Effect of Probiotics on Gastrointestinal Health Through the Aryl Hydrocarbon Receptor Pathway: A Systematic Review
Abstract
1. Introduction
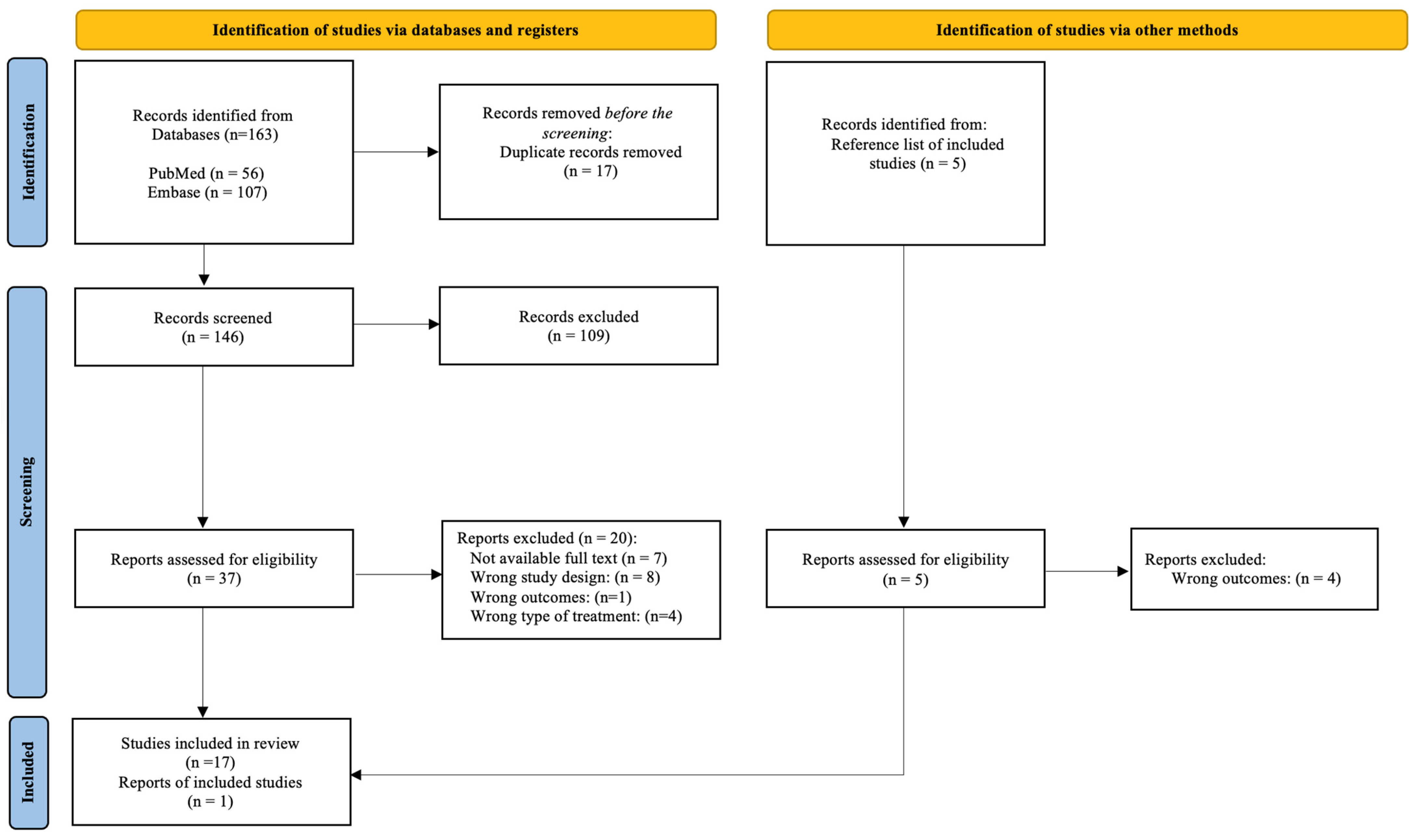
2. Methods of Searching
2.1. Eligibility Criteria
2.2. Search Strategy
2.3. Selection Process
2.4. Data Collection Process
2.5. Risk of Bias in Individual Studies
3. Results
3.1. Study Characteristics
3.2. Risk of Bias Assessment
3.3. Results of Individual Studies
3.3.1. Relationship Between Probiotics and AhR in Physiological Conditions
3.3.2. Relationship Between Probiotics and AhR in Gastrointestinal Pathologies
Inflammatory Bowel Disease
Celiac Disease
Necrotizing Enterocolitis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Borgeraas, H.; Johnson, L.K.; Skattebu, J.; Hertel, J.K.; Hjelmesæth, J. Effects of Probiotics on Body Weight, Body Mass Index, Fat Mass and Fat Percentage in Subjects with Overweight or Obesity: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Obes. Rev. 2018, 19, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Shinde, R.; McGaha, T.L. The Aryl Hydrocarbon Receptor: Connecting Immunity to the Microenvironment. Trends Immunol. 2018, 39, 1005. [Google Scholar] [CrossRef] [PubMed]
- Larigot, L.; Juricek, L.; Dairou, J.; Coumoul, X. AhR Signaling Pathways and Regulatory Functions. Biochim. Open 2018, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Neavin, D.R.; Liu, D.; Ray, B.; Weinshilboum, R.M. The Role of the Aryl Hydrocarbon Receptor (AHR) in Immune and Inflammatory Diseases. Int. J. Mol. Sci. 2018, 19, 3851. [Google Scholar] [CrossRef]
- Yue, B.; Yu, Z.L.; Lv, C.; Geng, X.L.; Wang, Z.T.; Dou, W. Regulation of the Intestinal Microbiota: An Emerging Therapeutic Strategy for Inflammatory Bowel Disease. World J. Gastroenterol. 2020, 26, 4378. [Google Scholar] [CrossRef]
- Cannon, A.R.; Shim, E.H.; Kuprys, P.V.; Choudhry, M.A. IL-22 and Lactobacillus Delbrueckii Mitigate Alcohol Induced Exacerbation of DSS-Induced Colitis. J. Leukoc. Biol. 2022, 112, 1471. [Google Scholar] [CrossRef]
- Wilkins, T.; Sequoia, J.; Jennings, W.; Dorn, B. Probiotics for Gastrointestinal Conditions: A Summary of the Evidence. Am. Fam. Physician 2017, 96, 170–178. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef]
- Hooijmans, C.R.; Rovers, M.M.; De Vries, R.B.M.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s Risk of Bias Tool for Animal Studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef]
- Takamura, T.; Harama, D.; Fukumoto, S.; Nakamura, Y.; Shimokawa, N.; Ishimaru, K.; Ikegami, S.; Makino, S.; Kitamura, M.; Nakao, A. Lactobacillus Bulgaricus OLL1181 Activates the Aryl Hydrocarbon Receptor Pathway and Inhibits Colitis. Immunol. Cell Biol. 2011, 89, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Burton, K.J.; Pimentel, G.; Zangger, N.; Vionnet, N.; Drai, J.; McTernan, P.G.; Pralong, F.P.; Delorenzi, M.; Vergères, G. Modulation of the Peripheral Blood Transcriptome by the Ingestion of Probiotic Yoghurt and Acidified Milk in Healthy, Young Men. PLoS ONE 2018, 13, e0192947. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tian, X.; He, B.; Hoang, T.K.; Taylor, C.M.; Blanchard, E.; Freeborn, J.; Park, S.; Luo, M.; Couturier, J.; et al. Microbiome and Host Interactions: Lactobacillus Reuteri DSM 17938 Feeding of Healthy Newborn Mice Regulates Immune Responses While Modulating Gut Microbiota and Boosting Beneficial Metabolites. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G824. [Google Scholar] [CrossRef]
