Association between Gut Microbiota Profiles, Dietary Intake, and Inflammatory Markers in Overweight and Obese Women
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Anthropometric and Blood Pressure Measurement
2.3. Dietary Intake Assessment
2.4. Biochemical Analyses
2.5. DNA Extraction, Sequencing, and Microbiome Data Analyses
2.6. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Study Participants
3.2. Comparison of Nutrient Intake in the Overweight or Obese and Healthy Groups
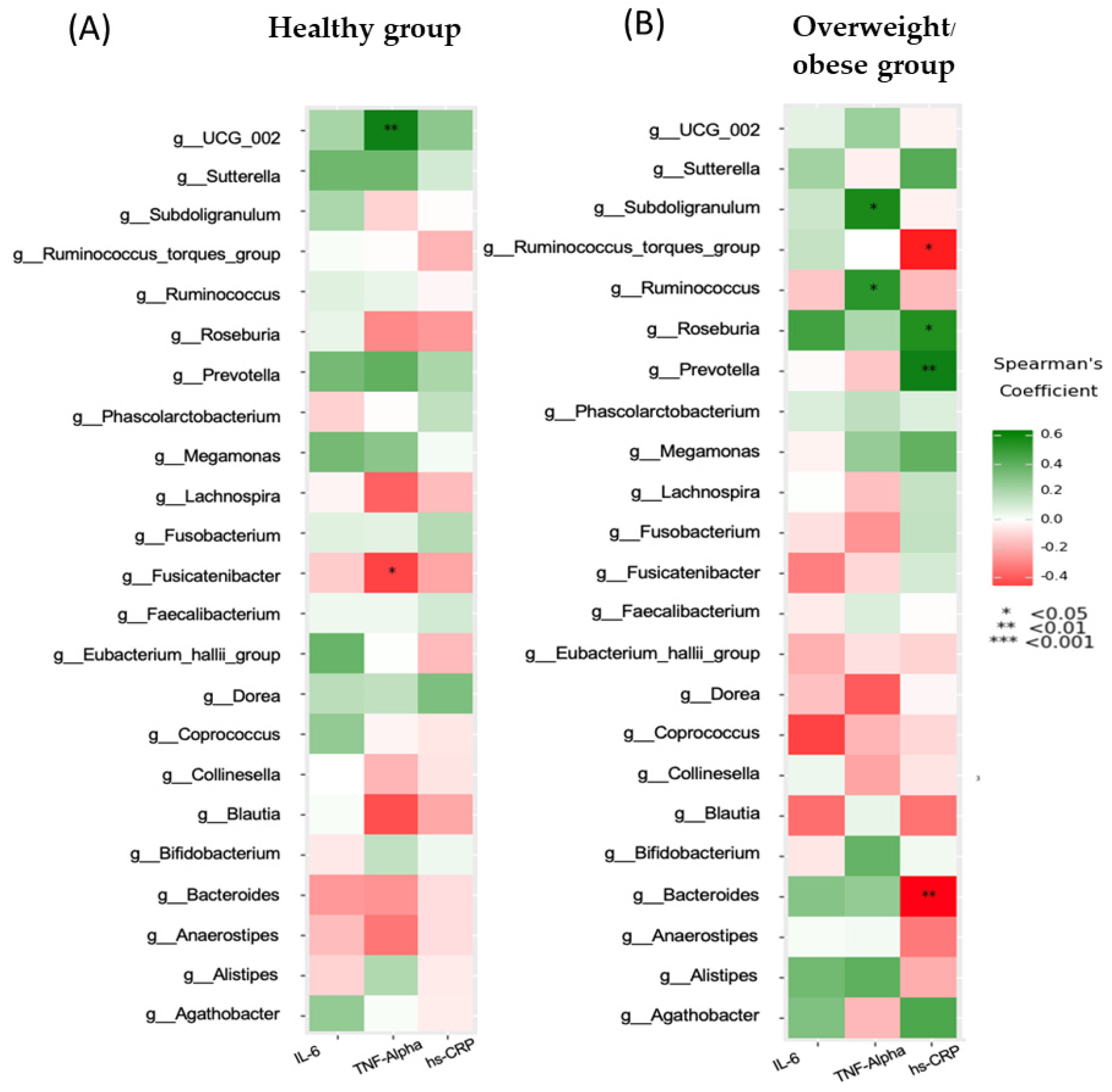
3.3. Association between Inflammatory Markers and Clinical, Biochemical, and Dietary Variables
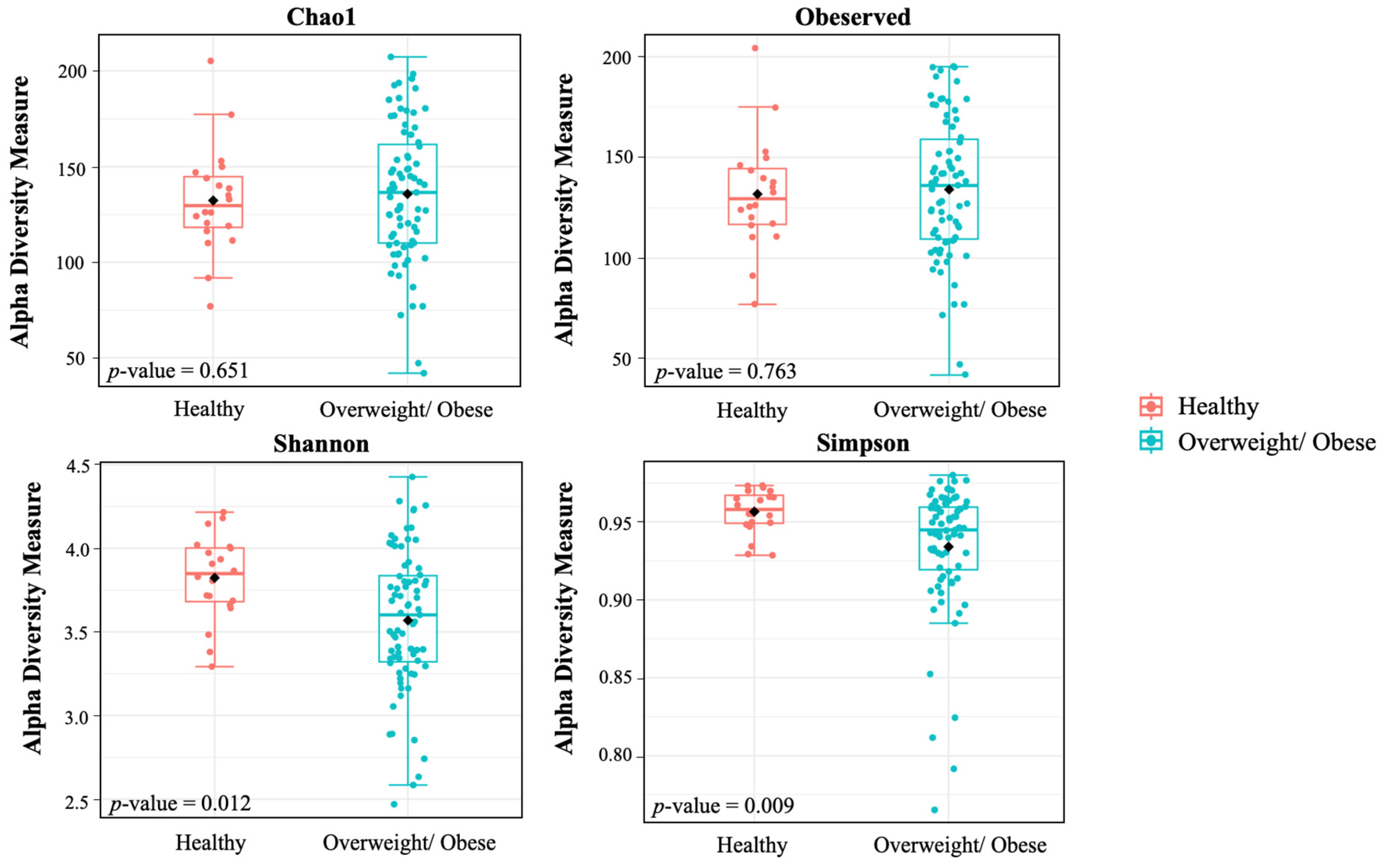
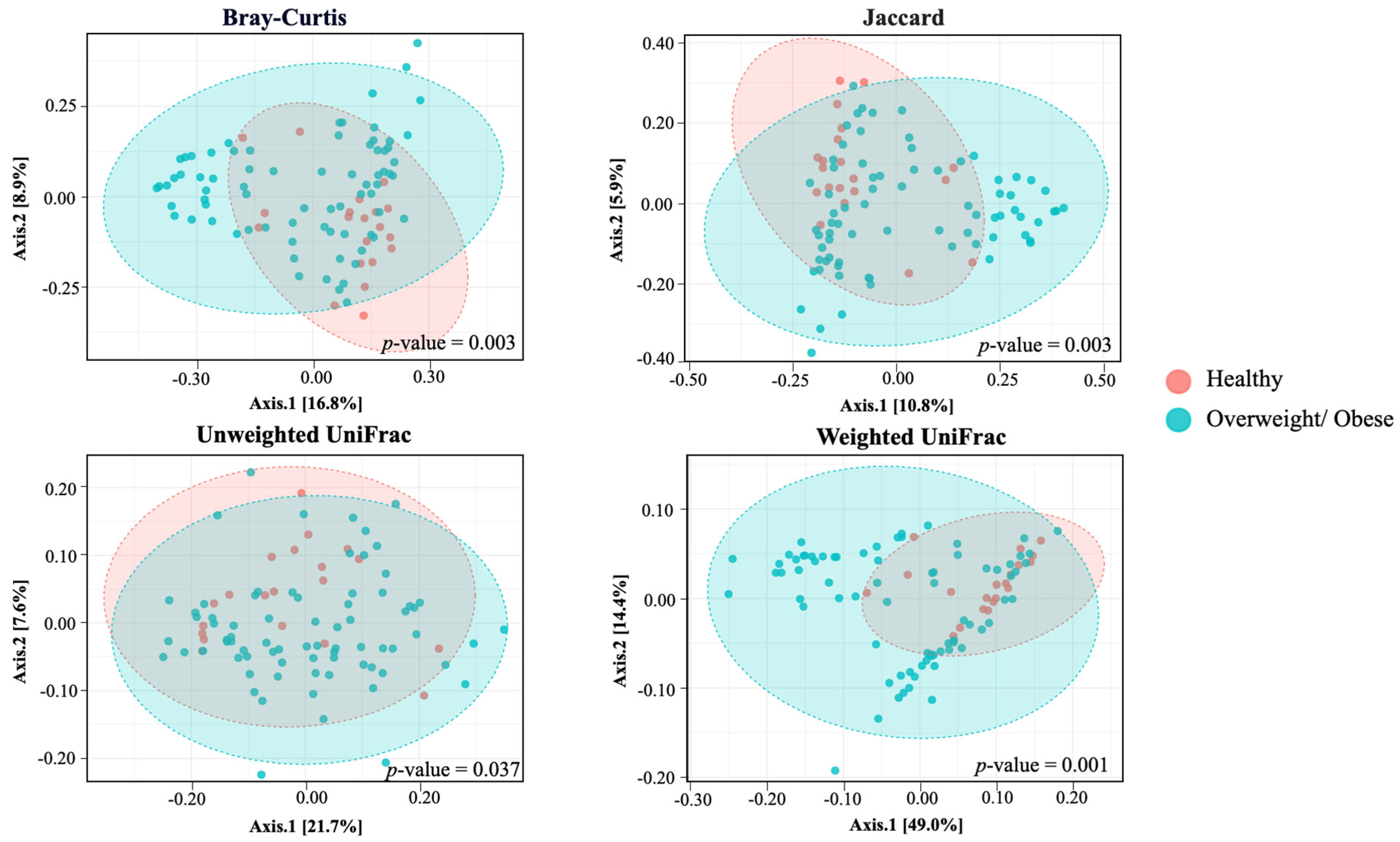
3.4. Alpha and Beta Diversity
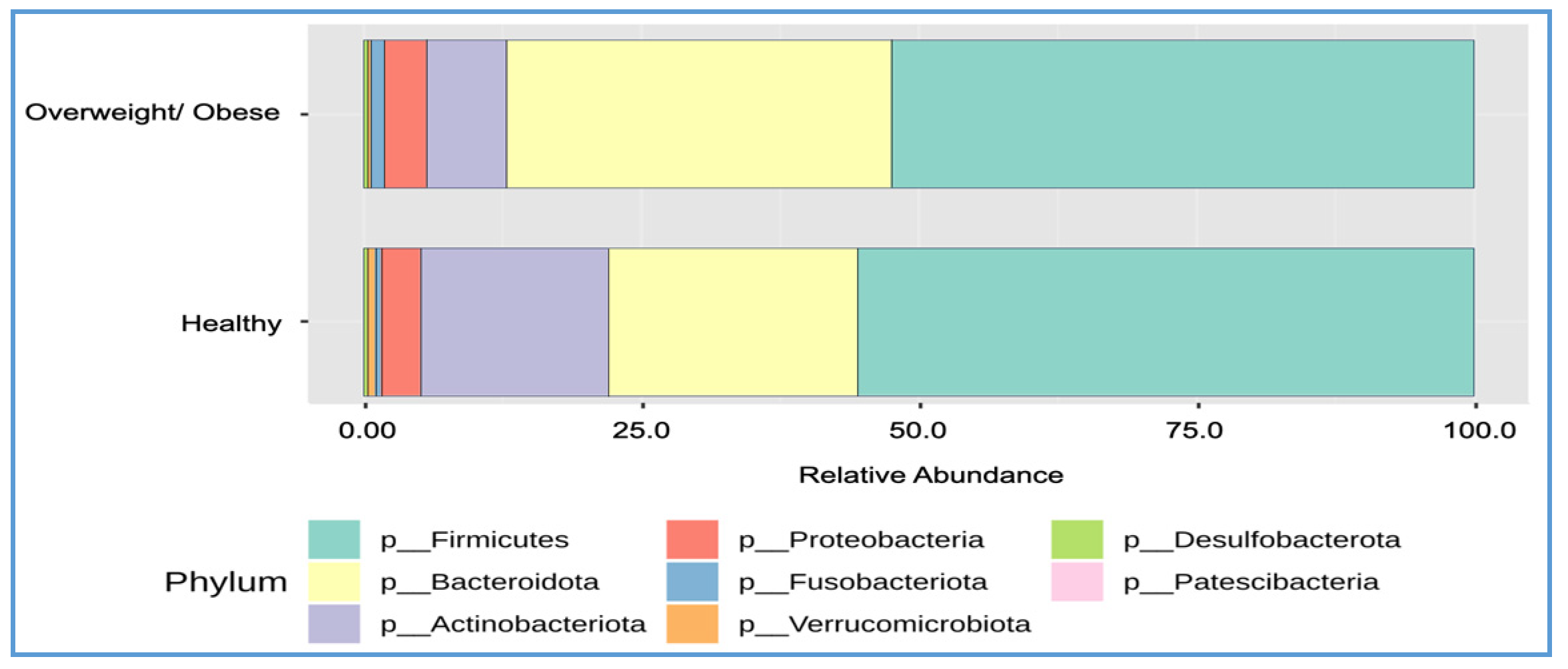
3.5. Differences in Bacterial Composition between the Overweight or Obese and Healthy Groups
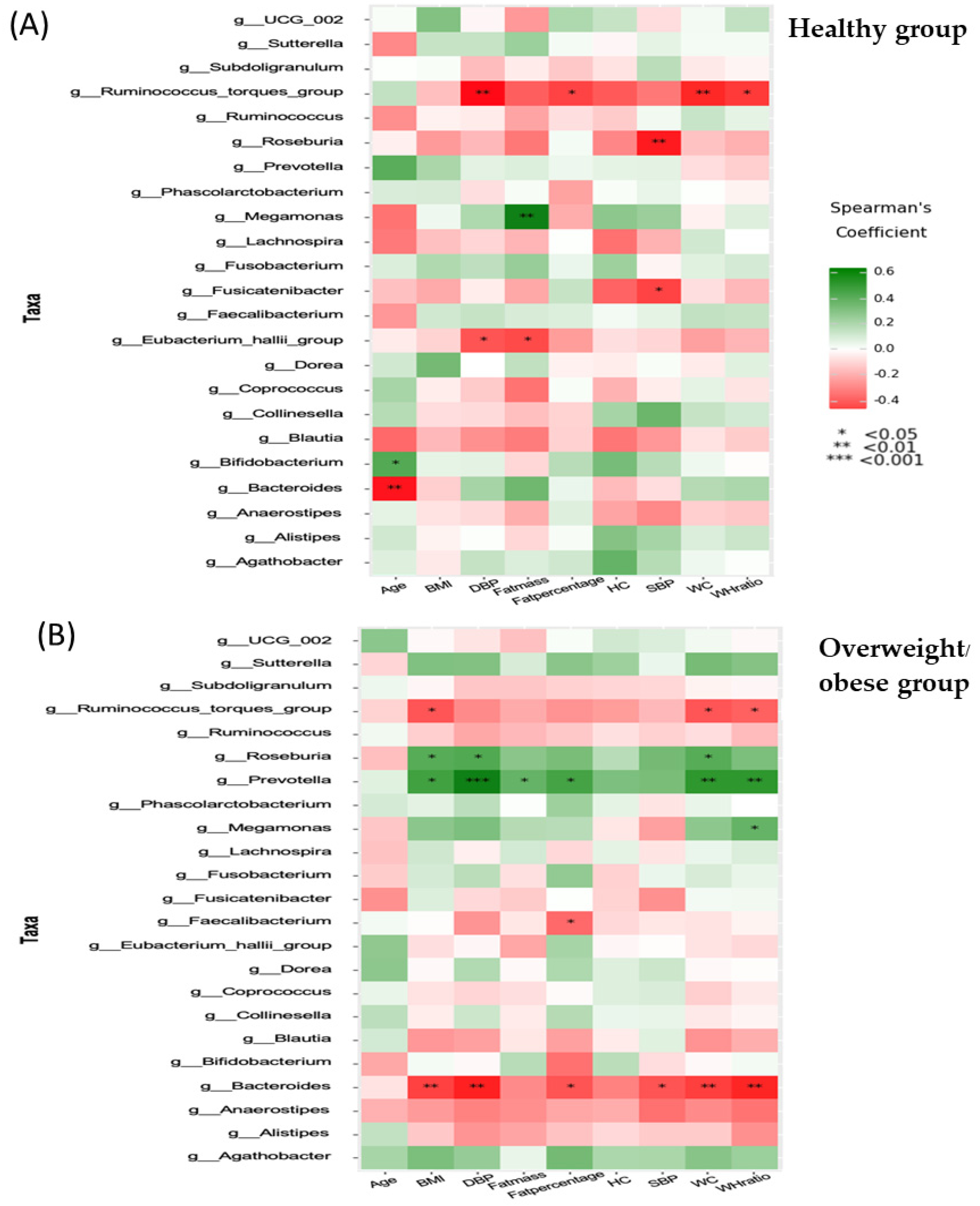
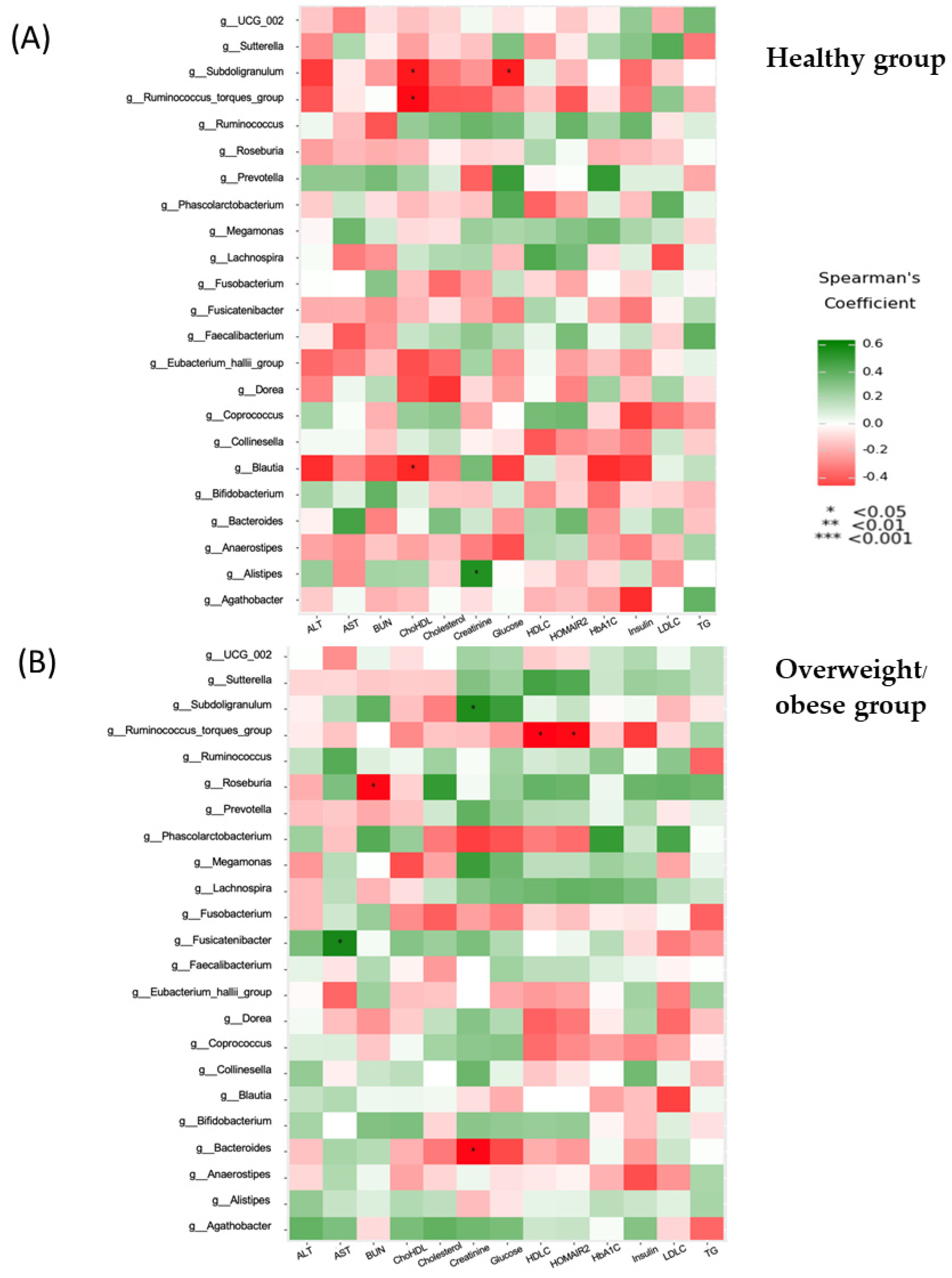
3.6. Associations between Anthropometric, Biochemical, Inflammatory, and Dietary Parameters and the Gut Microbiota (at the Genus Level)
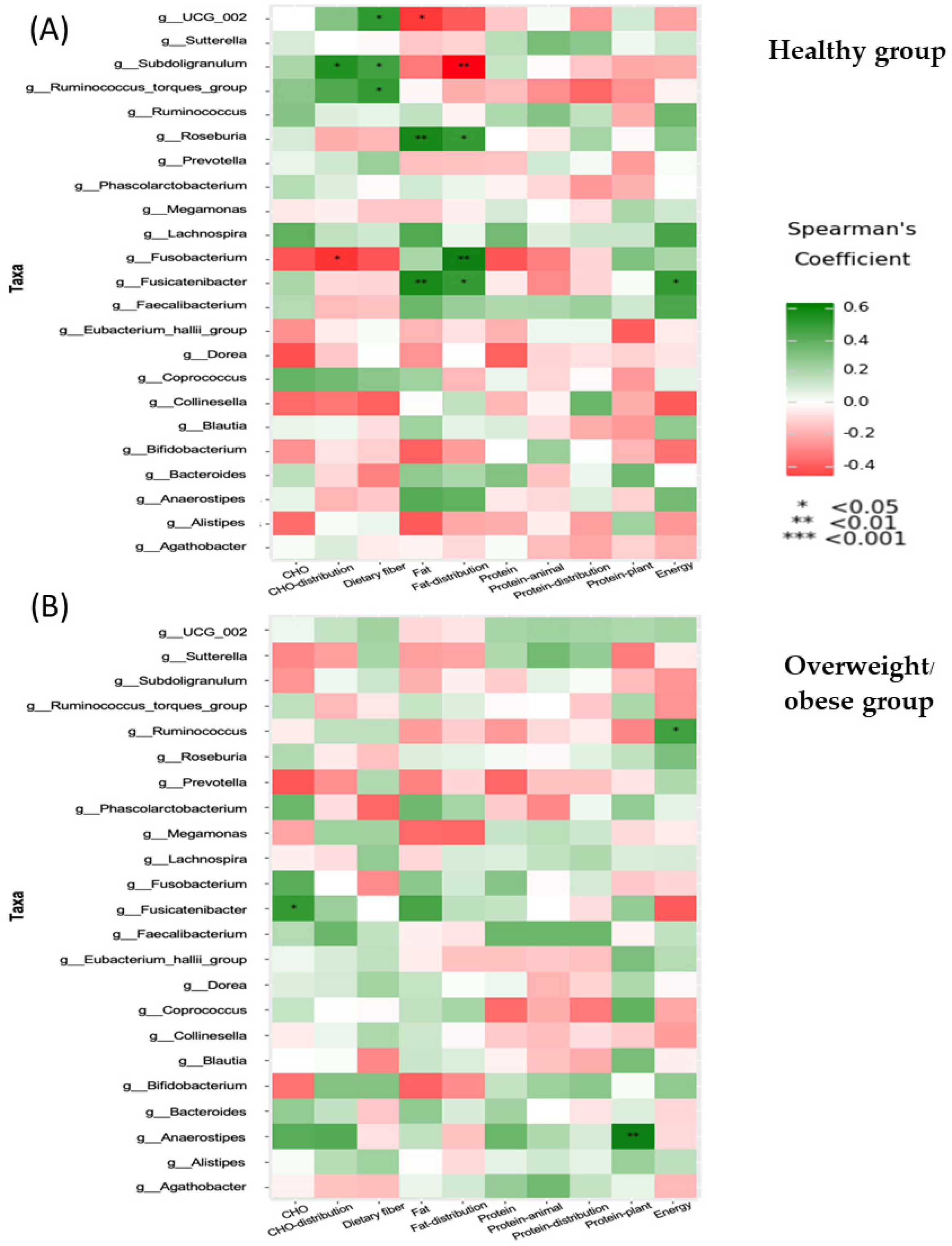
3.7. Functional Differences of Gut Microbiota in the Healthy and Overweight or Obese Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, M.; Liu, S.; Zhang, C. The related metabolic diseases and treatments of obesity. Healthcare 2022, 10, 1616. [Google Scholar] [CrossRef]
- Lin, X.; Li, H. Obesity: Epidemiology, pathophysiology, and therapeutics. Front. Endocrinol. 2021, 12, 706978. [Google Scholar] [CrossRef] [PubMed]
- Fujisaka, S.; Watanabe, Y.; Tobe, K. The gut microbiome: A core regulator of metabolism. J. Endocrinol. 2023, 256, e220111. [Google Scholar] [CrossRef]
- Candelli, M.; Franza, L.; Pignataro, G.; Ojetti, V.; Covino, M.; Piccioni, A.; Gasbarrini, A.; Franceschi, F. Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int. J. Mol. Sci. 2021, 22, 6242. [Google Scholar] [CrossRef]
- Mohammad, S.; Thiemermann, C. Role of metabolic endotoxemia in systemic inflammation and potential interventions. Front. Immunol. 2021, 11, 594150. [Google Scholar] [CrossRef]
- Yang, J.; Wu, J.; Li, Y.; Zhang, Y.; Cho, W.C.; Ju, X.; van Schothorst, E.M.; Zheng, Y. Gut bacteria formation and influencing factors. FEMS Microbiol. Ecol. 2021, 97, fiab043. [Google Scholar] [CrossRef] [PubMed]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.G.; Trevaskis, N.L.; Murphy, A.J.; Febbraio, M.A. Diet-induced gut dysbiosis and inflammation: Key drivers of obesity-driven NASH. iScience 2022, 26, 105905. [Google Scholar] [CrossRef]
