Fava Bean Protein Nanofibrils Modulate Cell Membrane Interfaces for Biomolecular Interactions as Unveiled by Atomic Force Microscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis of Protein Nanofibrils from Fava Bean Protein Isolate
2.2. Physiological Adaptation of PNFs
2.3. Thioflavin T Assay
2.4. MCC-13 Cell Culture
2.5. PNFs Interaction with Merkel (MCC-13) Cells
2.6. Formation of Phospholipid Liposomes
2.7. Interaction of PNFs with Supported Lipid Bilayer (SLBs)
2.8. Atomic Force Microscopy (AFM)
2.9. Data Analysis
2.10. Gene Response Data from PNFs-Cells Interaction
3. Results and Discussion
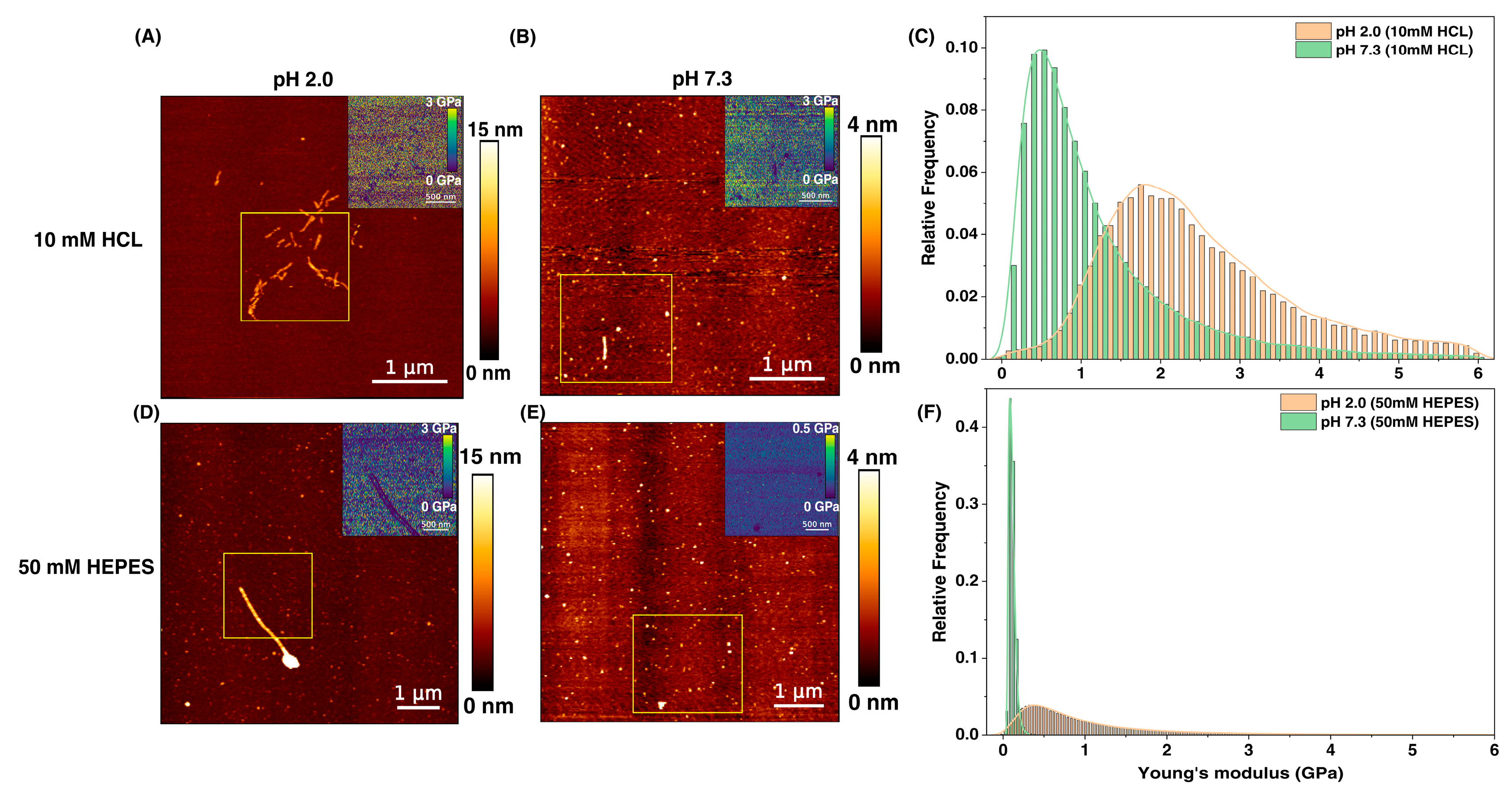
3.1. Influence of Microenvironment on Morphology and Elasticity of PNFs
3.2. Impact of PNFs on Quantitative Mechanics of MCC-13 Cells
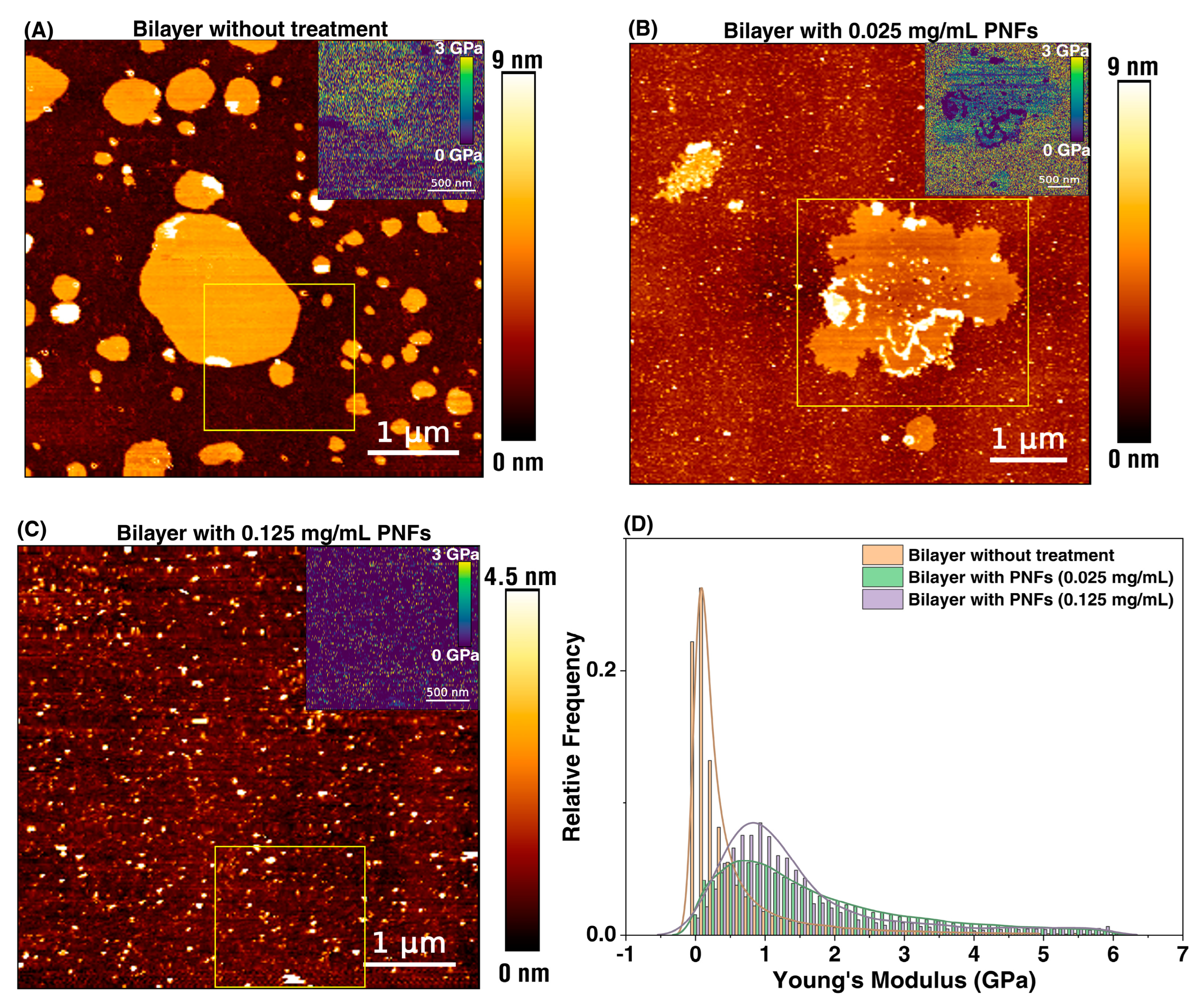
3.3. Supported Lipid Bilayer (SLBs)-PNFs Interaction
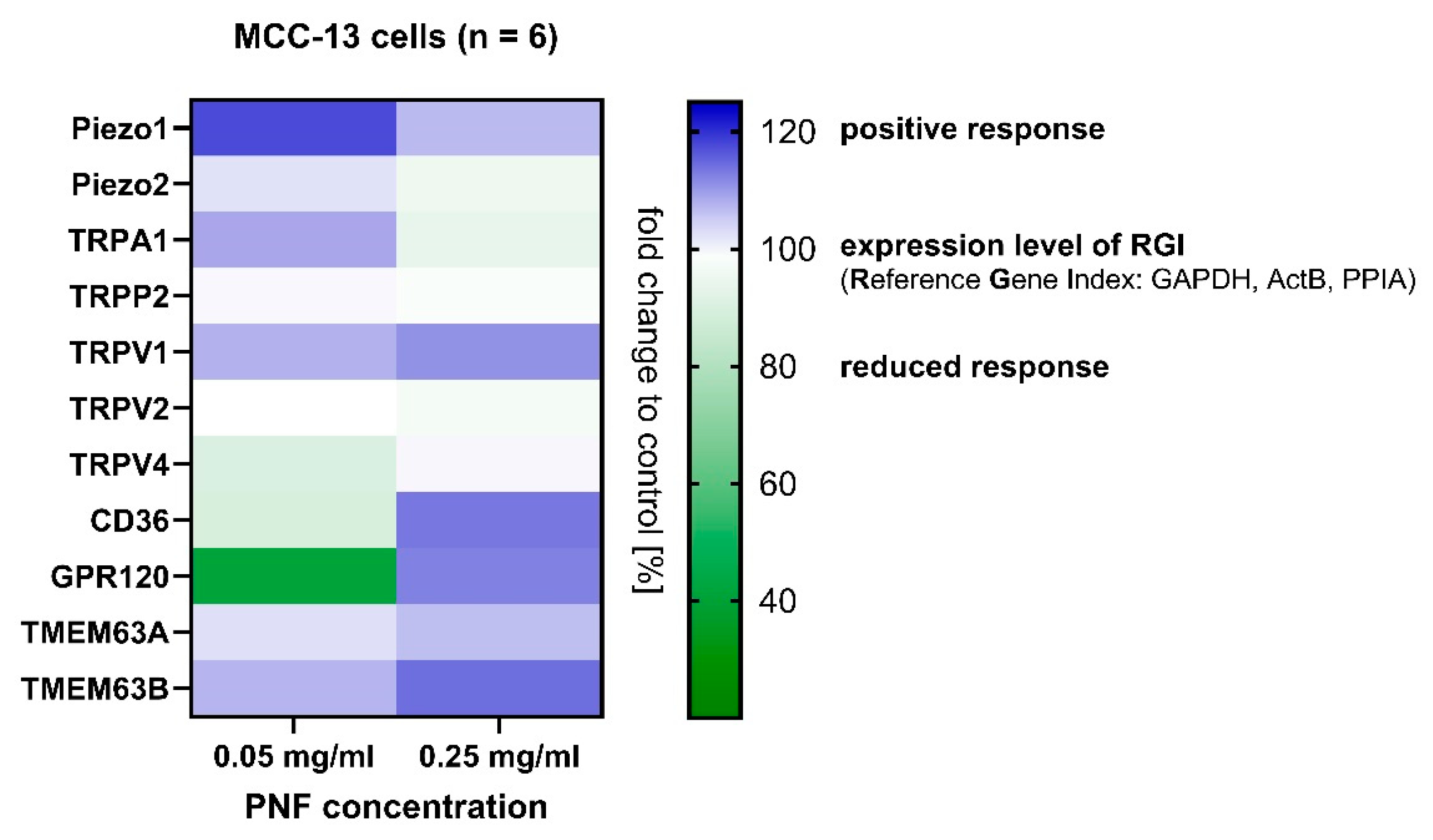
3.4. Gene Response from MCC-13 Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koehler, M.; Benthin, J.; Karanth, S.; Wiesenfarth, M.; Sebald, K.; Somoza, V. Biophysical investigations using atomic force microscopy can elucidate the link between mouthfeel and flavour perception. Nat. Food 2024, 5, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Mezzenga, R. Food protein amyloid fibrils: Origin, structure, formation, characterization, applications and health implications. Adv. Colloid Interface Sci. 2019, 269, 334–356. [Google Scholar] [CrossRef]
- Yue, J.; Yao, X.; Gou, Q.; Li, D.; Liu, N.; Yang, D.; Gao, Z.; Midgley, A.; Katsuyoshi, N.; Zhao, M. Recent advances of interfacial and rheological property based techno-functionality of food protein amyloid fibrils. Food Hydrocoll. 2022, 132, 107827. [Google Scholar] [CrossRef]
- Lassé, M.; Ulluwishewa, D.; Healy, J.; Thompson, D.; Miller, A.; Roy, N.; Chitcholtan, K.; Gerrard, J.A. Evaluation of protease resistance and toxicity of amyloid-like food fibrils from whey, soy, kidney bean, and egg white. Food Chem. 2016, 192, 491–498. [Google Scholar] [CrossRef]
- Goers, J.; Permyakov, S.E.; Permyakov, E.A.; Uversky, V.N.; Fink, A.L. Conformational prerequisites for α-lactalbumin fibrillation. Biochemistry 2002, 41, 12546–12551. [Google Scholar] [CrossRef] [PubMed]