- Özçam, M.; Tocmo, R.; Oh, J.H.; Afrazi, A.; Mezrich, J.D.; Roos, S.; Claesen, J.; van Pijkeren, J.P. Gut Symbionts Lactobacillus Reuteri R2lc and 2010 Encode a Polyketide Synthase Cluster That Activates the Mammalian Aryl Hydrocarbon Receptor. Appl. Environ. Microbiol. 2019, 85, e01661-18. [Google Scholar] [CrossRef]
- Xie, Z.; Li, M.; Qian, M.; Yang, Z.; Han, X. Co-Cultures of Lactobacillus Acidophilus and Bacillus Subtilis Enhance Mucosal Barrier by Modulating Gut Microbiota-Derived Short-Chain Fatty Acids. Nutrients 2022, 14, 4475. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; Angelo, C.D.; Massi-Benedetti, C.; Fallarino, F.; et al. Article Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Fukumoto, S.; Toshimitsu, T.; Matsuoka, S.; Maruyama, A.; Oh-Oka, K.; Takamura, T.; Nakamura, Y.; Ishimaru, K.; Fujii-Kuriyama, Y.; Ikegami, S.; et al. Identification of a Probiotic Bacteria-Derived Activator of the Aryl Hydrocarbon Receptor That Inhibits Colitis. Immunol. Cell Biol. 2014, 92, 460–465. [Google Scholar] [CrossRef]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 Impacts Colitis by Altering Gut Microbiota Metabolism of Tryptophan into Aryl Hydrocarbon Receptor Ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Ye, L.; Liu, H.; Huang, L.; Yang, Q.; Turner, J.; Yu, Q. Lactobacillus Accelerates ISCs Regeneration to Protect the Integrity of Intestinal Mucosa through Activation of STAT3 Signaling Pathway Induced by LPLs Secretion of IL-22. Cell Death Differ. 2018, 25, 1657. [Google Scholar] [CrossRef]
- Gu, Z.; Pei, W.; Shen, Y.; Wang, L.; Zhu, J.; Zhang, Y.; Fan, S.; Wu, Q.; Li, L.; Zhang, Z. Akkermansia Muciniphila and Its Outer Protein Amuc_1100 Regulates Tryptophan Metabolism in Colitis. Food Funct. 2021, 12, 10184–10195. [Google Scholar] [CrossRef]
- Li, K.; Hao, Z.; Du, J.; Gao, Y.; Yang, S.; Zhou, Y. Bacteroides Thetaiotaomicron Relieves Colon Inflammation by Activating Aryl Hydrocarbon Receptor and Modulating CD4+T Cell Homeostasis. Int. Immunopharmacol. 2020, 90, 107183. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.-Y.; Tian, X.-Y.; Liang, X.; Zhang, Z.; Wang, R.; Zhou, Y.; Yi, H.-X.; Gong, P.-M.; Lin, K.; Liu, T.-J.; et al. Bifidobacterium Bifidum Relieved DSS-Induced Colitis in Mice Potentially by Activating the Aryl Hydrocarbon Receptor. Food Funct. 2022, 13, 5115–5123. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Zhang, Z.; Tian, X.; Liang, X.; Lu, Y.; Shi, Y.; Kuerman, M.; Wang, R.; Zhou, Y.; Gong, P.; et al. Bifidobacterium Bifidum Ameliorates DSS-Induced Colitis in Mice by Regulating AHR/NRF2/NLRP3 Inflammasome Pathways through Indole-3-Lactic Acid Production. J. Agric. Food Chem. 2023, 71, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Park, I.S.; Kim, J.H.; Yu, J.; Shin, Y.J.; Kim, K.; Kim, T.I.; Kim, S.W.; Cheon, J.H. Bifidobacterium Breve CBT BR3 Is Effective at Relieving Intestinal Inflammation by Augmenting Goblet Cell Regeneration. J. Gastroenterol. Hepatol. 2023, 38, 1346–1354. [Google Scholar] [CrossRef]
