EjWRKY6 Is Involved in the ABA-Induced Carotenoid Biosynthesis in Loquat Fruit during Ripening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Fruit Firmness and Color Measurement
2.3. Extracting and Quantifying Carotenoid
2.4. Gene Expression Analysis by Real-Time Quantitative PCR (qPCR)
2.5. Transient Overexpression in Tobacco
2.6. Genetic Transformation of Tobacco
2.7. Dual-Luciferase Assays
2.8. Statistics Analysis
3. Results
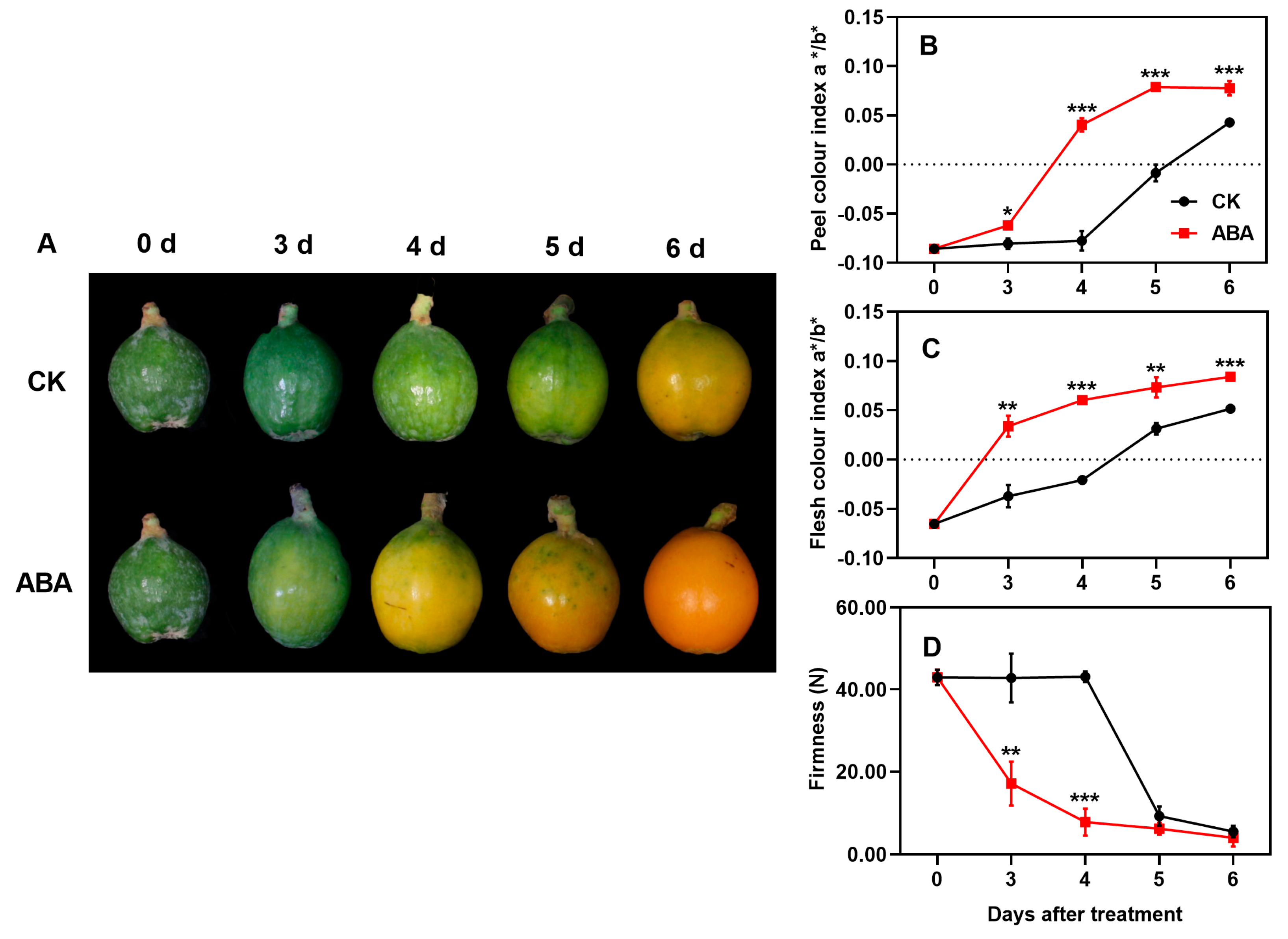
3.1. Effect of ABA Treatment on Color and Hardness of Loquat Fruits
3.2. Effect of ABA Treatment on Carotenoid Content and Composition of Loquat Fruits
3.3. Effects of ABA Treatment on the Expression of Carotenoid Biosynthesis Genes in Loquat Fruits
3.4. Effects of ABA Treatment on EjWRKY6 Expression in Loquat Fruits
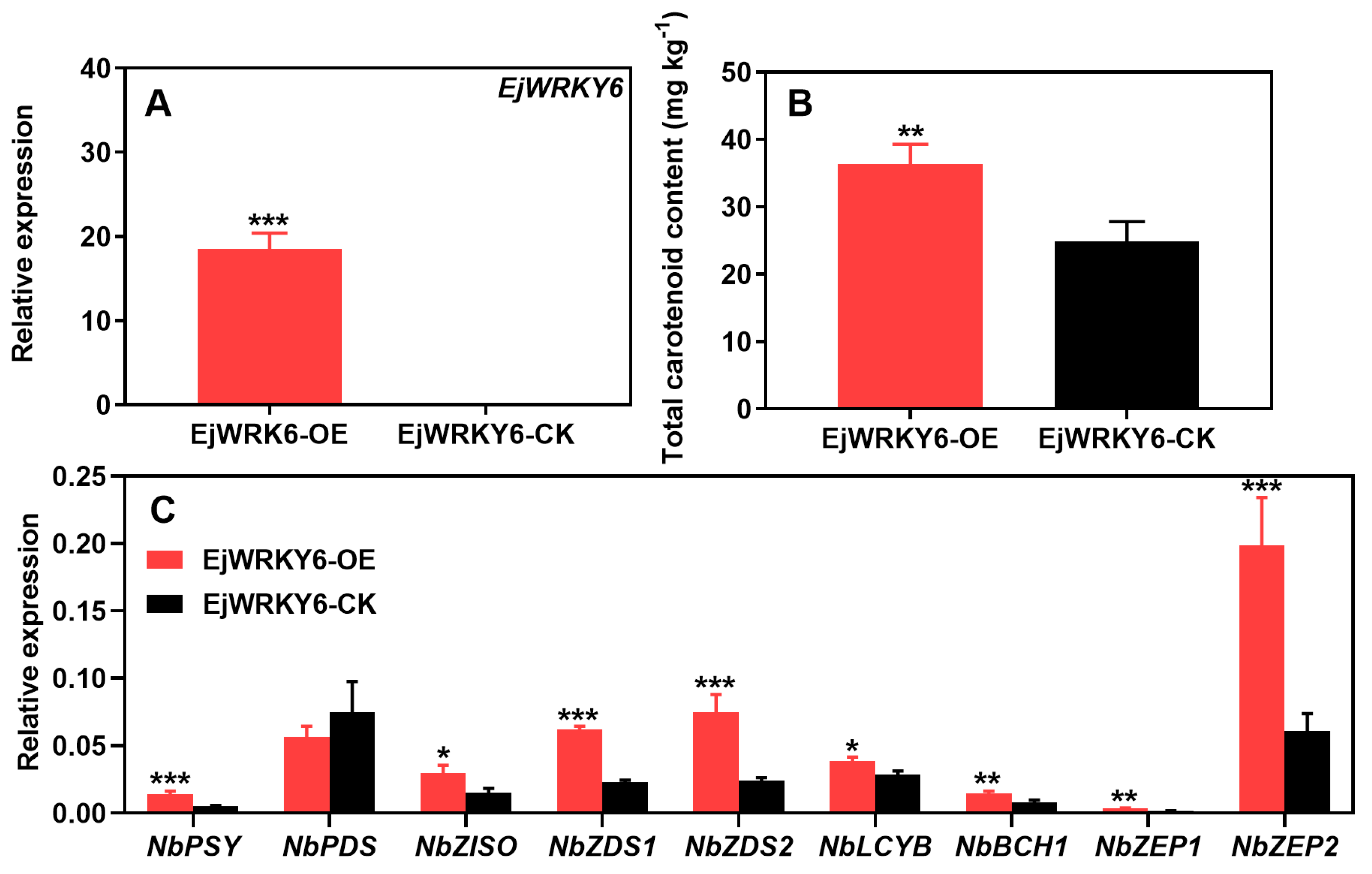
3.5. Transient Overexpression of EjWRKY6 in Nicotiana benthamiana
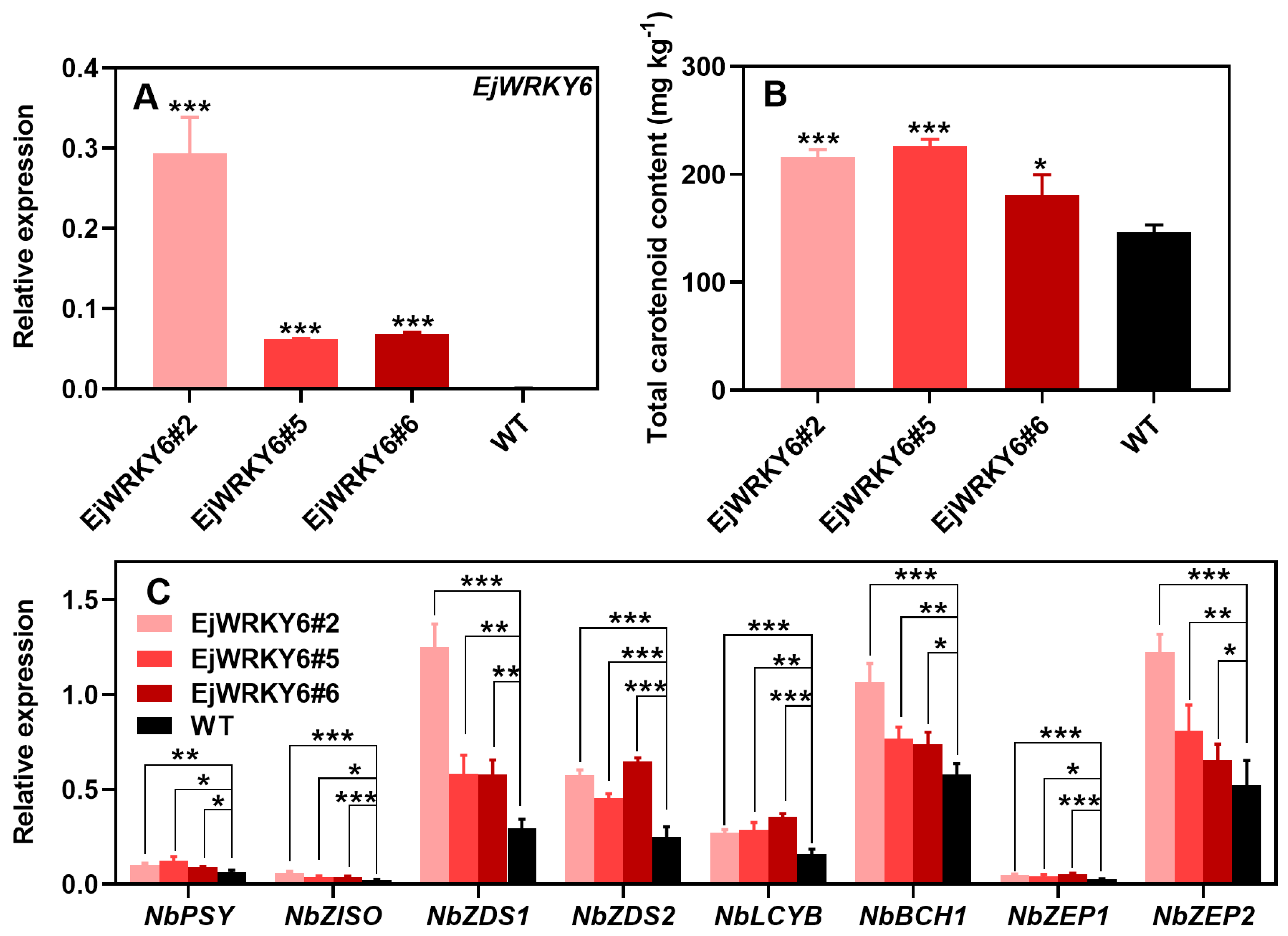
3.6. Stable Overexpression of EjWRKY6 in Nicotiana tabacum
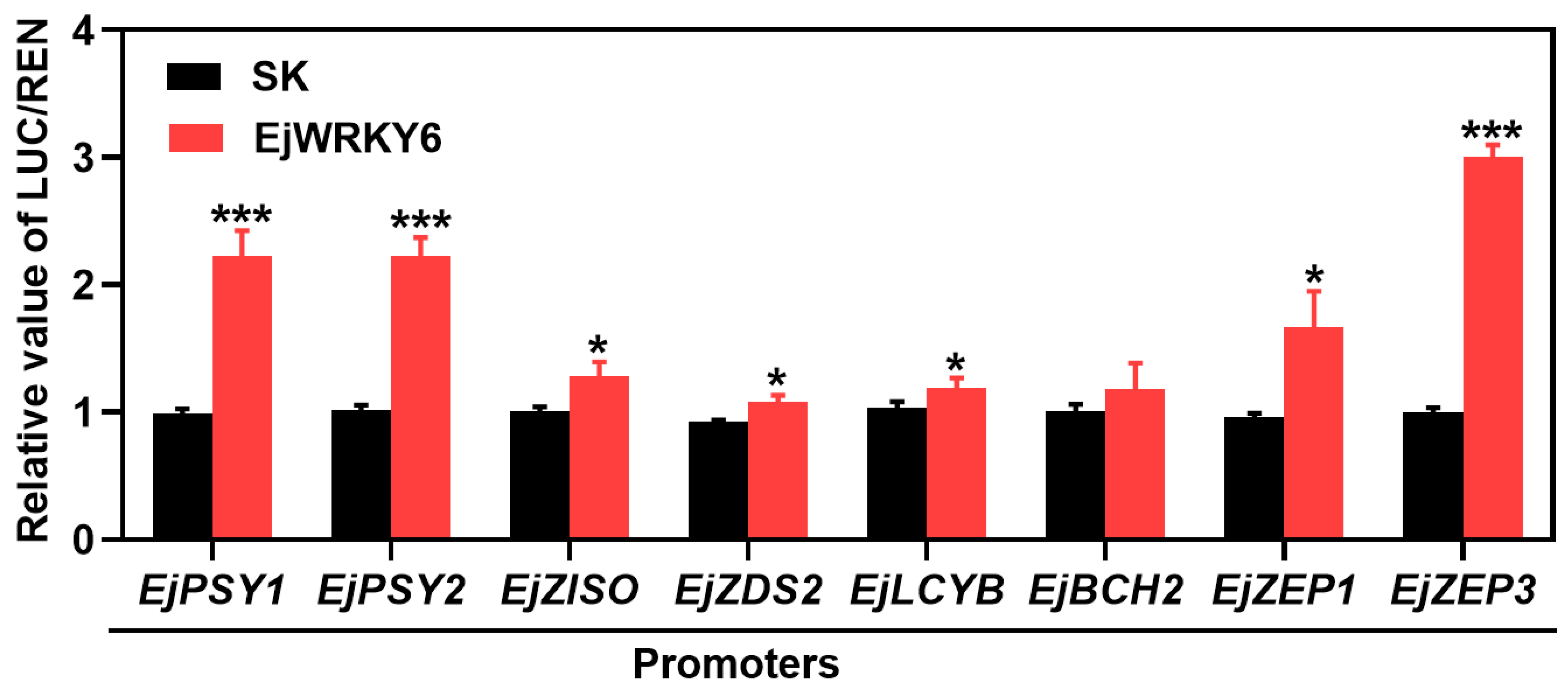
3.7. Transcriptional Regulation on EjWRKY6
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pareek, S.; Benkeblia, N.; Janick, J.; Cao, S.; Yahia, E.M. Postharvest physiology and technology of loquat (Eriobotrya japonica Lindl.) fruit. J. Sci. Food Agr. 2014, 94, 1495–1504. [Google Scholar] [CrossRef]
- Zhou, C.H.; Xu, C.J.; Sun, C.D.; Li, X.; Chen, K.S. Carotenoids in white- and red-fleshed loquat fruits. J. Agric. Food Chem. 2007, 55, 7822–7830. [Google Scholar] [CrossRef]