- Ahmad, M.A.; Karavetian, M.; Venema, K. Association of the gut microbiota with clinical variables in obese and lean Emirati subjects. Front. Microbiol. 2023, 14, 1182460. [Google Scholar] [CrossRef]
- Sugawara, Y.; Kanazawa, A.; Aida, M.; Yoshida, Y.; Yamashiro, Y.; Watada, H. Association of gut microbiota and inflammatory markers in obese patients with type 2 diabetes mellitus: Post hoc analysis of a synbiotic interventional study. Biosci. Microbiota Food Health 2022, 41, 103–111. [Google Scholar] [CrossRef]
- Karimi, E.; Yarizadeh, H.; Setayesh, L.; Sajjadi, S.F.; Ghodoosi, N.; Khorraminezhad, L.; Mirzaei, K. High carbohydrate intakes may predict more inflammatory status than high fat intakes in pre-menopause women with overweight or obesity: A cross-sectional study. BMC Res. Notes 2021, 14, 279. [Google Scholar] [CrossRef]
- Widjaja, N.A.; Arifani, R.; Irawan, R. Cut-off value of waist-to-hip ratio as a predictor of metabolic syndrome in adolescents with obesity. Acta Biomed. 2023, 94, e2023076. [Google Scholar]
- Zhang, X.; Wang, Y.; Li, Y.; Gui, J.; Mei, Y.; Yang, X.; Liu, H.; Guo, L.; Li, J.; Lei, Y. Four-years change of BMI and waist circumference are associated with metabolic syndrome in middle-aged and elderly Chinese. Sci. Rep. 2024, 14, 10220. [Google Scholar] [CrossRef] [PubMed]
- Bays, H.E.; Kirkpatrick, C.; Maki, K.C.; Toth, P.P.; Morgan, R.T.; Tondt, J.; Christensen, S.M.; Dixon, D.; Jacobson, T.A. Obesity, dyslipidemia, and cardiovascular disease: A joint expert review from the Obesity Medicine Association and the National Lipid Association 2024. J. Clin. Lipidol. 2024, 18, e320–e350. [Google Scholar] [CrossRef]
- Schmidt, F.M.; Weschenfelder, J.; Sander, C.; Minkwitz, J.; Thormann, J.; Chittka, T.; Mergl, R.; Kirkby, K.C.; Faßhauer, M.; Stumvoll, M. Inflammatory cytokines in general and central obesity and modulating effects of physical activity. PLoS ONE 2015, 10, e0121971. [Google Scholar] [CrossRef]
- Megawati, G.; Indraswari, N.; Johansyah, A.A.; Kezia, C.; Herawati, D.M.D.; Gurnida, D.A.; Musfiroh, I. Comparison of hs-CRP in Adult Obesity and Central Obesity in Indonesia Based on Omega-3 Fatty Acids Intake: Indonesian Family Life Survey 5 (IFLS 5) Study. Int. J. Environ. Res. Public Health 2023, 20, 6734. [Google Scholar] [CrossRef]
- Ellulu, M.S.; Patimah, I.; Khaza’ai, H.; Rahmat, A.; Abed, Y. Obesity and inflammation: The linking mechanism and the complications. Arch. Med. Sci. 2017, 13, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.-J.; Wang, H.-J.; Zhang, B.; Qi, S.-F.; Mi, Y.-J.; Pan, X.-B.; Wang, C.; Tian, Q.-B. Associations of fat and carbohydrate intake with becoming overweight and obese: An 11-year longitudinal cohort study. Br. J. Nutr. 2020, 124, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Shatwan, I.M.; Almoraie, N.M. Correlation between dietary intake and obesity risk factors among healthy adults. Clin. Nutr. Open Sci. 2022, 45, 32–41. [Google Scholar] [CrossRef]
