Unraveling Whole-Genome Sequence and Functional Characterization of P. megaterium PH3
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Kirby–Bauer Disk Diffusion Tests
2.3. DNA Extraction and Whole-Genome Sequencing
2.4. Genome Assembly and Optimization
2.5. Genome Assembly Genome Prediction
2.6. Annotation Strategy and Database Utilization
2.7. Analysis of Metabolic System
2.8. Analysis of Pathogenicity
2.9. Safety Evaluation
2.9.1. Experimental Analysis of Hemolysis
2.9.2. Experimental Analysis of Indigo Matrix
2.9.3. Amino Acid Decarboxylase Experiment
2.10. Experimental Analysis of Toxicology
2.10.1. Animal Experimental Design
2.10.2. Weight Measurement and Behavior Observation
2.10.3. Determination of Visceral Weight Coefficient
2.10.4. Pathological Section Analysis
2.10.5. Analysis of Serum Oxidative Stress
2.11. Statistical Analysis
3. Results and Discussion
3.1. P. megaterium PH3 Whole-Genome Analysis
3.1.1. Genome Evaluation
3.1.2. Genome Loading and Prediction
3.1.3. Genetic Annotations
3.2. Analysis of the Metabolic System of P. megaterium PH3 Genome
3.2.1. Annotated Analysis of CAZy
3.2.2. Analysis of Secondary Metabolite Synthetic Gene Clusters (DGCs)
3.3. Pathogenic System Analysis of P. megaterium PH3 Genome
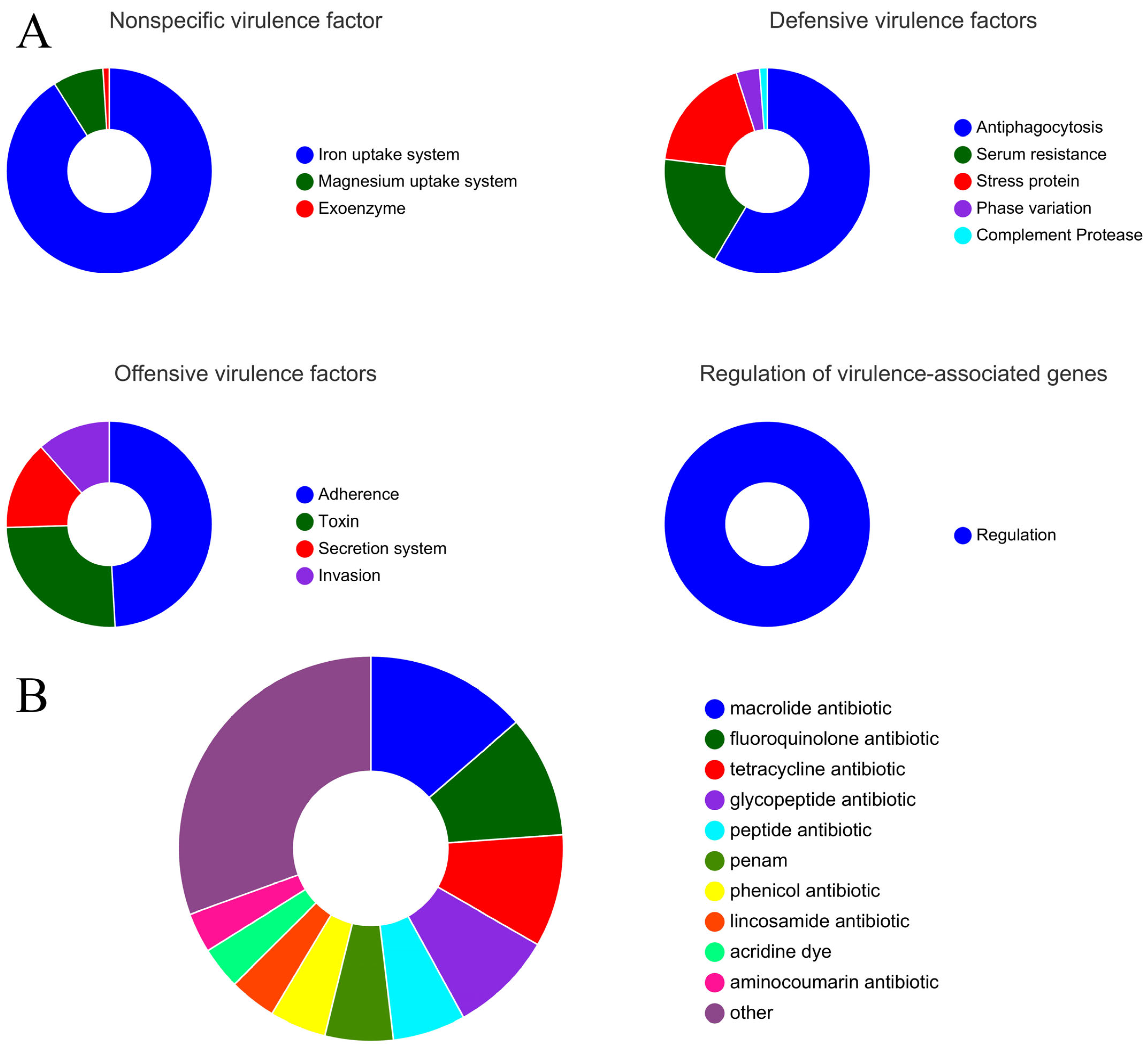
3.3.1. Prediction of Virulence Genes
3.3.2. Prediction of Drug-Resistance Genes
3.4. Internal Cell Regulation and Functional Protein Analysis
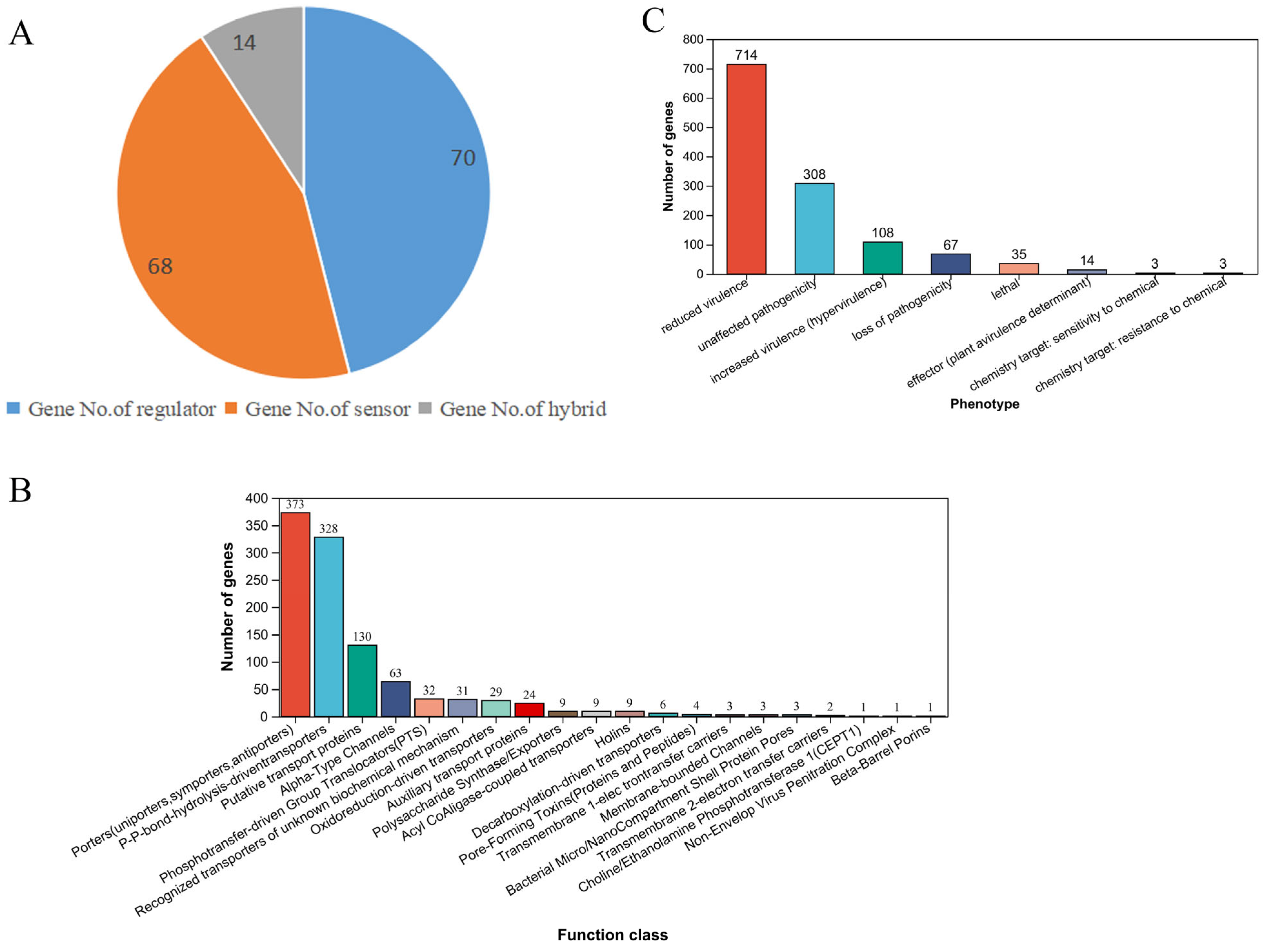
3.4.1. Analysis of a Two-Component Regulation System
3.4.2. Analysis of Transporter Protein
3.4.3. Analysis of the Interaction Between Pathogenic Microorganisms and Hosts
3.5. Physiological Characteristics and Drug-Sensitivity Analysis of P. megaterium PH3
3.5.1. Research on Hemolytic Characteristics
3.5.2. Experimental Detection of Indole
3.5.3. Detection of the Ability to Produce Bioamines
3.5.4. Research on Drug Sensitivity
3.6. Toxicological Analysis of P. megaterium PH3
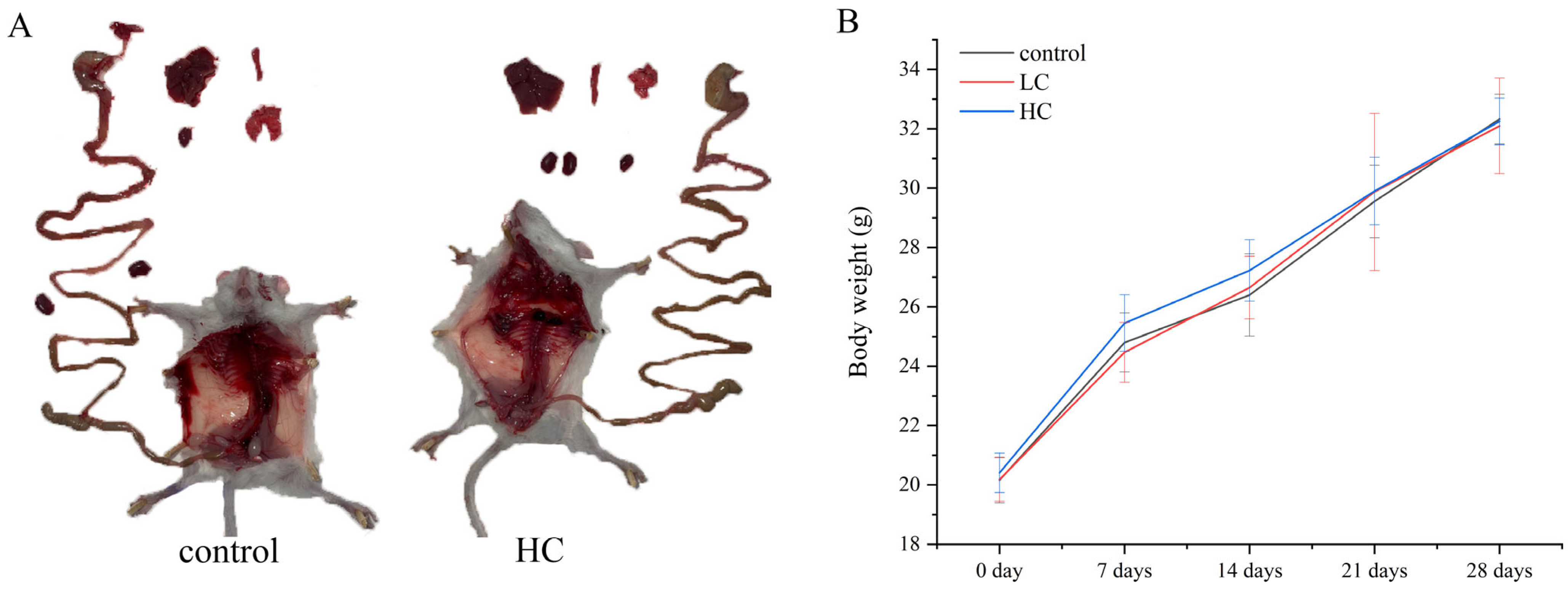
3.6.1. The Effect of P. megaterium PH3 on the Basic Indicators of Mice
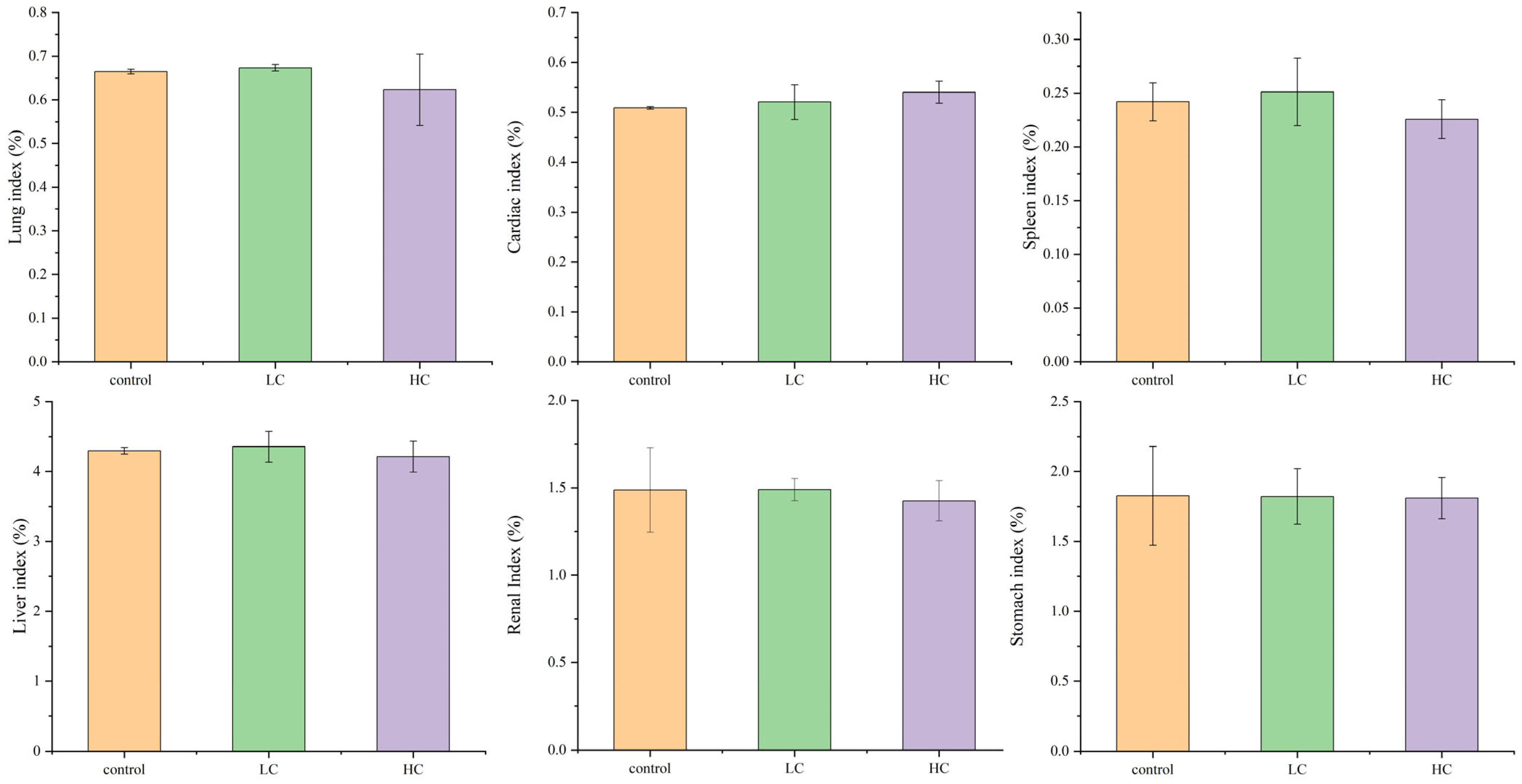
3.6.2. The Effect of P. megaterium PH3 on the Organ Index of Mice
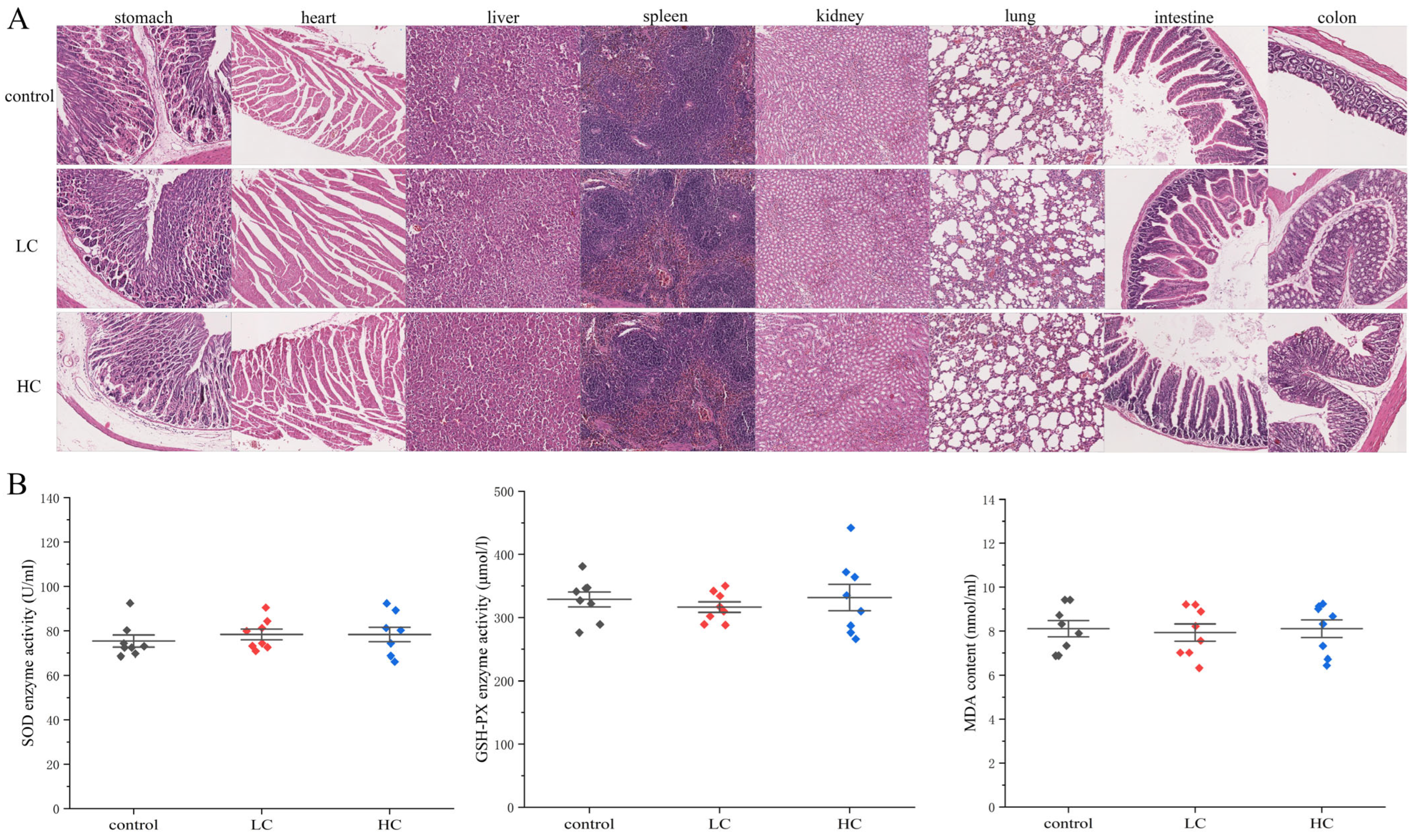