- Fu, X.; Kong, W.; Peng, G.; Zhou, J.; Azam, M.; Xu, C.; Grierson, D.; Chen, K. Plastid structure and carotenogenic gene expression in red- and white-fleshed loquat (Eriobotrya japonica) fruits. J. Exp. Bot. 2012, 63, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.T.; Zhang, Z.K.; Shahid, M.Q.; Wei, W.L.; Baloch, F.S.; Wu, J.C.; Lin, S.Q.; Yang, X.H. RNA-Seq reveals differential expression patterns of genes associated with carotenoid accumulation in loquat. Acta Physiol. Plant. 2017, 39, 168. [Google Scholar] [CrossRef]
- Sun, S.X.; Li, J.; Chen, D.; Xie, H.J.; Tu, M.Y.; Jia, L.; Jiang, G.L. Comparative transcriptomic analysis reveals a series of single nucleotide polymorphism between red and white-fleshed loquats (Eriobotrya japonica). Czech J. Genet. Plant Breed. 2017, 53, 97–106. [Google Scholar] [CrossRef]
- Hadjipieri, M.; Georgiadou, E.C.; Marin, A.; Diaz-Mula, H.M.; Goulas, V.; Fotopoulos, V.; Tomás-Barberán, F.A.; Manganaris, G.A. Metabolic and transcriptional elucidation of the carotenoid biosynthesis pathway in peel and flesh tissue of loquat fruit during on-tree development. BMC Plant Biol. 2017, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Wang, X.; Wang, Y.; Xiang, Y.; Chen, M.; Zhang, H.; Luo, X.; Xia, H.; Liang, D.; Lv, X.; et al. Dynamic changes in cell wall polysaccharides during fruit development and ripening of two contrasting loquat cultivars and associated molecular mechanisms. Foods 2023, 12, 309. [Google Scholar] [CrossRef]
- Howitt, C.A.; Pogson, B.J. Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ. 2006, 29, 435–445. [Google Scholar] [CrossRef]
- Hadley, C.W.; Miller, E.C.; Schwartz, S.J.; Clinton, S.K. Tomatoes, lycopene, and prostate cancer: Progress and promise. Exp. Biol. Med. 2002, 227, 869–880. [Google Scholar] [CrossRef]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef]