- Wehling, H.; Lusher, J. People with a body mass index ≥30 under-report their dietary intake: A systematic review. J. Health Psychol. 2019, 24, 2042–2059. [Google Scholar] [CrossRef]
- Mazidi, M.; Gao, H.-K.; Vatanparast, H.; Kengne, A.P. Impact of the dietary fatty acid intake on C-reactive protein levels in US adults. Medicine 2017, 96, e5736. [Google Scholar] [CrossRef] [PubMed]
- Chmurzynska, A.; Muzsik, A.; Krzyżanowska-Jankowska, P.; Walkowiak, J.; Bajerska, J. The effect of habitual fat intake, IL6 polymorphism, and different diet strategies on inflammation in postmenopausal women with central obesity. Nutrients 2019, 11, 1557. [Google Scholar] [CrossRef]
- Santos, E.W.; Oliveira, D.C.; Hastreiter, A.; Silva, G.B.; Beltran, J.S.d.O.; Rogero, M.M.; Fock, R.A.; Borelli, P. Short-term high-fat diet affects macrophages inflammatory response, early signs of a long-term problem. Braz. J. Pharm. Sci. 2019, 55, e17561. [Google Scholar] [CrossRef]
- Liu, B.N.; Liu, X.T.; Liang, Z.H.; Wang, J.H. Gut microbiota in obesity. World J. Gastroenterol. 2021, 27, 3837–3850. [Google Scholar] [CrossRef] [PubMed]
- Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [CrossRef]
- Pinart, M.; Dötsch, A.; Schlicht, K.; Laudes, M.; Bouwman, J.; Forslund, S.K.; Pischon, T.; Nimptsch, K. Gut microbiome composition in obese and non-obese persons: A systematic review and meta-analysis. Nutrients 2021, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Org, E.; Blum, Y.; Kasela, S.; Mehrabian, M.; Kuusisto, J.; Kangas, A.J.; Soininen, P.; Wang, Z.; Ala-Korpela, M.; Hazen, S.L. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017, 18, 70. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Armougom, F.; Henry, M.; Vialettes, B.; Raccah, D.; Raoult, D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS ONE 2009, 4, e7125. [Google Scholar] [CrossRef]
- Million, M.; Angelakis, E.; Paul, M.; Armougom, F.; Leibovici, L.; Raoult, D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb. Pathog. 2012, 53, 100–108. [Google Scholar] [CrossRef]
- Gong, H.; Gao, H.; Ren, Q.; He, J. The abundance of bifidobacterium in relation to visceral obesity and serum uric acid. Sci. Rep. 2022, 12, 13073. [Google Scholar] [CrossRef]