3.6.3. The Effect of P. megaterium PH3 on Organ Lesions in Mice
3.6.4. The Effect of P. megaterium PH3 on the Oxidative Stress Level of Mice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harirchi, S.; Sar, T.; Ramezani, M.; Aliyu, H.; Etemadifar, Z.; Nojoumi, S.A.; Yazdian, F.; Awasthi, M.K.; Taherzadeh, M.J. Bacillales: From Taxonomy to Biotechnological and Industrial Perspectives. Microorganisms 2022, 10, 2355. [Google Scholar] [CrossRef] [PubMed]
- Vary, P.S.; Biedendieck, R.; Fuerch, T.; Meinhardt, F.; Rohde, M.; Deckwer, W.-D.; Jahn, D. Bacillus Megaterium—From Simple Soil Bacterium to Industrial Protein Production Host. Appl. Microbiol. Biotechnol. 2007, 76, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Bacon, C.W.; Palencia, E.R.; Hinton, D.M. Abiotic and Biotic Plant Stress-Tolerant and Beneficial Secondary Metabolites Produced by Endophytic Bacillus Species. In Plant Microbes Symbiosis: Applied Facets; Springer: New Delhi, India, 2015; pp. 163–177. [Google Scholar] [CrossRef]
- Eppinger, M.; Bunk, B.; Johns, M.A.; Edirisinghe, J.N.; Kutumbaka, K.K.; Koenig, S.S.K.; Huot Creasy, H.; Rosovitz, M.J.; Riley, D.R.; Daugherty, S.; et al. Genome Sequences of the Biotechnologically Important Bacillus Megaterium Strains QM B1551 and DSM319. J. Bacteriol. 2011, 193, 4199–4213. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, I.D.A.; Bach, E.; da Silva Moreira, F.; Müller, A.R.; Rangel, C.P.; Wilhelm, C.M.; Barth, A.L.; Passaglia, L.M.P. Antifungal potential against Sclerotinia sclerotiorum (Lib.) de Bary and plant growth promoting abilities of Bacillus isolates from canola (Brassica napus L.) roots. Microbiol. Res. 2021, 248, 126754. [Google Scholar] [CrossRef]
- Shwed, P.S.; Crosthwait, J.; Weedmark, K.; Hoover, E.; Dussault, F. Complete genome sequences of Priestia megaterium type and clinical strains feature complex plasmid arrays. Microbiol. Resour. Announc. 2021, 10, 10–1128. [Google Scholar] [CrossRef]
- Krause, A.; Ramakumar, A.; Bartels, D.; Battistoni, F.; Bekel, T.; Boch, J.; Böhm, M.; Friedrich, F.; Hurek, T.; Krause, L.; et al. Complete Genome of the Mutualistic, N2-Fixing Grass Endophyte Azoarcus Sp. Strain BH72. Nat. Biotechnol. 2006, 24, 1384–1390. [Google Scholar] [CrossRef]