- Wurtzel, E.T.; Cuttriss, A.; Vallabhaneni, R. Maize provitamin a carotenoids, current resources, and future metabolic engineering challenges. Front. Plant Sci. 2012, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shao, Z.; Zhang, M.; Wang, Q. Regulation of carotenoid metabolism in tomato. Mol. Plant 2015, 8, 28–39. [Google Scholar] [CrossRef]
- Eisenreich, W.; Bacher, A.; Arigoni, D.; Rohdich, F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell. Mol. Life Sci. 2004, 61, 1401–1426. [Google Scholar] [CrossRef] [PubMed]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef]
- Lado, J.; Zacarías, L.; Rodrigo, M.J. Regulation of Carotenoid Biosynthesis During Fruit Development. Sub-Cell. Biochem. 2016, 79, 161–198. [Google Scholar] [CrossRef]
- Su, W.; Zhu, C.; Fan, Z.; Huang, M.; Lin, H.; Chen, X.; Deng, C.; Chen, Y.; Kou, Y.; Tong, Z.; et al. Comprehensive metabolome and transcriptome analyses demonstrate divergent anthocyanin and carotenoid accumulation in fruits of wild and cultivated loquats. Front. Plant Sci. 2023, 14, 1285456. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, T.; Xu, C.; Li, M.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X.; Han, Z.; Wu, T. MdMADS6 recruits histone deacetylase MdHDA19 to repress the expression of the carotenoid synthesis-related gene MdCCD1 during fruit ripening. Plants 2022, 11, 668. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef]
- Fu, C.C.; Han, Y.C.; Fan, Z.Q.; Chen, J.Y.; Chen, W.X.; Lu, W.J.; Kuang, J.F. The papaya transcription factor CpNAC1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during Fruit Ripening. J. Agric. Food Chem. 2016, 64, 5454–5463. [Google Scholar] [CrossRef]
