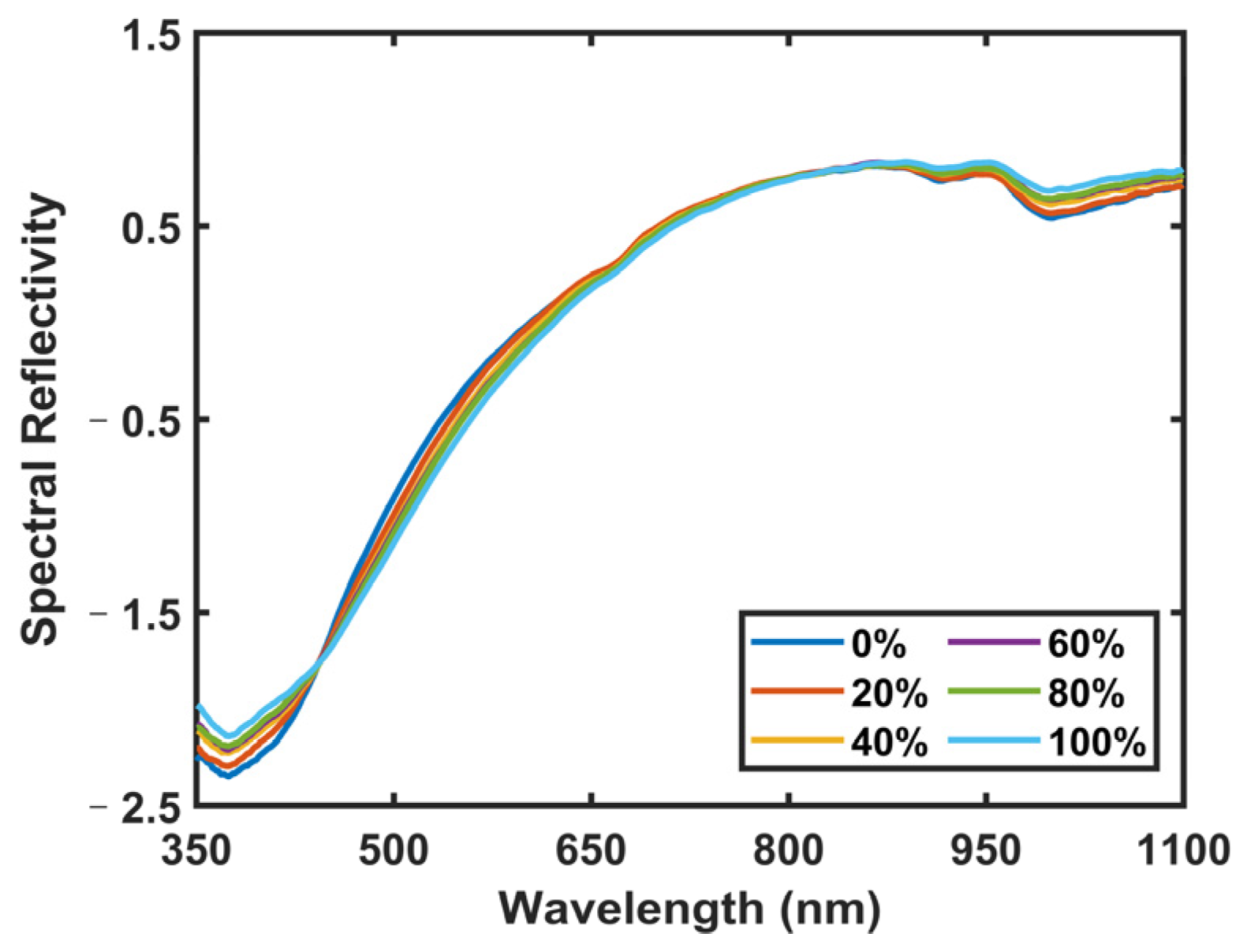
Figure 1. This experimental setup mainly consisted of two parts: one was the VNIR acquisition setup, and the other was the LIBS acquisition setup. The VNIR spectra of the samples were collected first. All VNIR spectra were collected using a VNIR spectrometer (QE65pro, spectral ranges: 350–1100 nm; Ocean Optics, Inc., Dunedin, USA) equipped with a Halogen lamp light source (Avalight-HAL-Mini, Avantes B.V., Apeldoom, Netherlands). The samples were placed on an X-Y-Z motion platform (DZY110TA-3Z, Beijing Jiangyun Juli Technology Co., Ltd., Beijing, China) to enable spectral collection at different positions. For the spectral collection process, a precision optical fiber probe was positioned perpendicularly above each sample, thereby enabling the acquisition of the diffuse reflectance spectra. The integration time for each scan was 10 milliseconds, and each spectrum was obtained by averaging ten consecutive scans at each spatial point. In total, 100 distinct spectra per pellet sample were collected at 100 different spatial points. Consequently, for each proportion (adulteration level) of the adulterated samples, we amassed 200 spectra, culminating in a comprehensive dataset comprising 1200 VNIR spectra for six adulteration levels. Each VNIR spectrum had a dimension of 1 × 997.