- Card, S.; Johnson, L.; Teasdale, S.; Caradus, J. Deciphering Endophyte Behaviour: The Link between Endophyte Biology and Efficacious Biological Control Agents. FEMS Microbiol. Ecol. 2016, 92, fiw114. [Google Scholar] [CrossRef]
- Tan, H.; Yu, Z.; Wang, C.; Zhang, Q.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Pilot Safety Evaluation of a Novel Strain of Bacteroides Ovatus. Front. Genet. 2018, 9, 539. [Google Scholar] [CrossRef]
- Rydell-Törmänen, K.; Johnson, J.R. The Applicability of Mouse Models to the Study of Human Disease. In Mouse Cell Culture: Methods and Protocols; Humana Press: New York, NY, USA, 2019; Volume 1940, pp. 3–22. [Google Scholar] [CrossRef]
- Longjam, L.A.; Tsering, D.C.; Das, D. A microbiological study of Acinetobacter calcoaceticus baumannii with special reference to multidrug resistance. J. Lab. Phys. 2022, 14, 169–174. [Google Scholar] [CrossRef]
- Delgado, E. Salmonella spp. Antibiotic Susceptibility Testing by the Kirby-Bauer Disk Diffusion Method, v1. 2021. Available online: https://www.protocols.io/view/salmonella-spp-antibiotic-susceptibility-testing-b-n92ld9p1og5b/v1 (accessed on 23 October 2023).
- M100-S25; Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Eighth Informational Supplement. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019.
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. GigaSci 2012, 1, 18. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Borodovsky, M. GeneMark: Web Software for Gene Finding in Prokaryotes, Eukaryotes and Viruses. Nucleic Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences; Springer: New York, NY, USA, 2019; Volume 1962, pp. 1–14. ISBN 978-1-4939-9172-3. [Google Scholar]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Darkoh, C.; Chappell, C.; Gonzales, C.; Okhuysen, P. A rapid and specific method for the detection of indole in complex biological samples. Appl. Environ. Microbiol. 2015, 81, 8093–8097. [Google Scholar] [CrossRef]
- Li, B.; Zhan, M.; Evivie, S.E.; Jin, D.; Zhao, L.; Chowdhury, S.; Sarker, S.K.; Huo, G.; Liu, F. Evaluating the Safety of Potential Probiotic Enterococcus Durans KLDS6.0930 Using Whole Genome Sequencing and Oral Toxicity Study. Front. Microbiol. 2018, 9, 1943. [Google Scholar] [CrossRef]
