Study on the Changes and Correlation of Microorganisms and Flavor in Different Processing Stages of Mianning Ham
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Sampling
2.2. Genomic DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
2.3. Illumina Miseq Sequencing
2.4. Processing and Calculation of Volatile Flavor Compounds in Samples
3. Results
3.1. Analysis of Microbial Diversity in Different Processing Stages
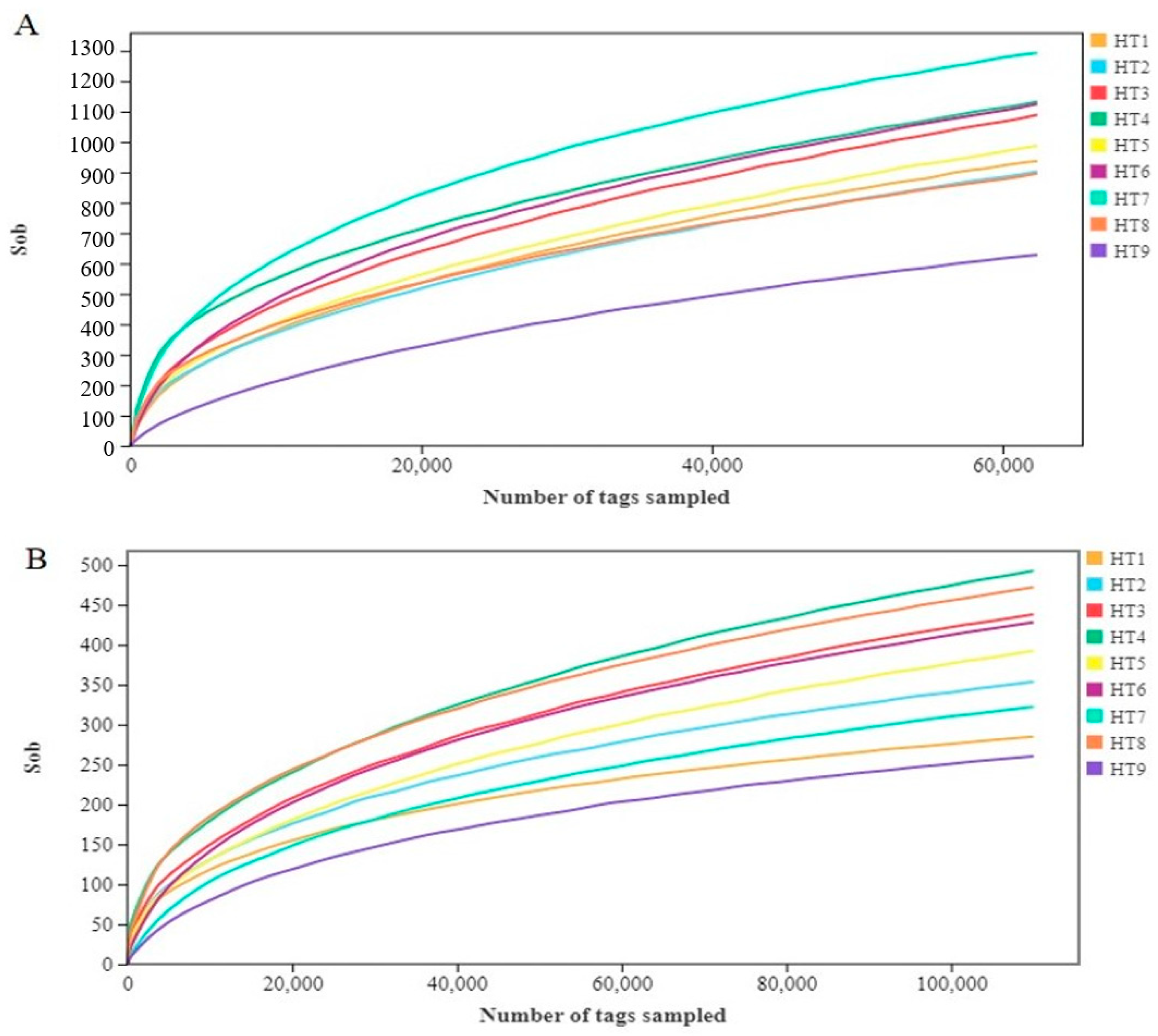
3.1.1. Microbial Sequencing Data
3.1.2. Microbial Diversity Analysis
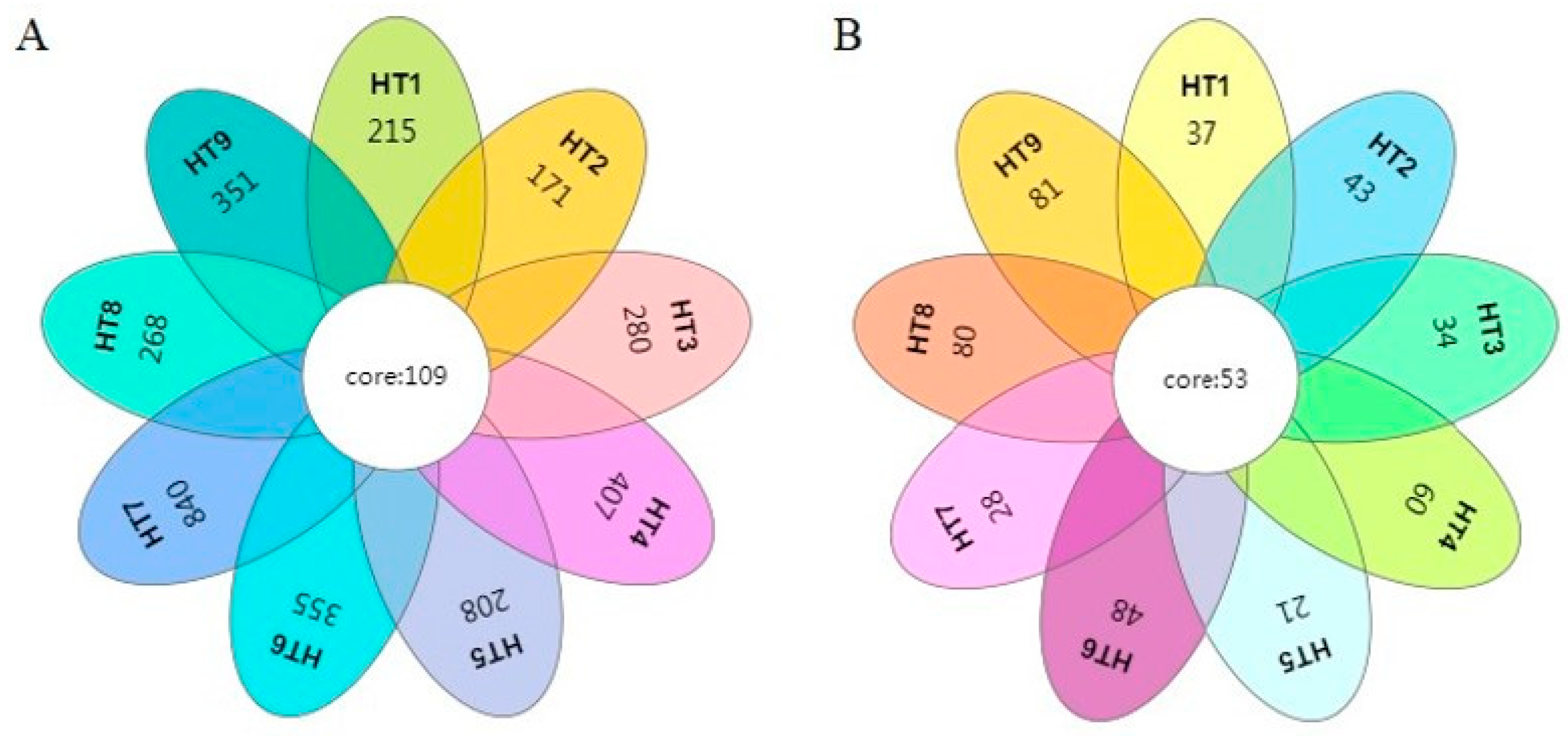
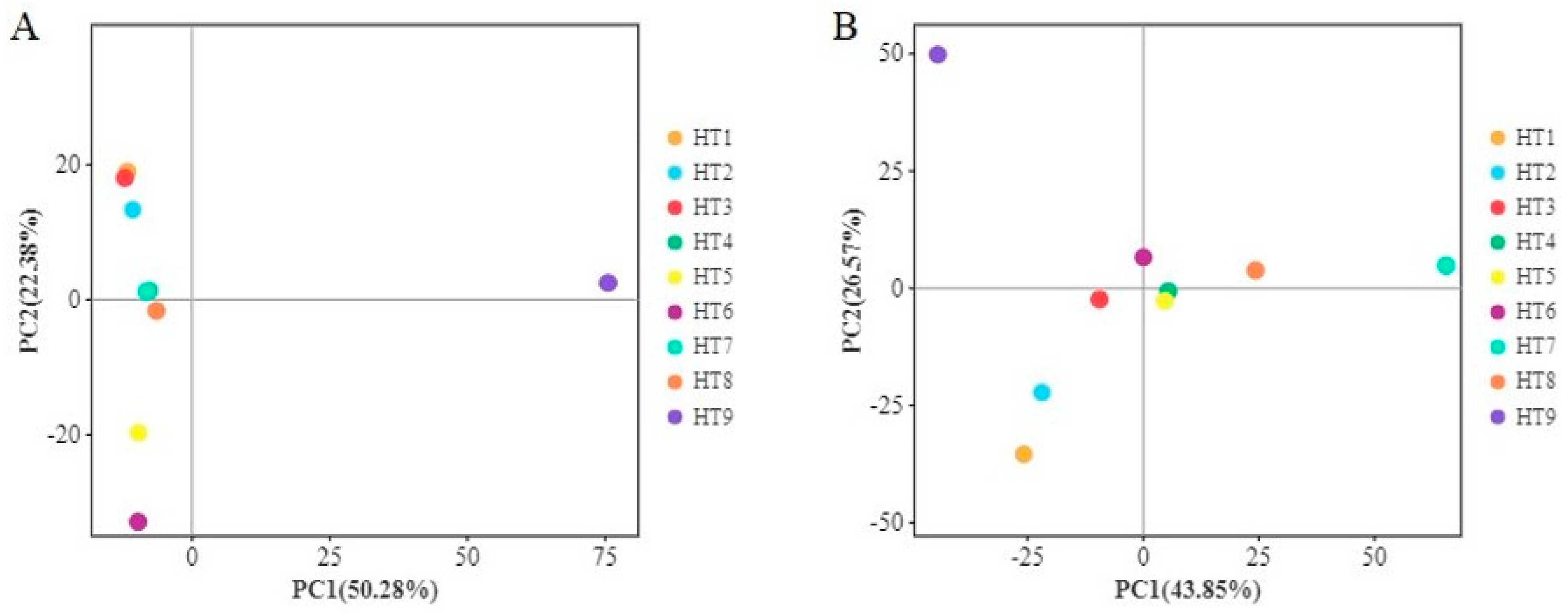
3.1.3. Analysis of Microbial Community Differences
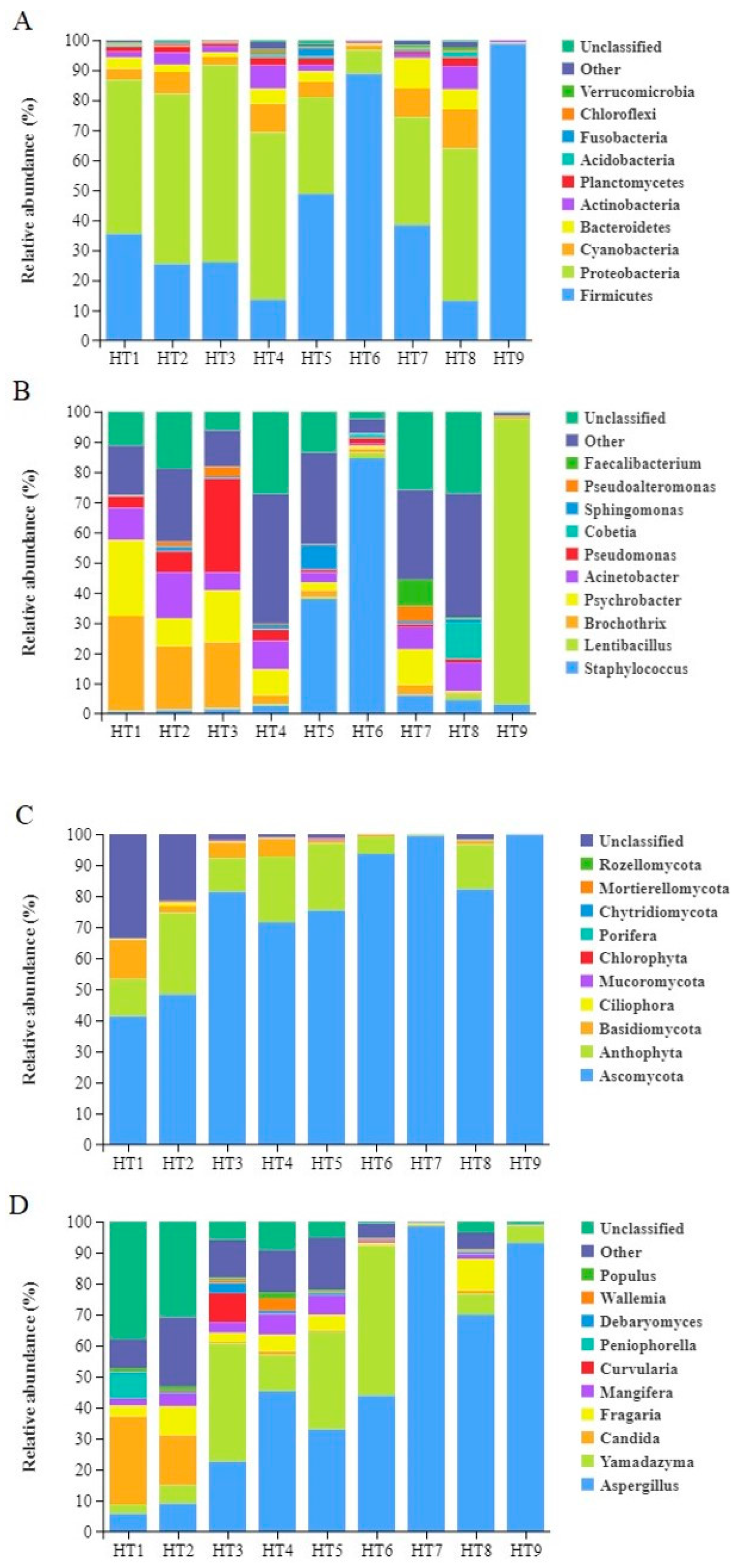
3.1.4. Analysis of Microbial Community Composition
3.2. Analysis of Volatile Flavor Compounds
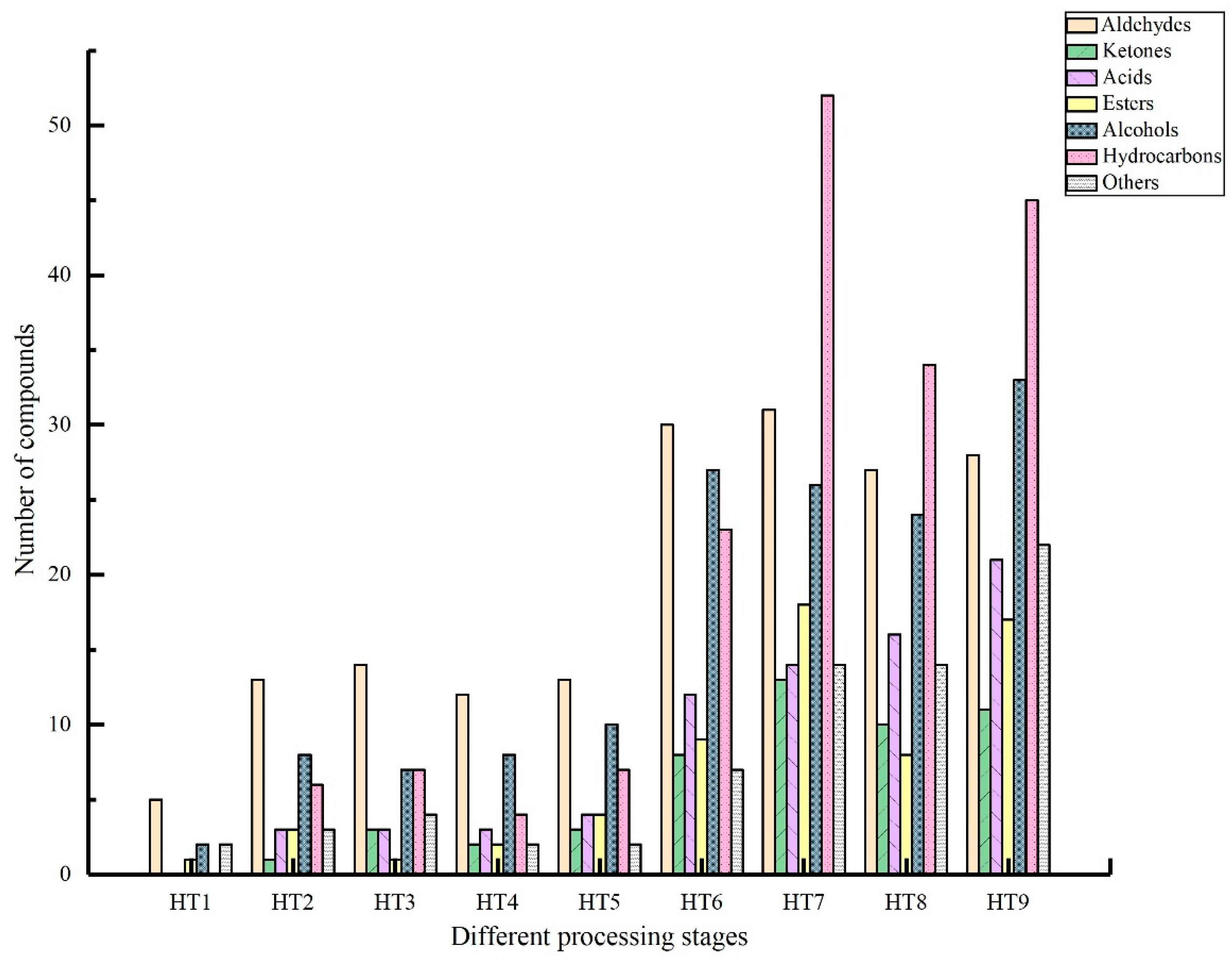
3.2.1. Analysis of Volatile Flavor Compounds in Different Processing Stages
3.2.2. Key Volatile Flavor Compounds
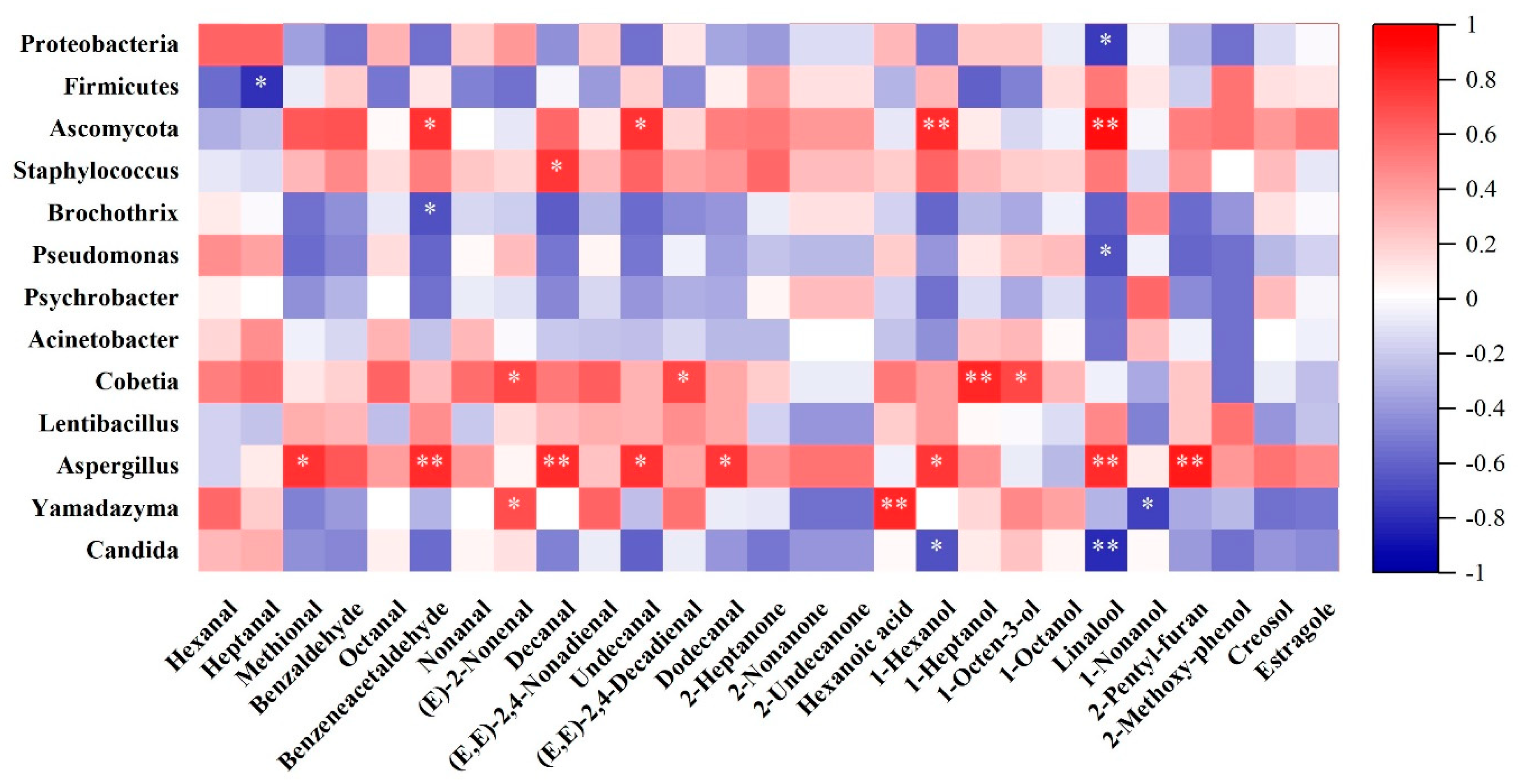
3.2.3. Correlation Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, X.H.; Ni, X.X.; Han, J.H.; Yao, W.H.; Fang, Y.J.; Zhu, Q.; Xu, M.F. High-intensity ultrasound modified the functional properties of Neosalanx taihuensis myofibrillar protein and improved its emulsion stability. Ultrason. Sonochem. 2023, 97, 106458. [Google Scholar] [CrossRef] [PubMed]
- Antequera, T.; López-Bote, C.J.; Córdoba, J.J.; García, C.; Asensio, M.A.; Ventanas, J.; García-Regueiro, J.A.; Díaz, I. Lipid oxidative changes in the processing of Iberian pig hams. Food Chem. 1992, 45, 105–110. [Google Scholar] [CrossRef]
- Petrova, I.; Aasen, I.M.; Rustad, T.; Eikevik, T.M. Manufacture of dry-cured ham: A review. Part 1. Biochemical changes during the technological process. Eur. Food Res. Technol. 2015, 241, 587–599. [Google Scholar] [CrossRef]
- Zhao, G.M.; Zhou, G.H.; Tian, W.; Xu, X.L.; Wang, Y.L.; Luo, X. Changes of alanyl aminopeptidase activity and free amino acid contents in biceps femoris during processing of Jinhua ham. Meat Sci. 2005, 71, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Z.; Ji, L.; Zhang, J.; Zhao, Z.; Zhang, R.; Bai, T.; Hou, B.; Wang, W. Flavor Composition and Microbial Community Structure of Mianning Ham. Front. Microbiol. 2021, 11, 623775. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Xu, H.; Li, X.; Wu, Y.; Xu, B.J.L. Correlation of characteristic flavor and microbial community in Jinhua ham during the post-ripening stage. LWT 2022, 171, 114067. [Google Scholar] [CrossRef]