- Xia, Y.; Liu, C.; Li, R.; Zheng, M.; Feng, B.; Gao, J.; Long, X.; Li, L.; Li, S.; Zuo, X.; et al. Lactobacillus-Derived Indole-3-Lactic Acid Ameliorates Colitis in Cesarean-Born Offspring via Activation of Aryl Hydrocarbon Receptor. iScience 2023, 26, 108279. [Google Scholar] [CrossRef]
- Lamas, B.; Hernandez-Galan, L.; Galipeau, H.J.; Constante, M.; Clarizio, A.; Jury, J.; Breyner, N.M.; Caminero, A.; Rueda, G.; Hayes, C.L.; et al. Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is Decreased in Celiac Disease Leading to Intestinal Inflammation. Sci. Transl. Med. 2020, 12, eaba0624. [Google Scholar] [CrossRef]
- Meng, D.; Sommella, E.; Salviati, E.; Campiglia, P.; Ganguli, K.; Djebali, K.; Zhu, W.; Walker, W.A. Indole-3-Lactic Acid, a Metabolite of Tryptophan, Secreted by Bifidobacterium Longum Subspecies Infantis Is Anti-Inflammatory in the Immature Intestine. Pediatr. Res. 2020, 88, 209–217. [Google Scholar] [CrossRef]
- Wang, A.; Guan, C.; Wang, T.; Mu, G.; Tuo, Y. Indole-3-Lactic Acid, a Tryptophan Metabolite of Lactiplantibacillus Plantarum DPUL-S164, Improved Intestinal Barrier Damage by Activating AhR and Nrf2 Signaling Pathways. J. Agric. Food Chem. 2023, 71, 18792–18801. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, K.; Han, B.; Sheng, B.; Yin, J.; Pu, A.; Li, L.; Sun, L.; Yu, M.; Qiu, Y.; et al. Aryl Hydrocarbon Receptor Inhibits Inflammation in DSS-Induced Colitis via the MK2/p-MK2/TTP Pathway. Int. J. Mol. Med. 2018, 41, 868. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The Role of Gut Microbiota in Immune Homeostasis and Autoimmunity. Gut Microbes 2012, 3, 4. [Google Scholar] [CrossRef]
- Özçam, M.; van Pijkeren, J.-P. Draft Genome Sequence of Aryl Hydrocarbon Receptor Activator Strains Lactobacillus Reuteri R2lc and 2010. Microbiol. Resour. Announc. 2019, 8, 67–86. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T. Pathophysiology of Inflammatory Bowel Diseases. N. Engl. J. Med. 2020, 383, 2652–2664. [Google Scholar] [CrossRef] [PubMed]
- Currò, D.; Ianiro, G.; Pecere, S.; Bibbò, S.; Cammarota, G. Probiotics, Fibre and Herbal Medicinal Products for Functional and Inflammatory Bowel Disorders. Br. J. Pharmacol. 2017, 174, 1426–1449. [Google Scholar] [CrossRef]
- Jeong, D.Y.; Kim, S.; Son, M.J.; Son, C.Y.; Kim, J.Y.; Kronbichler, A.; Lee, K.H.; Shin, J.I. Induction and Maintenance Treatment of Inflammatory Bowel Disease: A Comprehensive Review. Autoimmun. Rev. 2019, 18, 439–454. [Google Scholar] [CrossRef]
- De Souza, H.S.P.; Fiocchi, C. Immunopathogenesis of IBD: Current State of the Art. Nat. Rev. Gastroenterol. Hepatol. 2015, 13, 13–27. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, T.; Chen, W.; Chen, Y.; Li, M.; Ren, L.; Chen, J.; Cao, R.; Feng, Y.; Zhang, H.; et al. MicroRNA-124 Promotes Intestinal Inflammation by Targeting Aryl Hydrocarbon Receptor in Crohn’s Disease. J. Crohns Colitis 2016, 10, 703–712. [Google Scholar] [CrossRef]