- Mete, A.; Coşansu, S.; Demirkol, O.; Ayhan, K. Amino acid decarboxylase activities and biogenic amine formation abilities of lactic acid bacteria isolated from shalgam. Int. J. Food Prop. 2017, 20, 171–178. [Google Scholar] [CrossRef]
- Park, H.; Lee, M.; Jeong, D.; Park, S.; Ji, Y.; Todorov, S.D.; Holzapfel, W.H. Safety Evaluation and In Vivo Strain-Specific Functionality of Bacillus Strains Isolated from Korean Traditional Fermented Foods. Probiotics Antimicro. Prot. 2021, 13, 60–71. [Google Scholar] [CrossRef]
- Wang, A.; Li, P.; Ma, F.; Li, X.; Mu, G.; Tuo, Y. Mixed Lactiplantibacillus Plantarum Strains Alleviated DSS-Induced Intestinal Inflammation of Balb/c Mice via the 5-HT/5-HT7R/NF-κB Signaling Pathway. J. Funct. Foods 2023, 102, 105435. [Google Scholar] [CrossRef]
- Sahnoun, R.; Mnif, I.; Fetoui, H.; Gdoura, R.; Chaabouni, K.; Makni-Ayadi, F.; Kallel, C.; Ellouze-Chaabouni, S.; Ghribi, D. Evaluation of Bacillus Subtilis SPB1 Lipopeptide Biosurfactant Toxicity Towards Mice. Int. J. Pept. Res. Ther. 2014, 20, 333–340. [Google Scholar] [CrossRef]
- Yeh, Y.-H.; Hsieh, Y.-L.; Lee, Y.-T. Effects of Yam Peel Extract against Carbon Tetrachloride-Induced Hepatotoxicity in Rats. J. Agric. Food Chem. 2013, 61, 7387–7396. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zhou, T.; Wilke, C.O. A Universal Trend of Reduced mRNA Stability near the Translation-Initiation Site in Prokaryotes and Eukaryotes. PLoS Comput. Biol. 2010, 6, e1000664. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.F.; Dalgaard, M.D.; Bahl, M.I. Genomic GC-Content Affects the Accuracy of 16S rRNA Gene Sequencing Based Microbial Profiling Due to PCR Bias. Front. Microbiol. 2017, 8, 1934. [Google Scholar] [CrossRef] [PubMed]
- Plaza Onate, F.; Batto, J.-M.; Juste, C.; Fadlallah, J.; Fougeroux, C.; Gouas, D.; Pons, N.; Kennedy, S.; Levenez, F.; Dore, J.; et al. Quality Control of Microbiota Metagenomics by K-Mer Analysis. BMC Genom. 2015, 16, 183. [Google Scholar] [CrossRef]
- Fransz, P.; De Jong, H. From Nucleosome to Chromosome: A Dynamic Organization of Genetic Information. Plant J. 2011, 66, 4–17. [Google Scholar] [CrossRef]