- Zhang, X.M.; Dang, X.J.; Wang, Y.B.; Sun, T.; Wang, Y.; Yu, H.; Yang, W.S. Diversity and composition of microbiota during fermentation of traditional Nuodeng ham. J. Microbiol. 2021, 59, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Laranjo, M.; Elias, M.; Fraqueza, M.J. The Use of Starter Cultures in Traditional Meat Products. J. Food Qual. 2017, 2017, 9546026. [Google Scholar] [CrossRef]
- Ashaolu, T.J.; Khalifa, I.; Mesak, M.A.; Lorenzo, J.M.; Farag, M.A. A comprehensive review of the role of microorganisms on texture change, flavor and biogenic amines formation in fermented meat with their action mechanisms and safety. Crit. Rev. Food Sci. Nutr. 2023, 63, 3538–3555. [Google Scholar] [CrossRef]
- Comi, G.; Orlic, S.; Redzepovic, S.; Urso, R.; Iacumin, L. Moulds isolated from Istrian dried ham at the pre-ripening and ripening level. Int. J. Food Microbiol. 2004, 96, 29–34. [Google Scholar] [CrossRef]
- Li, P.; Bao, Z.; Wang, Y.; Su, X.; Zhou, H.; Xu, B. Role of microbiota and its ecological succession on flavor formation in traditional dry-cured ham: A review. Crit. Rev. Food Sci. Nutr. 2023, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Radulović, Z.; Živković, D.; Mirković, N.; Petrušić, M.; Stajić, S.; Perunović, M.; Paunović, D. Effect of probiotic bacteria on chemical composition and sensory quality of fermented sausages. Procedia Food Sci. 2011, 1, 1516–1522. [Google Scholar] [CrossRef]
- Zhong, A.; Chen, W.; Duan, Y.; Li, K.; Tang, X.; Tian, X.; Wu, Z.; Li, Z.; Wang, Y.; Wang, C. The potential correlation between microbial communities and flavors in traditional fermented sour meat. LWT 2021, 149, 111873. [Google Scholar] [CrossRef]