- Dang, Z.; Zhu, M.; Chen, H.; Zhang, Y.; Gao, A.; Ma, W.; Chen, Y.; Wei, Y.; Zhang, H. MiMYB10 transcription factor regulates biosynthesis and accumulation of carotenoid involved genes in mango fruit. Int. J. Biol. Macromol. 2023, 253, 127665. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.P.; Kracher, B.; Roccaro, M.; Somssich, I.E. Induced genome-wide binding of three arabidopsis WRKY transcription factors during early mamp-triggered immunity. Plant Cell 2017, 29, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Jiang, C.; Zhao, Y.; Gao, G.; Li, M.; Qi, H. Transcriptome and metabolomics analysis revealed that CmWRKY49 regulating CmPSY1 promotes β-carotene accumulation in orange fleshed oriental melon. Hortic. Plant J. 2022, 8, 650–666. [Google Scholar] [CrossRef]
- Giovannoni, J.J. Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 2007, 10, 283–289. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Yang, F.W.; Feng, X.Q. Abscisic acid biosynthesis and catabolism and their regulation roles in fruit ripening. Phyton-Int. J. Exp. Bot. 2015, 84, 444–453. [Google Scholar] [CrossRef]
- Pilati, S.; Bagagli, G.; Sonego, P.; Moretto, M.; Brazzale, D.; Castorina, G.; Simoni, L.; Tonelli, C.; Guella, G.; Engelen, K.; et al. Abscisic acid is a major regulator of grape berry ripening onset: New insights into ABA signaling network. Front. Plant Sci. 2017, 8, 1093. [Google Scholar] [CrossRef]
- Kou, X.; Yang, S.; Chai, L.; Wu, C.; Zhou, J.; Liu, Y.; Xue, Z. Abscisic acid and fruit ripening: Multifaceted analysis of the effect of abscisic acid on fleshy fruit ripening. Sci. Hortic. 2021, 281, 109999. [Google Scholar] [CrossRef]