- Medema, M.H.; Trefzer, A.; Kovalchuk, A.; Van Den Berg, M.; Müller, U.; Heijne, W.; Wu, L.; Alam, M.T.; Ronning, C.M.; Nierman, W.C.; et al. The Sequence of a 1.8-Mb Bacterial Linear Plasmid Reveals a Rich Evolutionary Reservoir of Secondary Metabolic Pathways. Genome Biol. Evol. 2010, 2, 212–224. [Google Scholar] [CrossRef]
- Rodríguez-Beltrán, J.; DelaFuente, J.; León-Sampedro, R.; MacLean, R.C.; San Millán, Á. Beyond Horizontal Gene Transfer: The Role of Plasmids in Bacterial Evolution. Nat. Rev. Microbiol. 2021, 19, 347–359. [Google Scholar] [CrossRef]
- Hoff, K.J.A.; Borodovsky, M.; Stanke, M. Whole-Genome Annotation with BRAKER. In Gene Prediction: Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 65–95. ISBN 978-1-4939-9172-3. [Google Scholar]
- Stein, L. Genome Annotation: From Sequence to Biology. Nat. Rev. Genet. 2001, 2, 493–503. [Google Scholar] [CrossRef]
- Wei, B.; Du, A.; Zhou, Z.; Lai, C.; Yu, W.; Yu, J.; Yu, Y.; Chen, J.; Zhang, H.; Xu, X.; et al. An Atlas of Bacterial Secondary Metabolite Biosynthesis Gene Clusters. Environ. Microbiol. 2021, 23, 6981–6992. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Henrissat, B.; Davies, G.J. Glycoside Hydrolases and Glycosyltransferases. Families, Modules, and Implications for Genomics. Plant Physiol. 2000, 124, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the Enzymatic Repertoire of the CAZy Database to Integrate Auxiliary Redox Enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liang, J.; Chen, H.; Ding, G.; Ma, B.; He, N. Evolutionary and Functional Analysis of Mulberry Type III Polyketide Synthases. BMC Genom. 2016, 17, 540. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-W.; Guo, H.-L.; Li, X.; Huang, L.-L.; Zhang, B.-N.; Pang, X.-B.; Liu, B.-Y.; Ma, L.-Q.; Wang, H. Two Type III Polyketide Synthases from Polygonum Cuspidatum: Gene Structure, Evolutionary Route and Metabolites. Plant Biotechnol. Rep. 2013, 7, 371–381. [Google Scholar] [CrossRef]
- Austin, M.B.; Noel, J.P. The Chalcone Synthase Superfamily of Type III Polyketide Synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef]
- Schröder, G.; Brown, J.W.; Schröder, J. Molecular Analysis of Resveratrol Synthase. cDNA, Genomic Clones and Relationship with Chalcone Synthase. Eur. J. Biochem. 1988, 172, 161–169. [Google Scholar] [CrossRef]