- Guo, M.; Wu, F.; Hao, G.; Qi, Q.; Li, R.; Li, N.; Wei, L.; Chai, T. Bacillus subtilis Improves Immunity and Disease Resistance in Rabbits. Front. Immunol. 2017, 8, 354. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, S.; Zhao, H. Unraveling microbial community diversity and succession of Chinese Sichuan sausages during spontaneous fermentation by high-throughput sequencing. J. Food Sci. Technol. 2019, 56, 3254–3263. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.L.; Liu, S.Y.; Wang, G.Y.; Pu, Y.H.; Ge, C.R.; Liao, G.Z. Analysis of bacterial community structure of Xuanwei ham based on PCR-DGGE. Food Res. Dev. 2020, 41, 192–198. [Google Scholar] [CrossRef]
- Wang, X.R.; Shi, Q.; Liu, B.Q.; Lei, Y.D.; Tang, H.H.; Zhang, S.Z.; Zhang, J.J.; Li, H. Bacterial dynamics during the processing of Nuodeng dry-cured ham. Sci. Technol. Food Ind. 2021, 42, 83–89+98. [Google Scholar] [CrossRef]
- Lin, D.Y.; Song, C.L.; Zhang, Y.; Zhang, Y.J.; Quang, W.; Shu, X.H. Comparative analysis of microbial diversity of five dry-cured hams in Yunnan. China Food Addit. 2023, 34, 233–241. [Google Scholar] [CrossRef]
- Yang, Z.; Liao, G.; Wan, D.; Kong, W.; Li, C.; Gu, D.; Pu, Y.; Ge, C.; Wang, G. Combined application of high-throughput sequencing and LC-MS/MS-based metabolomics to evaluate the formation of Zn-protoporphyrin in Nuodeng ham. Food Res. Int. 2022, 162, 112209. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, M.O.; Thapa, N.; Shangpliang, H.N.; Tamang, J.P. Metataxonomic profiling of bacterial communities and their predictive functional profiles in traditionally preserved meat products of Sikkim state in India. Food Res. Int. 2021, 140, 110002. [Google Scholar] [CrossRef] [PubMed]
- Betts, G. 23-Other spoilage bacteria. In Food Spoilage Microorganisms; Blackburn, C.d.W., Ed.; Woodhead Publishing: Cambridge, UK, 2006; pp. 668–693. [Google Scholar]
- Lippolis, R.; Rossi, C.; De Angelis, M.; Minervini, F.; Paparella, A.; Chaves-López, C. Adaptive remodelling of blue pigmenting Pseudomonas fluorescens pf59 proteome in response to different environmental conditions. Food Control 2021, 127, 108105. [Google Scholar] [CrossRef]
- Correa, A.G. CHAPTER 131-ACINETOBACTER. In Feigin and Cherry’s Textbook of Pediatric Infectious Diseases, 6th ed.; Feigin, R.D., Cherry, J.D., Demmler-Harrison, G.J., Kaplan, S.L., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2009; pp. 1638–1642. [Google Scholar]
- Wang, Y.; Li, F.; Chen, J.; Sun, Z.; Wang, F.; Wang, C.; Fu, L. High-throughput sequencing-based characterization of the predominant microbial community associated with characteristic flavor formation in Jinhua Ham. Food Microbiol. 2021, 94, 103643. [Google Scholar] [CrossRef]