- Adamcik, J.; Lara, C.; Usov, I.; Jeong, J.S.; Ruggeri, F.S.; Dietler, G.; Lashuel, H.A.; Hamley, I.W.; Mezzenga, R. Measurement of intrinsic properties of amyloid fibrils by the peak force QNM method. Nanoscale 2012, 4, 4426–4429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dee, D.R. Morphology, formation kinetics and core composition of pea and soy 7S and 11S globulin amyloid fibrils. J. Agric. Food Chem. 2023, 71, 4755–4765. [Google Scholar] [CrossRef]
- Labba, I.-C.M.; Frøkiær, H.; Sandberg, A.-S. Nutritional and antinutritional composition of fava bean (Vicia faba L., var. minor) cultivars. Food Res. Int. 2021, 140, 110038. [Google Scholar] [CrossRef]
- Warsame, A.O.; Michael, N.; O’Sullivan, D.M.; Tosi, P. Identification and quantification of major faba bean seed proteins. J. Agric. Food Chem. 2020, 68, 8535–8544. [Google Scholar] [CrossRef]
- Multari, S.; Stewart, D.; Russell, W.R. Potential of fava bean as future protein supply to partially replace meat intake in the human diet. Compr. Rev. Food Sci. Food Saf. 2015, 14, 511–522. [Google Scholar] [CrossRef]
- Żmudziński, D.; Goik, U.; Ptaszek, P. Functional and Rheological Properties of Vicia faba L. Protein Isolates. Biomolecules 2021, 11, 178. [Google Scholar]
- Chiti, F.; Taddei, N.; Baroni, F.; Capanni, C.; Stefani, M.; Ramponi, G.; Dobson, C.M. Kinetic partitioning of protein folding and aggregation. Nat. Struct. Biol. 2002, 9, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Lendel, C.; Langton, M.; Olsson, R.T.; Hedenqvist, M.S. Protein nanofibrils: Preparation, properties, and possible applications in industrial nanomaterials. In Industrial Applications of Nanomaterials; Elsevier: Amsterdam, The Netherlands, 2019; pp. 29–63. [Google Scholar]
- Herneke, A.; Karkehabadi, S.; Lu, J.; Lendel, C.; Langton, M. Protein nanofibrils from mung bean: The effect of pH on morphology and the ability to form and stabilise foams. Food Hydrocoll. 2023, 136, 108315. [Google Scholar] [CrossRef]