- Companys, J.; Gosalbes, M.J.; Pla-Pagà, L.; Calderón-Pérez, L.; Llauradó, E.; Pedret, A.; Valls, R.M.; Jiménez-Hernández, N.; Sandoval-Ramirez, B.A.; Del Bas, J.M. Gut microbiota profile and its association with clinical variables and dietary intake in overweight/obese and lean subjects: A cross-sectional study. Nutrients 2021, 13, 2032. [Google Scholar] [CrossRef]
- Duan, M.; Wang, Y.; Zhang, Q.; Zou, R.; Guo, M.; Zheng, H. Characteristics of gut microbiota in people with obesity. PLoS ONE 2021, 16, e0255446. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-J.; Park, S.-G.; Jang, H.B.; Choi, M.-G.; Park, K.-H.; Kang, J.H.; Park, S.I.; Lee, H.-J.; Cho, S.-H. Obesity alters the microbial community profile in Korean adolescents. PLoS ONE 2015, 10, e0134333. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Qin, Q.; Chen, J.; Yan, S.; Li, T.; Gao, X.; Yang, Y.; Li, A.; Ding, S. Gut microbiome alterations in patients with visceral obesity based on quantitative computed tomography. Front. Cell. Infect. Microbiol. 2022, 11, 823262. [Google Scholar] [CrossRef]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef]
- Moran-Ramos, S.; López-Contreras, B.E.; Canizales-Quinteros, S. Gut microbiota in obesity and metabolic abnormalities: A matter of composition or functionality? Arch. Med. Res. 2017, 48, 735–753. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Riezu-Boj, J.I.; Guruceaga, E.; Milagro, F.I.; Martínez, J.A. Sex-specific associations between gut Prevotellaceae and host genetics on adiposity. Microorganisms 2020, 8, 938. [Google Scholar] [CrossRef] [PubMed]
- Serena, C.; Ceperuelo-Mallafré, V.; Keiran, N.; Queipo-Ortuño, M.I.; Bernal, R.; Gomez-Huelgas, R.; Urpi-Sarda, M.; Sabater, M.; Pérez-Brocal, V.; Andrés-Lacueva, C. Elevated circulating levels of succinate in human obesity are linked to specific gut microbiota. ISME J. 2018, 12, 1642–1657. [Google Scholar] [CrossRef]
- Chambers, E.S.; Viardot, A.; Psichas, A.; Morrison, D.J.; Murphy, K.G.; Zac-Varghese, S.E.; MacDougall, K.; Preston, T.; Tedford, C.; Finlayson, G.S. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 2015, 64, 1744–1754. [Google Scholar] [CrossRef]
- van den Munckhof, I.C.; Kurilshikov, A.; ter Horst, R.; Riksen, N.P.; Joosten, L.; Zhernakova, A.; Fu, J.; Keating, S.T.; Netea, M.G.; De Graaf, J. Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: A systematic review of human studies. Obes. Rev. 2018, 19, 1719–1734. [Google Scholar] [CrossRef] [PubMed]