- Yang, X. Moraxellaceae. In Encyclopedia of Food Microbiology, 6th ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 826–833. [Google Scholar]
- Ventosa, A.; Arahal, D.R. Cobetia. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2020; pp. 1–9. [Google Scholar]
- Jung, M.J.; Roh, S.W.; Kim, M.S.; Whon, T.W.; Bae, J.W. Genome sequence of Lentibacillus jeotgali Grbi(T), isolated from traditional Korean salt-fermented seafood. J. Bacteriol. 2011, 193, 6414–6415. [Google Scholar] [CrossRef][Green Version]
- Heyrman, J.; Vos, P.D. Lentibacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Lin, F.; Cai, F.; Luo, B.; Gu, R.; Ahmed, S.; Long, C. Variation of Microbiological and Biochemical Profiles of Laowo Dry-Cured Ham, an Indigenous Fermented Food, during Ripening by GC-TOF-MS and UPLC-QTOF-MS. J. Agric. Food Chem. 2020, 68, 8925–8935. [Google Scholar] [CrossRef]
- Li, C.; Zheng, Z.; Wang, G.; Chen, G.; Zhou, N.; Zhong, Y.; Yang, Y.; Wu, H.; Yang, C.; Liao, G. Revealing the intrinsic relationship between microbial communities and physicochemical properties during ripening of Xuanwei ham. Food Res. Int. 2024, 186, 114377. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Shu, X.H.; Huang, X.; Lu, T.; Wang, Y.L.; Zhang, Y.L.; Zhang, Z.H.; Luo, G.; Li, X.H.; Li, X.N.; et al. Applying 16S rDNA sequencing to analyze the microbial diversity on the surface and in the interior of Saba ham, a traditional chinese fermented meat product. Meat Res. 2020, 34, 26–32. [Google Scholar] [CrossRef]
- Hong, S.-B.; Kim, D.-H.; Samson, R.A. Aspergillus Associated with Meju, a Fermented Soybean Starting Material for Traditional Soy Sauce and Soybean Paste in Korea. Mycobiology 2015, 43, 218–224. [Google Scholar] [CrossRef]
- Deng, X.Y.; Li, J.W.; He, L.C.; Zhang, Y.Y.; Huang, G.W.; Bao, X.L.; Qiu, C.K. Microbial community succession pattern on the surface of Xuanen ham during fermentation. Food Ferment. Ind. 2021, 47, 34–42. [Google Scholar] [CrossRef]
- Li, Y.F.; Shi, Y.N.; Wei, G.Q.; Li, X.; Huang, A.X. Changes of volatile flavor substances in the fermentation of Dahe black pig ham under standardized technological conditions. Mod. Food Sci. Technol. 2021, 37, 240–251. [Google Scholar] [CrossRef]