- Liu, C.; Wu, D.; Wang, P.; McClements, D.J.; Cui, S.; Liu, H.; Leng, F.; Sun, Q.; Dai, L. Study on the formation mechanism of pea protein nanofibrils and the changes of structural properties of fibril under different pH and temperature. Food Hydrocoll. 2024, 150, 109735. [Google Scholar] [CrossRef]
- Peng, D.; Yang, J.; Li, J.; Tang, C.; Li, B. Foams stabilized by β-lactoglobulin amyloid fibrils: Effect of pH. J. Agric. Food Chem. 2017, 65, 10658–10665. [Google Scholar] [CrossRef]
- Guo, S.; Akhremitchev, B.B. Packing density and structural heterogeneity of insulin amyloid fibrils measured by AFM nanoindentation. Biomacromolecules 2006, 7, 1630–1636. [Google Scholar] [CrossRef]
- Adamcik, J.; Mezzenga, R. Study of amyloid fibrils via atomic force microscopy. Curr. Opin. Colloid Interface Sci. 2012, 17, 369–376. [Google Scholar] [CrossRef]
- Adamcik, J.; Berquand, A.; Mezzenga, R. Single-step direct measurement of amyloid fibrils stiffness by peak force quantitative nanomechanical atomic force microscopy. Appl. Phys. Lett. 2011, 98, 193701. [Google Scholar] [CrossRef]
- Cao, Y.; Bolisetty, S.; Wolfisberg, G.; Adamcik, J.; Mezzenga, R. Amyloid fibril-directed synthesis of silica core–shell nanofilaments, gels, and aerogels. Proc. Natl. Acad. Sci. USA 2019, 116, 4012–4017. [Google Scholar] [CrossRef]
- Herneke, A.; Lendel, C.; Karkehabadi, S.; Lu, J.; Langton, M. Protein Nanofibrils from Fava Bean and Its Major Storage Proteins: Formation and Ability to Generate and Stabilise Foams. Foods 2023, 12, 521. [Google Scholar] [CrossRef]
- Aframian, D.; Davidowitz, T.; Benoliel, R. The distribution of oral mucosal pH values in healthy saliva secretors. Oral Dis. 2006, 12, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Leonard, J.H.; Dash, P.; Holland, P.; Kearsley, J.H.; Bell, J.R. Characterisation of four Merkel cell carcinoma adherent cell lines. Int. J. Cancer 1995, 60, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef]
- Herneke, A.; Lendel, C.; Johansson, D.; Newson, W.; Hedenqvist, M.; Karkehabadi, S.; Jonsson, D.; Langton, M. Protein nanofibrils for sustainable food–characterization and comparison of fibrils from a broad range of plant protein isolates. ACS Food Sci. Technol. 2021, 1, 854–864. [Google Scholar] [CrossRef]