- Austin, M.B.; Bowman, M.E.; Ferrer, J.-L.; Schröder, J.; Noel, J.P. An Aldol Switch Discovered in Stilbene Synthases Mediates Cyclization Specificity of Type III Polyketide Synthases. Chem. Biol. 2004, 11, 1179–1194. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Yan, Y. Bioproduction of Resveratrol. In Biotechnology of Natural Products; Schwab, W., Lange, B.M., Wüst, M., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 61–79. ISBN 978-3-319-67902-0. [Google Scholar]
- Lu, Y.; Ye, C.; Che, J.; Xu, X.; Shao, D.; Jiang, C.; Liu, Y.; Shi, J. Genomic Sequencing, Genome-Scale Metabolic Network Reconstruction, and in Silico Flux Analysis of the Grape Endophytic Fungus Alternaria Sp. MG1. Microb. Cell Fact. 2019, 18, 13. [Google Scholar] [CrossRef]
- Shmuleviz, R.; Amato, A.; Commisso, M.; D’Incà, E.; Luzzini, G.; Ugliano, M.; Fasoli, M.; Zenoni, S.; Tornielli, G.B. Temperature Affects Organic Acid, Terpene and Stilbene Metabolisms in Wine Grapes during Postharvest Dehydration. Front. Plant Sci. 2023, 14, 1107954. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Y.; Li, X.; Wang, X.; Li, H. Detection of Antibiotic Resistance, Virulence Gene, and Drug Resistance Gene of Staphylococcus Aureus Isolates from Bovine Mastitis. Microbiol. Spectr. 2022, 10, e00471-22. [Google Scholar] [CrossRef]
- Venkateswaran, P.; Vasudevan, S.; David, H.; Shaktivel, A.; Shanmugam, K.; Neelakantan, P.; Solomon, A.P. Revisiting ESKAPE Pathogens: Virulence, Resistance, and Combating Strategies Focusing on Quorum Sensing. Front. Cell. Infect. Microbiol. 2023, 13, 1159798. [Google Scholar] [CrossRef] [PubMed]
- Malik, B.; Bhattacharyya, S. Antibiotic Drug-Resistance as a Complex System Driven by Socio-Economic Growth and Antibiotic Misuse. Sci. Rep. 2019, 9, 9788. [Google Scholar] [CrossRef]
- Mitrophanov, A.Y.; Groisman, E.A. Signal Integration in Bacterial Two-Component Regulatory Systems. Genes. Dev. 2008, 22, 2601–2611. [Google Scholar] [CrossRef]
- Martín, J.F.; Van Den Berg, M.A.; Ver Loren Van Themaat, E.; Liras, P. Sensing and Transduction of Nutritional and Chemical Signals in Filamentous Fungi: Impact on Cell Development and Secondary Metabolites Biosynthesis. Biotechnol. Adv. 2019, 37, 107392. [Google Scholar] [CrossRef]