- Gao, Y.; Li, S.J.; Liang, D.N.; Hu, Y.J.; Xu, Q.L. Volatile flavor components of Sanchuan ham and air-dried ham. Meat Res. 2021, 35, 41–46. [Google Scholar] [CrossRef]
- Loury, M. Possible mechanisms of autoxidative rancidity. Lipids 1972, 7, 671–675. [Google Scholar] [CrossRef]
- Shahidi, F.; Rubin, L.J.; D’Souza, L.A. Meat flavor volatiles: A review of the composition, techniques of analysis, and sensory evaluation. Crit. Rev. Food Sci. Nutr. 1986, 24, 141–243. [Google Scholar] [CrossRef]
- Li, C.; Zou, Y.; Liao, G.; Zheng, Z.; Chen, G.; Zhong, Y.; Wang, G. Identification of characteristic flavor compounds and small molecule metabolites during the ripening process of Nuodeng ham by GC-IMS, GC–MS combined with metabolomics. Food Chem. 2024, 440, 138188. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.; Grimm, C.C.; Toldra, F.; Spanier, A.M. Correlations of sensory and volatile compounds of Spanish “Serrano” dry-cured ham as a function of two processing times. J. Agric. Food Chem. 1997, 45, 2178–2186. [Google Scholar] [CrossRef]
- Aparicio, R.; Morales, M.T.; Alonso, M.V. Relationship between volatile compounds and sensory attributes of olive oils by the sensory wheel. J. Am. Oil Chem. Soc. 1996, 73, 1253–1264. [Google Scholar] [CrossRef]
- Ramírez, R.; Cava, R. Volatile profiles of dry-cured meat products from three different Iberian x Duroc genotypes. J. Agric. Food Chem. 2007, 55, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Toldrá, F.; Aristoy, M.C. Dry-Cured Ham. In Handbook of Meat Processing; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 351–362. [Google Scholar]
- Vermeulen, N.; Czerny, M.; Gänzle, M.G.; Schieberle, P.; Vogel, R.F. Reduction of (E)-2-nonenal and (E,E)-2,4-decadienal during sourdough fermentation. J. Cereal Sci. 2007, 45, 78–87. [Google Scholar] [CrossRef]
- Feng, Y.; Su, G.; Sun-Waterhouse, D.; Cai, Y.; Zhao, H.; Cui, C.; Zhao, M. Optimization of Headspace Solid-Phase Micro-extraction (HS-SPME) for Analyzing Soy Sauce Aroma Compounds via Coupling with Direct GC-Olfactometry (D-GC-O) and Gas Chromatography-Mass Spectrometry (GC-MS). Food Anal. Methods 2017, 10, 713–726. [Google Scholar] [CrossRef]