- Gondalia, S.V.; Wymond, B.; Benassi-Evans, B.; Berbezy, P.; Bird, A.R.; Belobrajdic, D.P. Substitution of refined conventional wheat flour with wheat high in resistant starch modulates the intestinal microbiota and fecal metabolites in healthy adults: A randomized, controlled trial. J. Nutr. 2022, 152, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W.; Mao, Y.; Zhang, X.; Pang, X.; Wei, C. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Postigo, M.; Oliva-Olivera, W.; Coin-Aragüez, L.; Ramos-Molina, B.; Giraldez-Perez, R.M.; Lhamyani, S.; Alcaide-Torres, J.; Perez-Martinez, P.; El Bekay, R.; Cardona, F. Metabolic endotoxemia promotes adipose dysfunction and inflammation in human obesity. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E319–E332. [Google Scholar] [CrossRef] [PubMed]
- Omran, F.; Murphy, A.M.; Younis, A.Z.; Kyrou, I.; Vrbikova, J.; Hainer, V.; Sramkova, P.; Fried, M.; Ball, G.; Tripathi, G. The impact of metabolic endotoxaemia on the browning process in human adipocytes. BMC Med. 2023, 21, 154. [Google Scholar] [CrossRef]
- Yang, J.C.; Jacobs, J.P.; Hwang, M.; Sabui, S.; Liang, F.; Said, H.M.; Skupsky, J. Biotin deficiency induces intestinal dysbiosis associated with an inflammatory bowel disease-like phenotype. Nutrients 2023, 15, 264. [Google Scholar] [CrossRef]
- Liu, L.; Song, L.; Deng, R.; Lan, R.; Jin, W.; Tran Van Nhieu, G.; Cao, H.; Liu, Q.; Xiao, Y.; Li, X. Citrobacter freundii activation of NLRP3 inflammasome via the type VI secretion system. J. Infect. Dis. 2021, 223, 2174–2185. [Google Scholar] [CrossRef]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef]



| Healthy Group (n = 20) | Overweight/Obese Group (n = 75) | p-Value | |
|---|---|---|---|
| Age, year | 38.00 (35.00–50.00) | 41.00 (35.00–50.00) | 0.024 |
| BMI kg/m2 | 21.10 (19.10–22.80) | 27.30 (23.10–42.00) | 0.000 |
| Waist circumference, cm | 75.01 (63.50–89.50) | 92.00 (74.20–116.40) | 0.000 |
| Hip circumference, cm | 94.50 (87.00–101.10) | 106.03 (93.20–132.70) | 0.000 |
| Waist–hip ratio | 0.80 (0.72–0.89) | 0.86 (0.76–0.98) | 0.000 |
| Fat mass, kg | 15.05 (10.10–18.10) | 26.45 (18.80–58.10) | 0.000 |
| Body fat, % | 28.20 (22.30–31.40) | 38.14 (31.30–56.80) | 0.001 |
| SBP, mmHg | 115.00 (104.00–128.00) | 133.00 (102.00–165.00) | 0.002 |
| DBP, mmHg | 71.00 (59.00–82.00) | 82.00 (69.00–120.00) | 0.001 |
| Total cholesterol, mg/dL | 188.00 (142.00–204.00) | 231.00 (151.00–307.00) | 0.024 |
| Triglyceride, mg/dL | 75.04 (50.32–142.32) | 121.06 (66.36–232.19) | 0.005 |
| LDL-C, mg/dL | 128.32 (67.01–123.54) | 153.29 (96.87–232.56) | 0.002 |
| HDL-C, mg/dL | 65.02 (45.67–84.14) | 52.47 (32.51–97.26) | 0.000 |
| FPG, mg/dL | 82.09 (69.18–95.28) | 103.47 (72.00–168.44) | 0.003 |
| HbA1C, % | 5.10 (4.60–5.70) | 8.50 (4.70–8.69) | 0.000 |
| Insulin, µU/mL | 4.40 (3.20–9.10) | 8.45 (3.70–36.10) | 0.000 |
| HOMA-IR | 0.54 (0.39–0.94) | 1.11 (0.56–3.51) | 0.001 |