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; MacDonald, T.T.; Pallone, F.; Monteleone, G. Aryl Hydrocarbon Receptor-Induced Signals up-Regulate IL-22 Production and Inhibit Inflammation in the Gastrointestinal Tract. Gastroenterology 2011, 141, 237–248.e1. [Google Scholar] [CrossRef]
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut Microbiome and Health: Mechanistic Insights. Gut 2022, 71, 1020. [Google Scholar] [CrossRef] [PubMed]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.R.; Mearin, M.L.; Phillips, A.; Shamir, R.; Troncone, R.; Giersiepen, K.; Branski, D.; Catassi, C.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition Guidelines for the Diagnosis of Coeliac Disease. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 136–160. [Google Scholar] [CrossRef]
- Lebwohl, B.; Rubio-Tapia, A. Epidemiology, Presentation, and Diagnosis of Celiac Disease. Gastroenterology 2021, 160, 63–75. [Google Scholar] [CrossRef]
- Ray, K. Connecting Coeliac Disease to the AhR Pathway. Nat. Rev. Gastroenterol. Hepatol. 2020, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Chang, M.Y.; Mandik-Nayak, L.; Metz, R.; Muller, A.J. Indoleamine 2,3-Dioxygenase as a Modifier of Pathogenic Inflammation in Cancer and Other Inflammation-Associated Diseases. Curr. Med. Chem. 2011, 18, 2257. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, H.J.; Verdu, E.F. The Double-Edged Sword of Gut Bacteria in Celiac Disease and Implications for Therapeutic Potential. Mucosal Immunol. 2022, 15, 235–243. [Google Scholar] [CrossRef]
- Duess, J.W.; Sampah, M.E.; Lopez, C.M.; Tsuboi, K.; Scheese, D.J.; Sodhi, C.P.; Hackam, D.J. Necrotizing Enterocolitis, Gut Microbes, and Sepsis. Gut Microbes 2023, 15, 2221470. [Google Scholar] [CrossRef] [PubMed]
- Neu, J.; Walker, W.A. Necrotizing Enterocolitis. N. Engl. J. Med. 2011, 364, 255. [Google Scholar] [CrossRef]
- Yazji, I.; Sodhi, C.P.; Lee, E.K.; Good, M.; Egan, C.E.; Afrazi, A.; Neal, M.D.; Jia, H.; Lin, J.; Ma, C.; et al. Endothelial TLR4 Activation Impairs Intestinal Microcirculatory Perfusion in Necrotizing Enterocolitis via ENOS-NO-Nitrite Signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 9451–9456. [Google Scholar] [CrossRef] [PubMed]
- Nolan, L.S.; Wynn, J.L.; Good, M. Exploring Clinically-Relevant Experimental Models of Neonatal Shock and Necrotizing Enterocolitis. Shock 2020, 53, 596. [Google Scholar] [CrossRef]
- Lin, P.W.; Stoll, B.J. Necrotising Enterocolitis. Lancet 2006, 368, 1271–1283. [Google Scholar] [CrossRef]
- Hintz, S.R.; Kendrick, D.E.; Stoll, B.J.; Vohr, B.R.; Fanaroff, A.A.; Donovan, E.F.; Poole, W.K.; Blakely, M.L.; Wright, L.; Higgins, R. Neurodevelopmental and Growth Outcomes of Extremely Low Birth Weight Infants After Necrotizing Enterocolitis. Pediatrics 2005, 115, 696–703. [Google Scholar] [CrossRef]
- McNelis, K.; Goddard, G.; Jenkins, T.; Poindexter, A.; Wessel, J.; Helmrath, M.; Poindexter, B. Delay in Achieving Enteral Autonomy and Growth Outcomes in Very Low Birth Weight Infants with Surgical Necrotizing Enterocolitis. J. Perinatol. 2020, 41, 150–156. [Google Scholar] [CrossRef]