- Wang, W.; Feng, X.; Zhang, D.; Li, B.; Sun, B.; Tian, H.; Liu, Y. Analysis of volatile compounds in Chinese dry-cured hams by comprehensive two-dimensional gas chromatography with high-resolution time-of-flight mass spectrometry. Meat Sci. 2018, 140, 14–25. [Google Scholar] [CrossRef]
| Group | Observed OTUs | Shannon Index | Simpson | Chao Index | Ace Index | Good’s Coverage |
|---|---|---|---|---|---|---|
| HT1 | 1198.00 ± 74.67 | 4.61 ± 1.96 | 0.83 ± 0.18 | 1983.71 ± 114.59 | 2048.28 ± 136.15 | 0.99 ± 0.00 |
| HT2 | 1166.00 ± 84.12 | 5.28 ± 1.93 | 0.87 ± 0.17 | 1896.46 ± 77.46 | 2022.61 ± 102.18 | 1.00 ± 0.00 |
| HT3 | 1400.00 ± 78.48 | 4.78 ± 0.93 | 0.86 ± 0.09 | 2299.60 ± 132.60 | 2369.53 ± 102.40 | 0.99 ± 0.00 |
| HT4 | 1430.67 ± 60.87 | 7.22 ± 0.11 | 0.98 ± 0.00 | 2339.85 ± 80.57 | 2469.03 ± 47.70 | 0.99 ± 0.00 |
| HT5 | 1264.67 ± 128.59 | 5.27 ± 0.49 | 0.90 ± 0.05 | 2113.66 ± 197.84 | 2322.46 ± 170.39 | 0.99 ± 0.00 |
| HT6 | 1355.33 ± 20.21 | 4.30 ± 0.04 | 0.78 ± 0.00 | 2115.79 ± 49.11 | 2177.94 ± 33.50 | 0.99 ± 0.00 |
| HT7 | 1404.00 ± 277.70 | 6.35 ± 0.50 | 0.95 ± 0.02 | 2006.18 ± 219.88 | 2125.99 ± 235.03 | 0.99 ± 0.00 |
| HT8 | 1160.67 ± 98.57 | 6.26 ± 0.40 | 0.96 ± 0.03 | 1924.93 ± 127.87 | 2056.64 ± 126.76 | 1.00 ± 0.00 |
| HT9 | 832.67 ± 34.53 | 1.41 ± 0.19 | 0.28 ± 0.05 | 1293.89 ± 20.65 | 1411.36 ± 22.90 | 1.00 ± 0.00 |
| Group | Observed OTUs | Shannon Index | Simpson | Chao Index | Ace Index | Good’s Coverage |
|---|---|---|---|---|---|---|
| HT1 | 290.00 ± 117.48 | 2.84 ± 1.48 | 0.71 ± 0.17 | 418.71 ± 206.38 | 409.47 ± 209.09 | 1.00 ± 0.00 |
| HT2 | 371.00 ± 116.50 | 3.87 ± 1.86 | 0.82 ± 0.21 | 608.01 ± 179.59 | 562.92 ± 196.29 | 1.00 ± 0.00 |
| HT3 | 453.67 ± 35.50 | 3.56 ± 0.58 | 0.80 ± 0.05 | 681.33 ± 81.51 | 701.86 ± 77.60 | 1.00 ± 0.00 |
| HT4 | 517.00 ± 44.31 | 4.28 ± 0.34 | 0.87 ± 0.04 | 736.83 ± 39.29 | 780.05 ± 41.75 | 1.00 ± 0.00 |
| HT5 | 416.00 ± 25.12 | 3.49 ± 0.22 | 0.81 ± 0.02 | 637.21 ± 27.76 | 671.17 ± 42.81 | 1.00 ± 0.00 |
| HT6 | 443.67 ± 6.43 | 2.29 ± 0.03 | 0.67 ± 0.01 | 687.36 ± 40.14 | 666.42 ± 20.26 | 1.00 ± 0.00 |
| HT7 | 335.33 ± 38.03 | 0.57 ± 0.02 | 0.11 ± 0.01 | 534.35 ± 23.44 | 526.25 ± 60.44 | 1.00 ± 0.00 |
| HT8 | 493.67 ± 37.42 | 2.92 ± 0.34 | 0.69 ± 0.07 | 726.49 ± 31.17 | 741.44 ± 51.36 | 1.00 ± 0.00 |