| BUN, | 9.90 (6.60–16.60) | 10.65 (7.00–15.70) | 0.891 |
| Creatinine, | 0.69 (0.51–0.89) | 0.69 (0.44–0.98) | 0.534 |
| AST | 23.00 (16.00–28.00) | 22.00 (14.00–58.00) | 0.402 |
| ALT | 15.00 (7.00–26.00) | 21.00 (7.00–65.00) | 0.000 |
| hs-CRP, mg/L | 0.57 (0.40–3.48) | 3.37 (0.48–11.24) | 0.000 |
| IL-6, pg/mL | 2.74 (2.51–3.66) | 4.69 (4.03–6.04) | 0.000 |
| TNF-alpha, pg/mL | 11.34 (10.20–14.15) | 15.55 (13.94–19.31) | 0.000 |
| Healthy Group (n = 20) | Overweight /Obese Group (n = 75) | p-Value | |
|---|---|---|---|
| Total calories, kcal | 1531.00 (1179.00–1979.00) | 1417.00 (623.00–2298.00) | 0.563 |
| Carbohydrate, g | 175.00 (153.00–250.00) | 205.34 (127.00–375.00) | 0.005 |
| Energy from carbohydrates (%) | 35.04 (23.04–69.06) | 52.33 (26.32–69.39) | 0.000 |
| Protein, g/day | 54.34 (31.64–100.63) | 66.37 (24.60–144.33) | 0.058 |
| Energy from protein (%) | 14.72 (8.71–33.32) | 18.72 (9.05–28.33) | 0.028 |
| Protein—Animal, g | 35.21 (23.79–85.78) | 53.67 (22.00–117.90) | 0.008 |
| Protein—Vegetable, g | 16.40 (10.85–23.73) | 12.16 (10.04–96.55) | 0.103 |
| Fat, g | 78.77 (34.69–102.24) | 49.28 (24.60–144.33) | 0.001 |
| Energy from fat (%) | 43.53 (21.69–50.56) | 29.08 (17.65–50.46) | 0.000 |
| Cholesterol, mg | 228.00 (48.00–773.00) | 339.00 (56.00–1630.00) | 0.000 |
| Fiber, g | 10.38 (5.78–39.29) | 11.16 (5.18–35.40) | 0.579 |
| Calcium, mg | 225.00 (138.00–665.00) | 324.00 (142.00–956.00) | 0.005 |
| Phosphorus, mg | 628.00 (465.00–1070.00) | 584.00 (481.00–1123.00) | 0.963 |
| Iron, mg | 9.15 (5.02–38.00) | 11.08 (5.02–55.13) | 0.279 |
| Iron—Animal, mg | 2.61 (2.01–11.17) | 4.45 (2.05–27.07) | 0.044 |
| Iron—Vegetable, mg | 5.00 (2.13–8.61) | 5.08 (1.70–53.28) | 0.263 |
| Potassium, mg | 1611.00 (525.00–3177.00) | 1773.00 (819.00–3539.00) | 0.588 |
| Sodium, mg | 1946.00 (1379.00–3979.00) | 2445.00 (1202.00–4119.00) | 0.021 |
| Copper, mg | 0.70 (0.42–1.70) | 0.74 (0.42–4.68) | 0.266 |
| Magnesium, mg | 95.14 (12.98–259.83) | 43.71 (10.80–165.50) | 0.013 |
| Selenium, mcg | 37.57 (10.32–61.56) | 31.37 (5.01–120.72) | 0.544 |
| Zinc, mg | 2.69 (2.02–8.10) | 4.51 (2.13–8.28) | 0.037 |
| Vitamin A, RAE | 301.00 (262.00–1923.00) | 321.00 (210.00–3437.00) | 0.137 |
| Retinol, µg | 140.00 (88.00–1887.00) | 240.00 (29.00–2967.00) | 0.088 |
| Beta-Carotene, µg | 1826.00 (252.00–4372.00) | 530.00 (141.00–3787.00) | 0.038 |
| Thiamin-B1, mg | 1.38 (1.02–1.99) | 1.19 (1.09–14.72) | 0.335 |
| Riboflavin-B2, mg | 0.93 (0.71–3.42) | 1.17 (0.58–8.36) | 0.051 |
| Vitamin-B6, mg | 0.78 (0.16–1.13) | 0.53 (0.14–1.19) | 0.087 |