- Pammi, M.; Cope, J.; Tarr, P.I.; Warner, B.B.; Morrow, A.L.; Mai, V.; Gregory, K.E.; Simon Kroll, J.; McMurtry, V.; Ferris, M.J.; et al. Intestinal Dysbiosis in Preterm Infants Preceding Necrotizing Enterocolitis: A Systematic Review and Meta-Analysis. Microbiome 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Jilling, T.; Simon, D.; Lu, J.; Meng, F.J.; Li, D.; Schy, R.; Thomson, R.B.; Soliman, A.; Arditi, M.; Caplan, M.S. The Roles of Bacteria and TLR4 in Rat and Murine Models of Necrotizing Enterocolitis. J. Immunol. 2006, 177, 3273. [Google Scholar] [CrossRef] [PubMed]
- Eaton, S.; Rees, C.M.; Hall, N.J. Current Research on the Epidemiology, Pathogenesis, and Management of Necrotizing Enterocolitis. Neonatology 2017, 111, 423–430. [Google Scholar] [CrossRef]
- Afrazi, A.; Branca, M.F.; Sodhi, C.P.; Good, M.; Yamaguchi, Y.; Egan, C.E.; Lu, P.; Jia, H.; Shaffiey, S.; Lin, J.; et al. Toll-like Receptor 4-Mediated Endoplasmic Reticulum Stress in Intestinal Crypts Induces Necrotizing Enterocolitis. J. Biol. Chem. 2014, 289, 9584. [Google Scholar] [CrossRef] [PubMed]
- Jilling, T.; Lu, J.; Jackson, M.; Caplan, M.S. Intestinal Epithelial Apoptosis Initiates Gross Bowel Necrosis in an Experimental Rat Model of Neonatal Necrotizing Enterocolitis. Pediatr. Res. 2004, 55, 622–629. [Google Scholar] [CrossRef]
- Liu, Y.; Fatheree, N.Y.; Mangalat, N.; Rhoads, J.M. Lactobacillus Reuteri Strains Reduce Incidence and Severity of Experimental Necrotizing Enterocolitis via Modulation of TLR4 and NF-ΚB Signaling in the Intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G608. [Google Scholar] [CrossRef] [PubMed]
- Neal, M.D.; Sodhi, C.P.; Dyer, M.; Craig, B.T.; Good, M.; Jia, H.; Yazji, I.; Afrazi, A.; Richardson, W.M.; Beer-Stolz, D.; et al. A Critical Role for Toll-like Receptor-4 Induction of Autophagy in the Regulation of Enterocyte Migration and the Pathogenesis of Necrotizing Enterocolitis. J. Immunol. 2013, 190, 3541. [Google Scholar] [CrossRef]
- Hackam, D.J.; Sodhi, C.P. Toll-Like Receptor–Mediated Intestinal Inflammatory Imbalance in the Pathogenesis of Necrotizing Enterocolitis. Cell Mol. Gastroenterol. Hepatol. 2018, 6, 229. [Google Scholar] [CrossRef]
- Lu, P.; Yamaguchi, Y.; Fulton, W.B.; Wang, S.; Zhou, Q.; Jia, H.; Kovler, M.L.; Salazar, A.G.; Sampah, M.; Prindle, T.; et al. Maternal Aryl Hydrocarbon Receptor Activation Protects Newborns against Necrotizing Enterocolitis. Nat. Commun. 2021, 12, 1042. [Google Scholar] [CrossRef]
- Alfaleh, K.; Anabrees, J. Probiotics for Prevention of Necrotizing Enterocolitis in Preterm Infants. Cochrane Database Syst. Rev. 2014, 2014, CD005496. [Google Scholar] [CrossRef]