| HT9 | 266.00 ± 31.43 | 1.22 ± 0.20 | 0.38 ± 0.11 | 378.32 ± 44.55 | 387.73 ± 55.90 | 1.00 ± 0.00 |
| Compounds | Absolute Content (μg/kg) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HT1 | HT2 | HT3 | HT4 | HT5 | HT6 | HT7 | HT8 | HT9 | Total | |
| Aldehydes | 101.37 | 6224.53 | 7127.77 | 10,527.94 | 5487.13 | 4693.04 | 4977.81 | 7101.65 | 2306.42 | 48,547.65 |
| Ketones | - | 76.26 | 148.58 | 127.63 | 149.612 | 190.8 | 523.12 | 375.31 | 170.57 | 1761.95 |
| Acids | - | 242.94 | 450.29 | 735.17 | 478.75 | 413.1 | 210.3 | 417.21 | 655.4 | 3603.16 |
| Esters | 20.64 | 555.53 | 178.68 | 381.4 | 1016.84 | 444.41 | 659.7 | 654.64 | 649.04 | 4560.88 |
| Alcohols | 25.88 | 1544.72 | 1699.12 | 2519.4 | 2142.66 | 1655.45 | 1038.01 | 1319.42 | 970.99 | 12,915.65 |
| Hydrocarbons | - | 696.99 | 659.16 | 1048.83 | 781.82 | 597.69 | 815.12 | 1012.76 | 611.6 | 6223.97 |
| Others | 23.25 | 1373.82 | 1778.92 | 2015.28 | 927.68 | 1504.21 | 1138.2 | 981.63 | 515.97 | 10,258.96 |
| Total | 171.13 | 10,714.79 | 12,042.53 | 17,355.65 | 10,984.48 | 9498.69 | 9362.33 | 11,862.63 | 5879.98 | 87,872.22 |
| Compounds | Threshold Value (μg/kg) | OAV Value (OVA ≥ 1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HT1 | HT2 | HT3 | HT4 | HT5 | HT6 | HT7 | HT8 | HT9 | ||
| Hexanal | 7.5 | 4.48 | 409.86 | 459.84 | 671.5 | 345.3 | 181.33 | 115.49 | 337 | 111.62 |
| Heptanal | 10 | 0.69 | 34.21 | 32.95 | 47.18 | 22.47 | 18.8 | 24.78 | 33.1 | 9.27 |
| Methional | 0.04 | - | - | - | - | - | - | 525.33 | 807.55 | 334 |
| Benzaldehyde | 50 | 0.13 | - | - | - | - | 1.53 | 4.33 | 4.07 | 1.16 |
| Octanal | 0.1 | 148.04 | 5411.91 | 3805.37 | 6992.07 | 3188.86 | 3995.26 | 5535.58 | 5410.98 | 1268.06 |
| Benzeneacetaldehyde | 9 | - | - | - | - | - | 3.97 | 14.84 | 34.52 | 12.61 |
| Nonanal | 3.5 | 11.27 | 403.98 | 275.87 | 477.73 | 306.6 | 367.2 | 463.29 | 383.03 | 101.25 |
| (E)-2-octenal | 0.07 | - | 1577.11 | 3710.65 | 6882.78 | 2699.82 | 1707.69 | 1525.08 | 3295.86 | 780.82 |
| Decanal | 0.9 | - | 55.76 | 42.87 | 81.38 | 59.68 | 89.83 | 318.45 | 119.03 | 55.83 |
| (E,E)-2,4-Nonadienal | 0.06 | - | - | 1200.57 | 1830.18 | 663.47 | 389.68 | 212.68 | 852.52 | 137 |
| Undecanal | 14 | - | - | - | - | - | 0.82 | 1.82 | 1.3 | 0.29 |