- Almeida, Z.L.; Brito, R.M. Structure and aggregation mechanisms in amyloids. Molecules 2020, 25, 1195. [Google Scholar] [CrossRef] [PubMed]
- Gregori, M.; Cassina, V.; Brogioli, D.; Salerno, D.; De Kimpe, L.; Scheper, W.; Masserini, M.; Mantegazza, F. Stability of Aβ (1–42) peptide fibrils as consequence of environmental modifications. Eur. Biophys. J. 2010, 39, 1613–1623. [Google Scholar] [CrossRef]
- Jenkins, B.A.; Lumpkin, E.A. Developing a sense of touch. Development 2017, 144, 4078–4090. [Google Scholar] [CrossRef]
- Woo, S.-H.; Lumpkin, E.A.; Patapoutian, A. Merkel cells and neurons keep in touch. Trends Cell Biol. 2015, 25, 74–81. [Google Scholar] [CrossRef]
- Maksimovic, S.; Nakatani, M.; Baba, Y.; Nelson, A.M.; Marshall, K.L.; Wellnitz, S.A.; Firozi, P.; Woo, S.-H.; Ranade, S.; Patapoutian, A. Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature 2014, 509, 617–621. [Google Scholar] [CrossRef]
- Romero, L.O.; Caires, R.; Nickolls, A.R.; Chesler, A.T.; Cordero-Morales, J.F.; Vásquez, V. A dietary fatty acid counteracts neuronal mechanical sensitization. Nat. Commun. 2020, 11, 2997. [Google Scholar] [CrossRef]
- Delmas, P.; Parpaite, T.; Coste, B. PIEZO channels and newcomers in the mammalian mechanosensitive ion channel family. Neuron 2022, 110, 2713–2727. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.C.; Park, H.J.; Kim, J.G.; Lee, I.H.; Cho, H.; Park, C.; Sung, T.S.; Koh, S.D.; Park, S.W.; Bae, Y.M. The Piezo2 ion channel is mechanically activated by low-threshold positive pressure. Sci. Rep. 2019, 9, 6446. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.; Chia, S.; Klimont, E.; Knowles, T.P.; Vendruscolo, M.; Ruggeri, F.S. Maturation-dependent changes in the size, structure and seeding capacity of Aβ42 amyloid fibrils. Commun. Biol. 2024, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Lee, H.-H.; Lee, C.-H. Substrate properties modulate cell membrane roughness by way of actin filaments. Sci. Rep. 2017, 7, 9068. [Google Scholar] [CrossRef]
- Escamilla-García, M.; Ríos-Romo, R.A.; Melgarejo-Mancilla, A.; Díaz-Ramírez, M.; Hernández-Hernández, H.M.; Amaro-Reyes, A.; Pierro, P.D.; Regalado-González, C. Rheological and antimicrobial properties of chitosan and quinoa protein filmogenic suspensions with thyme and rosemary essential oils. Foods 2020, 9, 1616. [Google Scholar] [CrossRef]