- Li, D.; Luo, Z.; Mou, W.; Wang, Y.; Ying, T.; Mao, L. ABA and UV-C effects on quality, antioxidant capacity and anthocyanin contents of strawberry fruit (Fragaria ananassa Duch.). Postharvest Biol. Technol. 2014, 90, 56–62. [Google Scholar] [CrossRef]
- Wu, Q.; Bai, J.; Tao, X.; Mou, W.; Luo, Z.; Mao, L.; Ban, Z.; Ying, T.; Li, L. Synergistic effect of abscisic acid and ethylene on color development in tomato (Solanum lycopersicum L.) fruit. Sci. Hortic. 2018, 235, 169–180. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, S.; Jiao, B.; Duan, M.; Meng, Q.; Ma, N.; Lv, W. SlSGRL, a tomato SGR-like protein, promotes chlorophyll degradation downstream of the ABA signaling pathway. Plant Physiol. Biochem. 2020, 157, 316–327. [Google Scholar] [CrossRef]
- Alos, E.; Martinez-Fuentes, A.; Reig, C.; Mesejo, C.; Zacarías, L.; Agustí, M.; Rodrigo, M.J. Involvement of ethylene in color changes and carotenoid biosynthesis in loquat fruit (Eriobotrya japonica Lindl. cv. Algerie). Postharvest Biol. Technol. 2019, 149, 129–138. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, Y.; Su, D.; Lu, W.; Li, Z. SlGRAS4 accelerates fruit ripening by regulating ethylene biosynthesis genes and SlMADS1 in tomato. Hortic. Res. 2021, 8, 3. [Google Scholar] [CrossRef]
- Tuan, P.A.; Thwe, A.A.; Kim, Y.B.; Kim, J.K.; Kim, S.; Lee, S.; Chung, S.; Park, S.U. Effects of white, blue, and red light-emitting diodes on carotenoid biosynthetic gene expression levels and carotenoid accumulation in sprouts of tartary buckwheat (Fagopyrum tataricum Gaertn.). J. Agric. Food Chem. 2013, 61, 12356–12361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Wang, J.; Zhang, R.X.; Huang, R.F. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, X.; Hu, X.; Wu, T.; Wang, Y.; Xu, X.; Zhang, X.; Han, Z. High miR156 expression is required for auxin-induced adventitious root formation via MxSPL26 independent of PINs and ARFs in Malus xiaojinensis. Front. Plant Sci. 2017, 8, 1059. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Zhang, J.L.; Ji, N.A.; Zuo, X.X.; Ru, X.Y.; Jin, P.; Wang, K.T.; Zheng, Y.H. PpMYB44 positively affects salicylic acid biosynthesis in Pichia guilliermondii-induced peach fruit resistance against Rhizopus stolonifer. Postharvest Biol. Technol. 2023, 202, 10. [Google Scholar] [CrossRef]
- Kapoor, L.; Simkin, A.J.; George Priya Doss, C.; Siva, R. Fruit ripening: Dynamics and integrated analysis of carotenoids and anthocyanins. BMC Plant Biol. 2022, 22, 27. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, L.; Zhang, L.; Lv, H.; He, Q.; Guo, L.; Zhang, X.; He, H.; Ren, S.; Zhang, N.; et al. Melatonin promotes carotenoid biosynthesis in an ethylene-dependent manner in tomato fruits. Plant Sci. 2020, 298, 110580. [Google Scholar] [CrossRef]
- Su, L.; Diretto, G.; Purgatto, E.; Danoun, S.; Zouine, M.; Li, Z.; Roustan, J.P.; Bouzayen, M.; Giuliano, G.; Chervin, C. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol. 2015, 15, 114. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Zhang, L.; Kudaka, R.; Inaba, H.; Furuya, T.; Kitamura, M.; Kitaya, Y.; Yamamoto, R.; Yahata, M.; Matsumoto, H.; et al. Exogenous application of ABA and NAA alleviates the delayed coloring caused by puffing inhibitor in citrus fruit. Cells 2021, 10, 308. [Google Scholar] [CrossRef]