- Olsen, R.; Greisen, G.; Schrøder, M.; Brok, J. Prophylactic Probiotics for Preterm Infants: A Systematic Review and Meta-Analysis of Observational Studies. Neonatology 2016, 109, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Dermyshi, E.; Wang, Y.; Yan, C.; Hong, W.; Qiu, G.; Gong, X.; Zhang, T. The “Golden Age” of Probiotics: A Systematic Review and Meta-Analysis of Randomized and Observational Studies in Preterm Infants. Neonatology 2017, 112, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Robertson, C.; Robertson, C.; Savva, G.M.; Clapuci, R.; Jones, J.; Maimouni, H.; Brown, E.; Minocha, A.; Hall, L.J.; Clarke, P.; et al. Incidence of Necrotising Enterocolitis before and after Introducing Routine Prophylactic Lactobacillus and Bifidobacterium Probiotics. Arch. Dis. Child. Fetal Neonatal Ed. 2020, 105, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.Y.; Chen, J.H.; Chang, J.H.; Lin, H.C.; Lin, C.Y.; Peng, C.C. Multiple Strains Probiotics Appear to Be the Most Effective Probiotics in the Prevention of Necrotizing Enterocolitis and Mortality: An Updated Meta-Analysis. PLoS ONE 2017, 12, e0171579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Q.; Zhang, F.; Sun, C. Probiotics for Preventing Necrotizing Enterocolitis: A Meta-Analysis with Trial Sequential Analysis. J. Clin. Pharm. Ther. 2023, 2023, 8626191. [Google Scholar] [CrossRef]
- Ganguli, K.; Meng, D.; Rautava, S.; Lu, L.; Walker, W.A.; Nanthakumar, N. Probiotics Prevent Necrotizing Enterocolitis by Modulating Enterocyte Genes That Regulate Innate Immune-Mediated Inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, 132–141. [Google Scholar] [CrossRef]
- Thänert, R.; Keen, E.C.; Dantas, G.; Warner, B.B.; Tarr, P.I. Necrotizing Enterocolitis and the Microbiome: Current Status and Future Directions. J. Infect. Dis. 2021, 223, S257–S263. [Google Scholar] [CrossRef]
- Wu, W.; Wang, Y.; Zou, J.; Long, F.; Yan, H.; Zeng, L.; Chen, Y. Bifidobacterium Adolescentis Protects against Necrotizing Enterocolitis and Upregulates TOLLIP and SIGIRR in Premature Neonatal Rats. BMC Pediatr. 2017, 17, 1. [Google Scholar] [CrossRef]

| Study | Item 1 | Item 2 | Item 3 | Item 4 | Item 5 | Item 6 | Item 7 | Item 8 | Item 9 | Item 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Takamura, et al., 2011 [11] | No | Yes | Yes | Yes | Yes | Yes | No | Yes | No | Unclear |
| Ozçam et al., 2019 [14] | No | Yes | Yes | Yes | Unclear | Unclear | No | No | Unclear | Yes |
| Zelante et al., 2013 [16] | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Unclear | Unclear |
| Fukumoto et al., 2014 [17] | No | Yes | Yes | Unclear | Yes | Unclear | No | Unclear | Yes | No |
| Hou et al., 2018 [19] | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Meng et al., 2020 [27] | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Cui et al., 2023 [23] | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Unclear | Yes |
| Park et al., 2023 [24] | No | Yes | Yes | No | Yes | Yes | No | Yes | Yes | Yes |
| Wang et al., 2023 [28] | No | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Study | Item 1 | Item 2 | Item 3 | Item 4 | Item 5 | Item 6 | Item 7 | Item 8 | Item 9 | Item 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Takamura et al., 2011 [11] | No | No | No | No | No | No | No | Unclear | No | Unclear |
| Liu et al., 2019 [13] | No | Yes | Yes | Yes | Yes | Yes | No | Unclear | Yes | Yes |