- Anselme, K.; Ploux, L.; Ponche, A. Cell/material interfaces: Influence of surface chemistry and surface topography on cell adhesion. J. Adhes. Sci. Technol. 2010, 24, 831–852. [Google Scholar] [CrossRef]
- Jacob, R.S.; George, E.; Singh, P.K.; Salot, S.; Anoop, A.; Jha, N.N.; Sen, S.; Maji, S.K. Cell adhesion on amyloid fibrils lacking integrin recognition motif. J. Biol. Chem. 2016, 291, 5278–5298. [Google Scholar] [CrossRef]
- Kimura, A.; Fukuda, T.; Zhang, M.; Motoyama, S.; Maruyama, N.; Utsumi, S. Comparison of Physicochemical Properties of 7S and 11S Globulins from Pea, Fava Bean, Cowpea, and French Bean with Those of Soybean—French Bean 7S Globulin Exhibits Excellent Properties. J. Agric. Food Chem. 2008, 56, 10273–10279. [Google Scholar] [CrossRef]
- Pan, H.-J.; Wang, R.-L.; Xiao, J.-L.; Chang, Y.-J.; Cheng, J.-Y.; Chen, Y.-R.; Lee, C.-H. Using optical profilometry to characterize cell membrane roughness influenced by amyloid-beta 42 aggregates and electric fields. J. Biomed. Opt. 2014, 19, 011009. [Google Scholar] [CrossRef]
- Dang, S.; Feng, S.; Tien, J.; Peters, C.J.; Bulkley, D.; Lolicato, M.; Zhao, J.; Zuberbühler, K.; Ye, W.; Qi, L. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 2017, 552, 426–429. [Google Scholar] [CrossRef]
- Shao, Y.; Gao, Z.; Feldman, T.; Jiang, X. Stimulation of ATG12-ATG5 conjugation by ribonucleic acid. Autophagy 2007, 3, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Karanth, S.; Azinfar, A.; Helm, C.A.; Delcea, M. Identification of a critical lipid ratio in raft-like phases exposed to nitric oxide: An AFM study. Biophys. J. 2021, 120, 3103–3111. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.; Mukhopadhyay, A.; Brisson, A. Pathways of lipid vesicle deposition on solid surfaces: A combined QCM-D and AFM study. Biophys. J. 2003, 85, 3035–3047. [Google Scholar] [CrossRef]
- Sparr, E.; Engel, M.F.; Sakharov, D.V.; Sprong, M.; Jacobs, J.; de Kruijff, B.; Höppener, J.W.; Antoinette Killian, J. Islet amyloid polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. FEBS Lett. 2004, 577, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Mao, F.; Yang, Y.; Jiang, H. Electromechanical model for object roughness perception during finger sliding. Biophys. J. 2022, 121, 4740–4747. [Google Scholar] [CrossRef]