- Wisutiamonkul, A.; Ampomah-Dwamena, C.; Allan, A.C.; Ketsa, S. Carotenoid accumulation in durian (Durio zibethinus) fruit is affected by ethylene via modulation of carotenoid pathway gene expression. Plant Physiol. Biochem. 2017, 115, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wei, J.; Zhang, M.; Zhang, L.; Li, C.; Wang, Q. Ethylene independent induction of lycopene biosynthesis in tomato fruits by jasmonates. J. Exp. Bot. 2012, 63, 5751–5761. [Google Scholar] [CrossRef] [PubMed]
- Keawmanee, N.; Ma, G.; Zhang, L.; Yahata, M.; Murakami, K.; Yamamoto, M.; Kojima, N.; Kato, M. Exogenous gibberellin induced regreening through the regulation of chlorophyll and carotenoid metabolism in Valencia oranges. Plant Physiol. Biochem. 2022, 173, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 controls arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, Z.; Wang, L.; Kim, S.G.; Seo, P.J.; Qiao, M.; Wang, N.; Li, S.; Cao, X.; Park, C.M.; et al. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016, 85, 96–106. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, L.; Yu, D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 2018, 176, 790–803. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Li, L.Q.; Xu, Q.; Kong, Y.H.; Wang, H.; Wu, W.H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in arabidopsis. Plant Cell 2009, 21, 3554–3566. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, X.; Wang, L.; Tian, Y.; Jia, N.; Chen, S.; Shi, N.; Huang, X.; Zhou, C.; Yu, Y.; et al. Regulation of ethylene-responsive SlWRKYs involved in color change during tomato fruit ripening. Sci. Rep. 2017, 7, 16674. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Qu, Y.; Sha, G.; Zhang, S.; Ma, Y.; Chen, M.; Zhai, R.; Yang, C.; Xu, L.; Wang, Z. PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT, and PbMYB10b in pear. Physiol. Plantarum. 2021, 173, 1841–1849. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, J.; Hu, K.; Wei, S.; Sun, H.; Hu, L.; Han, Z.; Yao, G.; Zhang, H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 2020, 7, 37. [Google Scholar] [CrossRef]
- Zhu, Q.; Gong, Z.; Huang, J.; Grierson, D.; Chen, K.; Yin, X. High-CO2/hypoxia-responsive transcription factors DkERF24 and DkWRKY1 interact and activate DkPDC2 promoter. Plant Physiol. 2019, 180, 621–633. [Google Scholar] [CrossRef]
- Cheng, M.; Huang, Z.; Hua, Q.; Shan, W.; Kuang, J.; Lu, W.; Qin, Y.; Chen, J. The WRKY transcription factor HpWRKY44 regulates CytP450-like1 expression in red pitaya fruit (Hylocereus polyrhizus). Hortic. Res. 2017, 4, 17039. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Bao, Z.; Zhou, Q.; Wu, W.; Chen, W.; Yang, Z.; Wang, L.; Li, X.; Cao, S.; Shi, L. EjWRKY6 Is Involved in the ABA-Induced Carotenoid Biosynthesis in Loquat Fruit during Ripening. Foods 2024, 13, 2829. https://doi.org/10.3390/foods13172829
Yu Y, Bao Z, Zhou Q, Wu W, Chen W, Yang Z, Wang L, Li X, Cao S, Shi L. EjWRKY6 Is Involved in the ABA-Induced Carotenoid Biosynthesis in Loquat Fruit during Ripening. Foods. 2024; 13(17):2829. https://doi.org/10.3390/foods13172829
Chicago/Turabian StyleYu, Yan, Zeyang Bao, Qihang Zhou, Wei Wu, Wei Chen, Zhenfeng Yang, Li Wang, Xuewen Li, Shifeng Cao, and Liyu Shi. 2024. "EjWRKY6 Is Involved in the ABA-Induced Carotenoid Biosynthesis in Loquat Fruit during Ripening" Foods 13, no. 17: 2829. https://doi.org/10.3390/foods13172829