- Akhtar, A.A.; Turner, D.P. The role of bacterial ATP-binding cassette (ABC) transporters in pathogenesis and virulence: Therapeutic and vaccine potential. Microb. Pathog. 2022, 171, 105734. [Google Scholar] [CrossRef]
- Méndez, C.; Salas, J.A. The role of ABC transporters in antibiotic-producing organisms: Drug secretion and resistance mechanisms. Res. Microbiol. 2001, 1152, 341–350. [Google Scholar] [CrossRef]
- Zafar, H.; Saier, M.H., Jr. Comparative genomics of the transport proteins of ten Lactobacillus strains. Genes 2020, 11, 1234. [Google Scholar] [CrossRef]
- Barzana, G.; Rios, J.J.; Lopez-Zaplana, A.; Nicolas-Espinosa, J.; Yepes-Molina, L.; Garcia-Ibañez, P.; Carvajal, M. Interrelations of Nutrient and Water Transporters in Plants under Abiotic Stress. Physiol. Plant. 2021, 171, 595–619. [Google Scholar] [CrossRef]
- Steinert, M.; Hentschel, U.; Hacker, J. Symbiosis and Pathogenesis: Evolution of the Microbe-Host Interaction. Naturwissenschaften 2000, 87, 1–11. [Google Scholar] [CrossRef]
- Meena, M.; Swapnil, P.; Zehra, A.; Aamir, M.; Dubey, M.K.; Patel, C.B.; Upadhyay, R.S. Virulence Factors and Their Associated Genes in Microbes. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2019; pp. 181–208. [Google Scholar] [CrossRef]
- Minasyan, H. Sepsis: Mechanisms of bacterial injury to the patient. Scand. J. Trauma Resusc. Emerg. Med. 2019, 27, 19. [Google Scholar] [CrossRef]
- Huang, B.; Liang, Y.; Pan, H.; Xie, L.; Jiang, T.; Jiang, T. Hemolytic and Cytotoxic Activity from Cultures of Aureococcus Anophagefferens—a Causative Species of Brown Tides in the North-Western Bohai Sea, China. Chemosphere 2020, 247, 125819. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, M.; Greathouse, K.L. Targeting Dietary and Microbial Tryptophan-Indole Metabolism as Therapeutic Approaches to Colon Cancer. Nutrients 2021, 13, 1189. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Li, J.; Ying, S. Tryptophan Metabolism and Gut Microbiota: A Novel Regulatory Axis Integrating the Microbiome, Immunity, and Cancer. Metabolites 2023, 13, 1166. [Google Scholar] [CrossRef] [PubMed]