| Niacin, mg | 13.31 (7.37–18.42) | 11.26 (5.77–28.32) | 0.66 |
| Vitamin-B12, µg | 0.59 (0.40–3.21) | 0.83 (0.14–19.94) | 0.877 |
| Vitamin C, mg | 65.12 (14.46–450.65) | 62.09 (9.59–488.22) | 0.686 |
| Vitamin E, mg | 0.87 (0.39–32.14) | 1.19 (0.38–6.84) | 0.074 |
| Variable | hs-CRP | IL-6 | TNF-α | |||
|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | |
| Age | 0.142 | 0.183 | 0.128 | 0.268 | 0.192 | 0.085 |
| SBP (mmHg) | 0.350 | 0.001 | 0.335 | 0.003 | 0.250 | 0.025 |
| DBP (mmHg) | 0.249 | 0.018 | 0.358 | 0.001 | 0.264 | 0.018 |
| BMI (kg/m2) | 0.632 | 0.000 | 0.467 | 0.000 | 0.402 | 0.000 |
| WC (cm) | 0.548 | 0.000 | 0.377 | 0.001 | 0.376 | 0.001 |
| HC (cm) | 0.575 | 0.000 | 0.434 | 0.000 | 0.422 | 0.000 |
| Waist–hip ratio | 0.306 | 0.003 | 0.142 | 0.216 | 0.166 | 0.140 |
| Fat (%) | 0.638 | 0.000 | 0.526 | 0.000 | 0.476 | 0.000 |
| Fat mass(kg) | 0.613 | 0.000 | 0.459 | 0.000 | 0.395 | 0.000 |
| Total cholesterol (mg/dL) | 0.252 | 0.017 | 0.222 | 0.052 | 0.288 | 0.009 |
| Triglyceride (mg/dL) | 0.306 | 0.004 | 0.205 | 0.077 | 0.253 | 0.023 |
| HDL-Cholesterol (mg/dL) | −0.410 | 0.000 | −0.203 | 0.076 | −0.124 | 0.271 |
| LDL-Cholesterol (mg/dL) | 0.418 | 0.000 | 0.289 | 0.011 | 0.249 | 0.025 |
| FPG (mg/dL) | 0.275 | 0.010 | 0.064 | 0.582 | 0.091 | 0.426 |
| HbA1C (%) | 0.132 | 0.218 | 0.090 | 0.438 | 0.174 | 0.121 |
| Insulin (uU/mL) | 0.451 | 0.000 | 0.360 | 0.001 | 0.240 | 0.033 |
| HOMA-IR | 0.348 | 0.001 | 0.304 | 0.008 | 0.244 | 0.029 |
| AST (U/L) | 0.338 | 0.001 | 0.055 | 0.635 | 0.107 | 0.210 |
| ALT (U/L) | 0.462 | 0.000 | 0.156 | 0.179 | 0.195 | 0.083 |
| Creatinine (mg/dL) | −0.095 | 0.378 | −0.021 | 0.859 | 0.059 | 0.598 |
| BUN (mg/dL) | 0.049 | 0.647 | −0.010 | 0.931 | −0.052 | 0.642 |
| Total calories, kcal | 0.388 | 0.000 | 0.058 | 0.622 | 0.098 | 0.386 |
| Carbohydrate, g/day | 0.124 | 0.667 | 0.109 | 0.486 | 0.108 | 0.342 |
| Energy from carbohydrates (%) | 0.097 | 0.215 | 0.118 | 0.078 | 0.098 | 0.094 |
| Protein, g/day | −0.026 | 0.811 | −0.075 | 0.521 | 0.109 | 0.336 |
| Energy from protein (%) | 0.121 | 0.081 | 0.109 | 0.124 | 0.209 | 0.106 |
| Fat, g/day | 0.248 | 0.020 | 0.210 | 0.069 | 0.173 | 0.125 |
| Energy from fat (%) | 0.368 | 0.014 | 0.249 | 0.006 | 0.261 | 0.029 |
| Cholesterol, mg | 0.317 | 0.007 | 0.127 | 0.097 | 0.171 | 0.109 |
| Phylum | Healthy (%) | Overweight/Obese (%) | p-Value | q-Value |
|---|---|---|---|---|
| Firmicutes | 55.60 | 52.50 | 0.2938 | 0.4433 |
| Bacteroidota | 22.40 | 34.60 | 9.17 × 10−5 | 0.0004 |
| Actinobacteriota | 17.00 | 7.30 | 1.64 × 10−5 | 0.0001 |
| Proteobacteria | 3.50 | 3.80 | 0.2855 | 0.4433 |
| Verrucomicrobiota | 0.70 | 0.20 | 0.0528 | 0.1585 |