| Xie Z et al., 2022 [15] | No | Yes | Yes | Yes | Yes | Yes | No | No | Unclear | Yes |
| Zelante et al., 2013 [16] | No | Yes | Yes | Yes | Yes | No | No | No | Yes | Yes |
| Fukumoto et al., 2014 [17] | No | Unclear | Unclear | Unclear | Yes | Unclear | No | No | Yes | No |
| Lamas et al., 2016 [18] | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes |
| Hou et al., 2018 [19] | No | Yes | Yes | Yes | Yes | Unclear | No | Yes | Yes | Yes |
| Lamas et al., 2020 [26] | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No |
| Gu et al., 2021 [20] | No | Yes | Yes | Yes | Yes | Unclear | No | Unclear | Yes | Yes |
| Li et al., 2021 [21] | No | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Cui et al., 2022 [22] | No | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Cui et al., 2023 [23] | No | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Park et al., 2023 [24] | No | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Xia et al., 2023 [25] | No | Yes | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Study | Probiotic | Sample/Model | Main Results |
|---|---|---|---|
| Takamura et al., 2011 [11] |
| Human Caco2 cells Colon from female C57BL/6 mice Age = 4 to 6 weeks old |
|
| Burton et al., 2018 [12] |
| Serum, plasma, and whole blood collected postprandially from young male patients Age = 24.6 ± 4.7 years |
|
| Liu et al., 2019 [13] | Lactobacillus reuteri DSM 17938 (107 CFU/day) | Plasma and fecal samples from female and male C57BL/6J mice Age = 8 days to 2 weeks |
|
| Ozçam et al., 2019 [14] | Lactobacillus reuteri R2lc and 2010 | Murine hepatoma cell line H1L6.1c3 |
|
| Xie Z et al., 2022 [15] | Lactobacillus acidophilus and Bacillus subtilis (106 CFU/g) | Colon and colonic contents from piglets Age = 28 days |
|
| Study | Pathology | Probiotic | Sample/Model | Main Results |
|---|---|---|---|---|
| Zelante et al., 2013 [16] | Colitis | Lactobacillus reuteri (108 CFU) | Stomachs from C57BL/6 Colon and colonic NKp46+ cells from C57BL/6 and AhR−/− mice with DSS-induced colitis Age = 8 to 10 weeks old |
|
| Fukumoto et al., 2014 [17] | Colitis | DHNA (derived of Propionibacterium freudenreichii ET-3 strain) | Human Caco2 cells Small intestine and large intestine from male C57BL/6, AhR−/− and DSS-induced colitis mice Age = 6 to 8 weeks old |
|
| Lamas et al., 2016 [18] | Colitis | Lactobacillus. murinus CNCM I-5020 Lactobacillus. reuteri CNCM I-5022 Lactobacillus taiwanensis CNCM I-5019 | Colon from male C57BL/6J mice and Card9−/− with DSS-induced colitis Age = 8 weeks old |
|
| Hou et al., 2018 [19] | Colitis | Lactobacillus reuteri D8 | Jejunum and colon from C57BL/6 mice with DSS-induced colitis Age = 4 weeks old Co-cultured system of mouse intestinal organoids with lamina propria lymphocytes from small intestine |
|
| Lamas et al., 2020 [26] | Celiac disease | Lactobacillus reuteri CNCM-I5022 and CNCM-I5429 | Duodenum, feces, and plasma from male and female gluten-treated NOD/DQ8 mice Age = 8 to 12 weeks old |
|