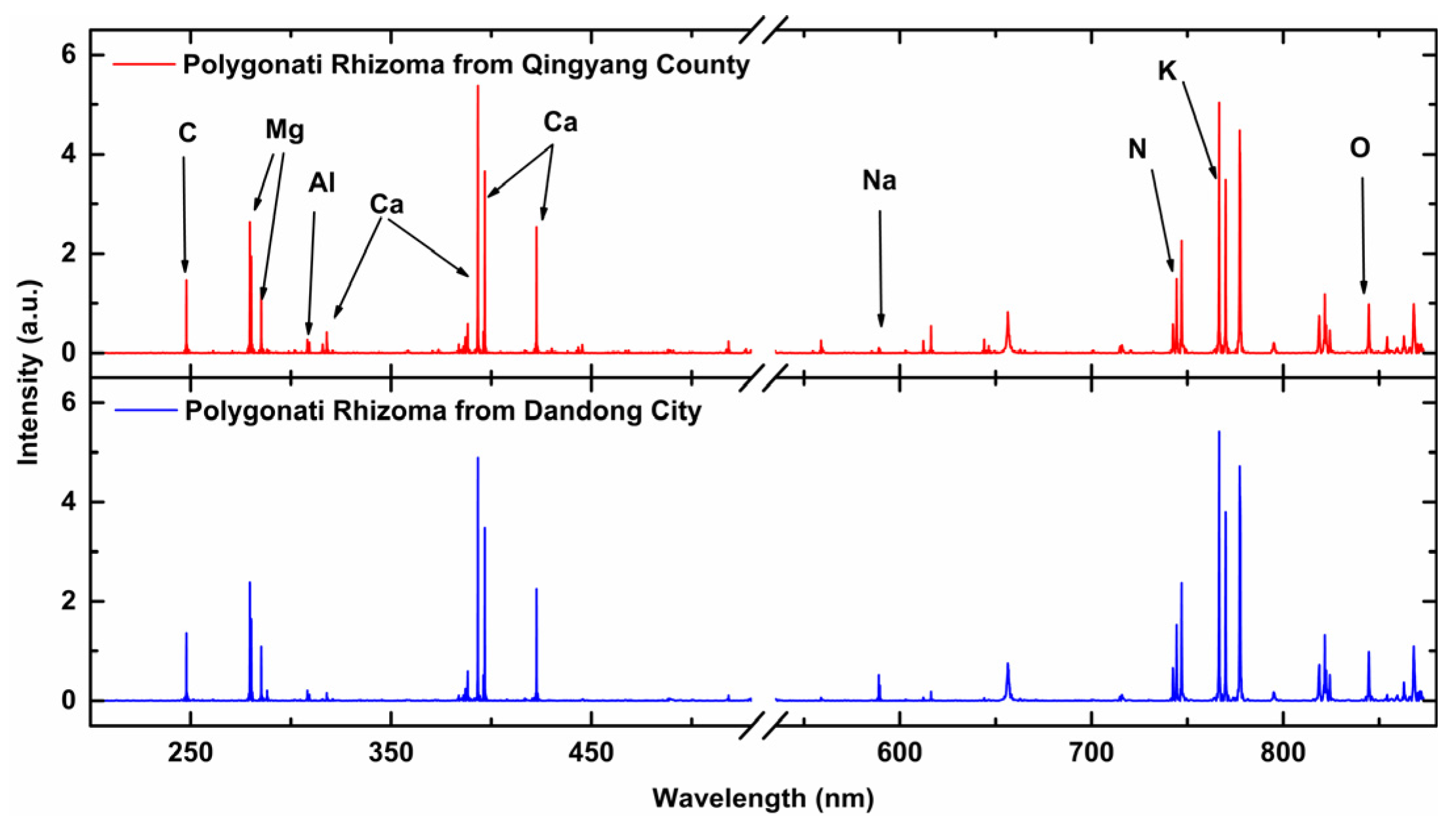
After the VNIR acquisition, the samples were moved to the LIBS acquisition setup through the displacement platform. For the LIBS acquisition setup, a Q-switched Nd: YAG laser (Beamtech Optronics, Nimma-400; pulse duration: 8 ns; flattened Gaussian beam; Beamtech Optronics Co., Ltd., Beijing, China) operating at 532 nm, 1 Hz, and 130 mJ was used as the ablation source. The laser beam was reflected by a 45° mirror and focused by a quartz lens (focal length: 150 mm) onto the sample surface to generate plasmas. The plasma emission was collected by a collector and transmitted by fiber to a six-channel spectrometer (AvaSpec-ULS4096CL-EVO; spectral ranges: 196–874 nm; minimum gate width: 9 μs; Avantes B.V., Apeldoom, Netherlands). The gate delay and width were set to 2 μs, and 9 μs, respectively. A digital delay generator (LDG 3.0, Wuhan NRD Laser Engineering Co., Ltd., Wuhan, China) synchronized the laser and spectrograph. The experiment was conducted in the air atmosphere. For each pellet sample, 400 spectra were obtained and then averaged to 100 spectra to improve the stability of spectral intensity. Thus, 200 spectra for each proportion and 1200 spectra in total were obtained. Each LIBS spectrum had a dimension of 1 × 24,564.