- Zheng, W.; Rawson, S.; Shen, Z.; Tamilselvan, E.; Smith, H.E.; Halford, J.; Shen, C.; Murthy, S.E.; Ulbrich, M.H.; Sotomayor, M. TMEM63 proteins function as monomeric high-threshold mechanosensitive ion channels. Neuron 2023, 111, 3195–3210.e3197. [Google Scholar] [CrossRef]
- Murthy, S.E.; Dubin, A.E.; Whitwam, T.; Jojoa-Cruz, S.; Cahalan, S.M.; Mousavi, S.A.R.; Ward, A.B.; Patapoutian, A. OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 2018, 7, e41844. [Google Scholar] [CrossRef]
- Aiello, B.R.; Stewart, T.A.; Hale, M.E. Mechanosensation in an adipose fin. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152794. [Google Scholar] [CrossRef]
- Kefauver, J.; Ward, A.; Patapoutian, A. Discoveries in structure and physiology of mechanically activated ion channels. Nature 2020, 587, 567–576. [Google Scholar] [CrossRef]
- Pepino, M.Y.; Kuda, O.; Samovski, D.; Abumrad, N.A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014, 34, 281–303. [Google Scholar] [CrossRef]
- Karakuła-Juchnowicz, H.; Róg, J.; Juchnowicz, D.; Morylowska-Topolska, J. GPR120: Mechanism of action, role and potential for medical applications. Postep. Hig. I Med. Dosw. (Online) 2017, 71, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.F.; Nilius, B. Transient receptor potential channels in mechanosensing and cell volume regulation. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2007; Volume 428, pp. 183–207. [Google Scholar]
- Zhang, Y.; Lv, X.; Abker, A.M.; Oh, D.-H.; Kassem, J.M.; Salama, M.; Fu, X. Research progress of protein fibrils: A review of formation mechanism, characterization and applications in the food field. Food Hydrocoll. 2024, 155, 110199. [Google Scholar] [CrossRef]
- An, D.; Ban, Q.; Du, H.; Wang, Q.; Teng, F.; Li, L.; Xiao, H. Nanofibrils of food-grade proteins: Formation mechanism, delivery systems, and application evaluation. Compr. Rev. Food Sci. Food Saf. 2022, 21, 4847–4871. [Google Scholar] [CrossRef] [PubMed]