| Fusobacteriota | 0.60 | 1.30 | 0.3210 | 0.4433 |
| Desulfobacterota | 0.30 | 0.40 | 0.8802 | 0.8802 |
| F–B ratio | 2.80 | 1.96 | 0.0351 | 0.0012 |
| Genus | Phylum | Log2FC | p-Value |
|---|---|---|---|
| More prevalent in healthy subjects | |||
| Acidaminococcus | Firmicutes | −4.040 | 0.017 |
| Coprobacillus | Firmicutes | −3.400 | 0.039 |
| Lactobacillus | Firmicutes | −2.650 | 0.000 |
| CAG_352 | Firmicutes | −1.880 | 0.000 |
| Bifidobacterium | Actinomycetota | −1.500 | 0.003 |
| Akkermansia | Verrucomicrobiota | −1.450 | 0.000 |
| Collinsella | Actinomycetota | −0.605 | 0.037 |
| Megasphaera | Firmicutes | −0.442 | 0.000 |
| Senegalimassilia | Actinomycetota | −0.402 | 0.000 |
| Eubacterium_ruminantium_group | Firmicutes | −0.398 | 0.000 |
| Faecalibacterium | Firmicutes | −0.056 | 0.000 |
| More prevalent in overweight/obese subjects | |||
| Megamonas | Firmicutes | 0.213 | 0.000 |
| Sutterella | Pseudomonadota | 0.305 | 0.000 |
| Dialister | Firmicutes | 0.356 | 0.000 |
| Holdemanella | Firmicutes | 0.454 | 0.000 |
| Lachnospiraceae_UCG_008 | Firmicutes | 0.459 | 0.000 |
| Butyricicoccus | Firmicutes | 0.517 | 0.015 |
| Bacteroides | Bacteroidota | 0.636 | 0.007 |
| Prevotella | Bacteroidota | 0.898 | 0.000 |
| Lachnospiraceae_UCG_004 | Firmicutes | 1.040 | 0.001 |
| Lachnoclostridium | Firmicutes | 1.330 | 0.000 |
| Sellimonas | Firmicutes | 1.490 | 0.000 |
| Lactococcus | Firmicutes | 1.800 | 0.025 |
| Ruminococcus_gnavus_group | Firmicutes | 2.100 | 0.030 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chansa, O.; Shantavasinkul, P.C.; Monsuwan, W.; Sirivarasai, J. Association between Gut Microbiota Profiles, Dietary Intake, and Inflammatory Markers in Overweight and Obese Women. Foods 2024, 13, 2592. https://doi.org/10.3390/foods13162592
Chansa O, Shantavasinkul PC, Monsuwan W, Sirivarasai J. Association between Gut Microbiota Profiles, Dietary Intake, and Inflammatory Markers in Overweight and Obese Women. Foods. 2024; 13(16):2592. https://doi.org/10.3390/foods13162592
Chicago/Turabian StyleChansa, Orada, Prapimporn Chattranukulchai Shantavasinkul, Wutarak Monsuwan, and Jintana Sirivarasai. 2024. "Association between Gut Microbiota Profiles, Dietary Intake, and Inflammatory Markers in Overweight and Obese Women" Foods 13, no. 16: 2592. https://doi.org/10.3390/foods13162592
APA StyleChansa, O., Shantavasinkul, P. C., Monsuwan, W., & Sirivarasai, J. (2024). Association between Gut Microbiota Profiles, Dietary Intake, and Inflammatory Markers in Overweight and Obese Women. Foods, 13(16), 2592. https://doi.org/10.3390/foods13162592