5. Conclusions
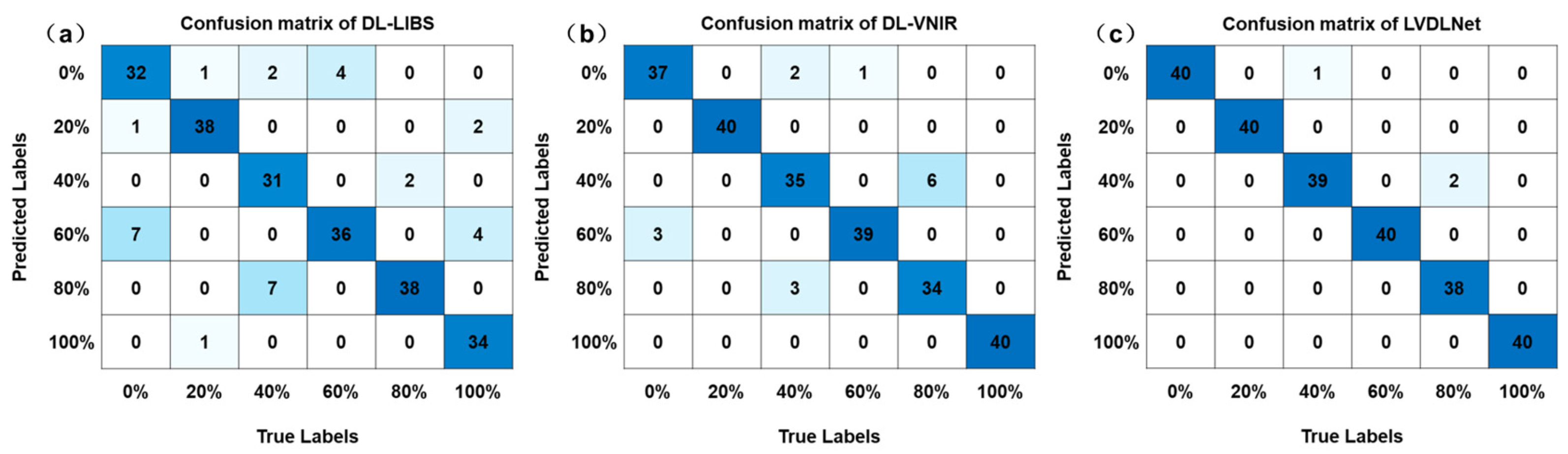
Food adulteration identification is essential for protecting consumers’ interests, but no universal method has been widely adopted, especially in industrial scenarios. This study presented a novel deep learning framework, LIBS-VNIR fusion based on a deep learning network (LVDLNet), for identifying adulterated Polygonati Rhizoma (PR). In the LVDLNet model, an interval point standardization strategy in LIBS and a refined peak and trough focus in VNIR data processing improved signal clarity and extraction efficiency. By integrating LIBS elemental information with VNIR molecular information, we enhanced the accuracy of authentication. The LVDLNet model achieved good results, with the accuracy (Acc), macro-F measure (Mac_F), macro-precision (Mac_P), and macro-recall (Mac_R) being 98.75%, 98.50%, 98.78%, and 98.75%, respectively. It significantly enhanced the classification evaluation metrics, increasing them from approximately 87% for LIBS and 93% for VNIR to over 98%. Additionally, tests on various adulterated source samples further confirmed the efficacy of the LVDLNet model, with all four classification metrics improved from about 87% for LIBS and 86% for VNIR to above 96%. In addition, this work confirmed the classification effect of the proposed method from different feature inputs and conventional machine learning models. All in all, this study presented a pioneering deep learning framework that synergizes LIBS and VNIR to effectively detect adulterated PR, offering a novel perspective and methodology for identifying food adulteration. Future work can apply this deep learning framework to a wider range of samples, including more different origins and different types of foods, which may be affected by adulteration.