| (E,E)-2,4-Decadienal | 0.03 | - | - | 1725.5 | 3698.97 | 1358.97 | 1208.43 | 1005.7 | 3887.7 | 379.63 |
| Dodecanal | 1.07 | - | - | - | 38.66 | - | 13.34 | 30.71 | 11.13 | 11.59 |
| 2-Heptanone | 70 | - | - | - | - | - | 1 | 1.73 | - | - |
| 2-Nonanone | 25 | - | - | - | - | - | - | 8.1 | - | - |
| 2-Undecanone | 10 | - | - | - | - | - | - | 1.39 | - | - |
| Hexanoic acid | 200 | - | 0.21 | 1.48 | 2.63 | 1.34 | 0.37 | 0.05 | 0.24 | 0.11 |
| 1-Hexanol | 200 | - | 0.24 | 0.16 | 0.32 | 0.13 | 1.06 | 0.7 | 0.34 | 0.37 |
| 1-Heptanol | 200 | - | 0.57 | 0.54 | 0.95 | 0.42 | 0.61 | 0.62 | 0.66 | - |
| 1-Octen-3-ol | 2 | - | 381.75 | 143.25 | 233.43 | 212.81 | 295.18 | 136.22 | 297.03 | 94.81 |
| 1-Octanol | 54 | - | 3.52 | - | - | - | 4.24 | - | - | - |
| Linalool | 1.5 | - | - | - | - | - | 9.69 | 66.56 | 6.66 | 87.89 |
| 1-Nonanol | 2 | 2.15 | - | - | - | - | - | 16.26 | 0 | 0 |
| 2-Pentyl-furan | 4.8 | - | 17.74 | 16.51 | 25.73 | 19.21 | 17.03 | 42.63 | 36.78 | 21.26 |
| 2-Methoxy-phenol | 0.17 | - | - | - | - | - | - | - | - | 124.76 |
| Creosol | 10 | - | - | - | - | - | - | 1.07 | - | - |
| Estragole | 7.5 | - | 7.99 | 6.39 | - | - | - | 18.67 | 3.54 | 12.32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Wang, Z.; Gan, L.; Zhang, J.; Wang, W.; Ji, L.; Chen, L. Study on the Changes and Correlation of Microorganisms and Flavor in Different Processing Stages of Mianning Ham. Foods 2024, 13, 2587. https://doi.org/10.3390/foods13162587
Huang Y, Wang Z, Gan L, Zhang J, Wang W, Ji L, Chen L. Study on the Changes and Correlation of Microorganisms and Flavor in Different Processing Stages of Mianning Ham. Foods. 2024; 13(16):2587. https://doi.org/10.3390/foods13162587
Chicago/Turabian StyleHuang, Yue, Zhengli Wang, Ling Gan, Jiamin Zhang, Wei Wang, Lili Ji, and Lin Chen. 2024. "Study on the Changes and Correlation of Microorganisms and Flavor in Different Processing Stages of Mianning Ham" Foods 13, no. 16: 2587. https://doi.org/10.3390/foods13162587
APA StyleHuang, Y., Wang, Z., Gan, L., Zhang, J., Wang, W., Ji, L., & Chen, L. (2024). Study on the Changes and Correlation of Microorganisms and Flavor in Different Processing Stages of Mianning Ham. Foods, 13(16), 2587. https://doi.org/10.3390/foods13162587