- Lendel, C.; Solin, N. Protein nanofibrils and their use as building blocks of sustainable materials. RSC Adv. 2021, 11, 39188–39215. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, P.; Natarajan, A.; Salmen, S.H.; Alharbi, S.A.; Shavrov, V.; Lega, P.; Subramani, R.; Pushparaj, C. Utilizing protein nanofibrils as a scaffold for enhancing nutritional value in toned milk. Environ. Res. 2023, 239, 117420. [Google Scholar] [CrossRef] [PubMed]
- Romani, P.; Valcarcel-Jimenez, L.; Frezza, C.; Dupont, S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 22–38. [Google Scholar] [CrossRef]
- Martineau-Côté, D.; Achouri, A.; Karboune, S.; L’Hocine, L. Faba bean: An untapped source of quality plant proteins and bioactives. Nutrients 2022, 14, 1541. [Google Scholar] [CrossRef]
- Sharan, S.; Zanghelini, G.; Zotzel, J.; Bonerz, D.; Aschoff, J.; Saint-Eve, A.; Maillard, M.N. Fava bean (Vicia faba L.) for food applications: From seed to ingredient processing and its effect on functional properties, antinutritional factors, flavor, and color. Compr. Rev. Food Sci. Food Saf. 2021, 20, 401–428. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karanth, S.; Wiesenfarth, M.; Benthin, J.; Koehler, M. Fava Bean Protein Nanofibrils Modulate Cell Membrane Interfaces for Biomolecular Interactions as Unveiled by Atomic Force Microscopy. Foods 2024, 13, 3411. https://doi.org/10.3390/foods13213411
Karanth S, Wiesenfarth M, Benthin J, Koehler M. Fava Bean Protein Nanofibrils Modulate Cell Membrane Interfaces for Biomolecular Interactions as Unveiled by Atomic Force Microscopy. Foods. 2024; 13(21):3411. https://doi.org/10.3390/foods13213411
Chicago/Turabian StyleKaranth, Sanjai, Marina Wiesenfarth, Julia Benthin, and Melanie Koehler. 2024. "Fava Bean Protein Nanofibrils Modulate Cell Membrane Interfaces for Biomolecular Interactions as Unveiled by Atomic Force Microscopy" Foods 13, no. 21: 3411. https://doi.org/10.3390/foods13213411
APA StyleKaranth, S., Wiesenfarth, M., Benthin, J., & Koehler, M. (2024). Fava Bean Protein Nanofibrils Modulate Cell Membrane Interfaces for Biomolecular Interactions as Unveiled by Atomic Force Microscopy. Foods, 13(21), 3411. https://doi.org/10.3390/foods13213411