| Meng et al., 2020 [27] | Necrotizing enterocolitis (NEC) | Bifidobacterium longum subsp. Infantis (B. infantis) | H4 cells Enterocytes with NEC from the viable margins of resected ileal NEC tissues from a NEC neonate at 25-week gestation Human immature intestinal organoids from gestational age 15 and 22 weeks are therapeutically aborted |
|
| Gu et al., 2021 [20] | Colitis | Akkermansia muciniphila (Akk) | Plasma and colon of male C57BL/6J mice with DSS-induced colitis Age = 6–8 weeks old |
|
| Li et al., 2021 [21] | Colitis | Bacteroides thetaiotaomicron (B. thetaiotaomicron) | Colon from male KM mice Age = 6–8 weeks old Weight= 20 ± 2 g |
|
| Cui et al., 2022 [22] | Colitis | Bifidobacterium bifidum FL-276.1 and FL-228.1 | Colon from C57BL/6N male mice with DSS-induced colitis Age = 4 weeks old |
|
| Cui et al., 2023 [23] | Colitis | Bifidobacterium bifidum FL-276.1 and FL-228.1 | Caco2 cells exposed to lipopolysaccharide (LPS) Serum and colon of male C57BL/6N mice with DSS-induced colitis Age = 4 weeks old |
|
| Park et al., 2023 [24] | Colitis | Bifidobacterium breve CBT BR3 (B. breve) | Colon of C57BL/6 male mice with DSS or dinitrobenzene sulfonic acid (DNBS)-induced colitis Age = 8 weeks old Caco2 and HT29-LuciaTM cells exposed to TNF-α and H2O2 |
|
| Wang et al., 2023 [28] | Intestinal barrier damage | Lactiplantibacillus plantarum (L. plantarum) DPUL-S164 Isolated from human feces. | HT-29 cells with LPS-induced barrier damage |
|
| Xia et al., 2023 [25] | Colitis | Lactobacillus acidophilus | Colon of mice with DSS-induced colitis divided into two groups: Cesarean section (CS); N = 6 Vaginal delivery (VD); N = 6 Age = 6–8 weeks old |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De la Rosa González, A.; Guerra-Ojeda, S.; Camacho-Villa, M.A.; Valls, A.; Alegre, E.; Quintero-Bernal, R.; Martorell, P.; Chenoll, E.; Serna-García, M.; Mauricio, M.D.; et al. Effect of Probiotics on Gastrointestinal Health Through the Aryl Hydrocarbon Receptor Pathway: A Systematic Review. Foods 2024, 13, 3479. https://doi.org/10.3390/foods13213479
De la Rosa González A, Guerra-Ojeda S, Camacho-Villa MA, Valls A, Alegre E, Quintero-Bernal R, Martorell P, Chenoll E, Serna-García M, Mauricio MD, et al. Effect of Probiotics on Gastrointestinal Health Through the Aryl Hydrocarbon Receptor Pathway: A Systematic Review. Foods. 2024; 13(21):3479. https://doi.org/10.3390/foods13213479
Chicago/Turabian StyleDe la Rosa González, Adrián, Sol Guerra-Ojeda, María Alejandra Camacho-Villa, Alicia Valls, Eva Alegre, Ronald Quintero-Bernal, Patricia Martorell, Empar Chenoll, Marta Serna-García, Maria D. Mauricio, and et al. 2024. "Effect of Probiotics on Gastrointestinal Health Through the Aryl Hydrocarbon Receptor Pathway: A Systematic Review" Foods 13, no. 21: 3479. https://doi.org/10.3390/foods13213479
APA StyleDe la Rosa González, A., Guerra-Ojeda, S., Camacho-Villa, M. A., Valls, A., Alegre, E., Quintero-Bernal, R., Martorell, P., Chenoll, E., Serna-García, M., Mauricio, M. D., & Serna, E. (2024). Effect of Probiotics on Gastrointestinal Health Through the Aryl Hydrocarbon Receptor Pathway: A Systematic Review. Foods, 13(21), 3479. https://doi.org/10.3390/foods13213479