- Önal, A.; Tekkeli, S.E.; Önal, C. A review of the liquid chromatographic methods for the determination of biogenic amines in foods. Food Chem. 2013, 138, 509–515. [Google Scholar] [CrossRef]
- Jeon, A.R.; Lee, J.H.; Mah, J.H. Biogenic amine formation and bacterial contribution in Cheonggukjang, a Korean traditional fermented soybean food. LWT 2018, 92, 282–289. [Google Scholar] [CrossRef]
- Arellano, K.; Vazquez, J.; Park, H.; Lim, J.; Ji, Y.; Kang, H.-J.; Cho, D.; Jeong, H.W.; Holzapfel, W.H. Safety Evaluation and Whole-Genome Annotation of Lactobacillus Plantarum Strains from Different Sources with Special Focus on Isolates from Green Tea. Probiotics Antimicro. Prot. 2020, 12, 1057–1070. [Google Scholar] [CrossRef]
- Ashaolu, T.J.; Khalifa, I.; Mesak, M.A.; Lorenzo, J.M.; Farag, M.A. A Comprehensive Review of the Role of Microorganisms on Texture Change, Flavor and Biogenic Amines Formation in Fermented Meat with Their Action Mechanisms and Safety. Crit. Rev. Food Sci. Nutr. 2023, 63, 3538–3555. [Google Scholar] [CrossRef]
- Yang, W.; Ji, X. Analysis of the Microbial Species, Antimicrobial Sensitivity and Drug Resistance in 2652 Patients of Nursing Hospital. Heliyon 2020, 6, e03965. [Google Scholar] [CrossRef]
- Jirkof, P. Burrowing and Nest Building Behavior as Indicators of Well-Being in Mice. J. Neurosci. Methods 2014, 234, 139–146. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Xia, K.; Dai, J.; Huang, J.; Wang, Y.; Zhu, G.; Hu, Z.; Zeng, Z.; Jia, Y. Effects of Dietary Selenium on Immune Function of Spleen in Mice. J. Funct. Foods 2022, 89, 104914. [Google Scholar] [CrossRef]
- Wang, Y. Safety Evaluation of a Novel Strain of Bacteroides Fragilis. Front. Microbiol. 2017, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wen, X.; Liu, J.; Kan, J.; Qian, C.; Wu, C.; Jin, C. Protective Effect of an Arabinogalactan from Black Soybean against Carbon Tetrachloride-Induced Acute Liver Injury in Mice. Int. J. Biol. Macromol. 2018, 117, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Leal, C.A.M.; Schetinger, M.R.C.; Leal, D.B.R.; Morsch, V.M.; Da Silva, A.S.; Rezer, J.F.P.; De Bairros, A.V.; Jaques, J.A.D.S. Oxidative Stress and Antioxidant Defenses in Pregnant Women. Redox Rep. 2011, 16, 230–236. [Google Scholar] [CrossRef] [PubMed]
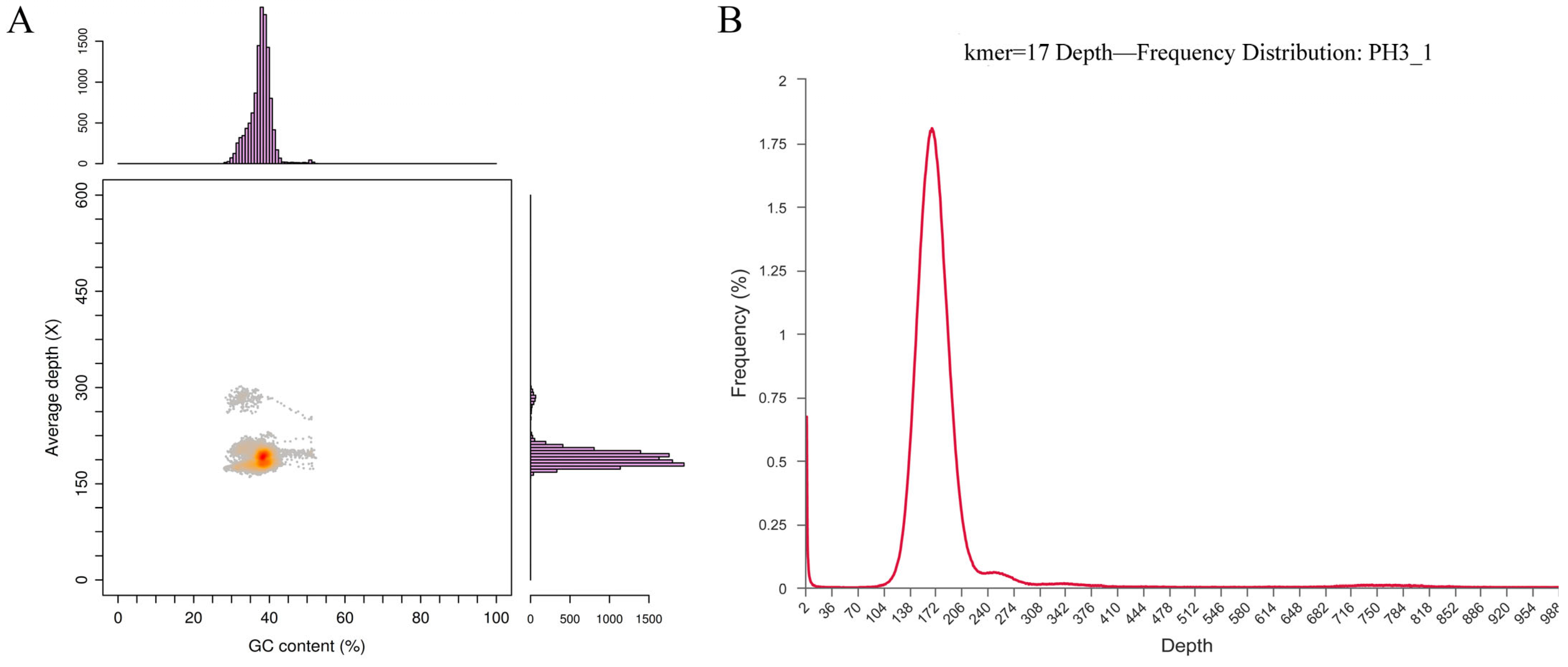
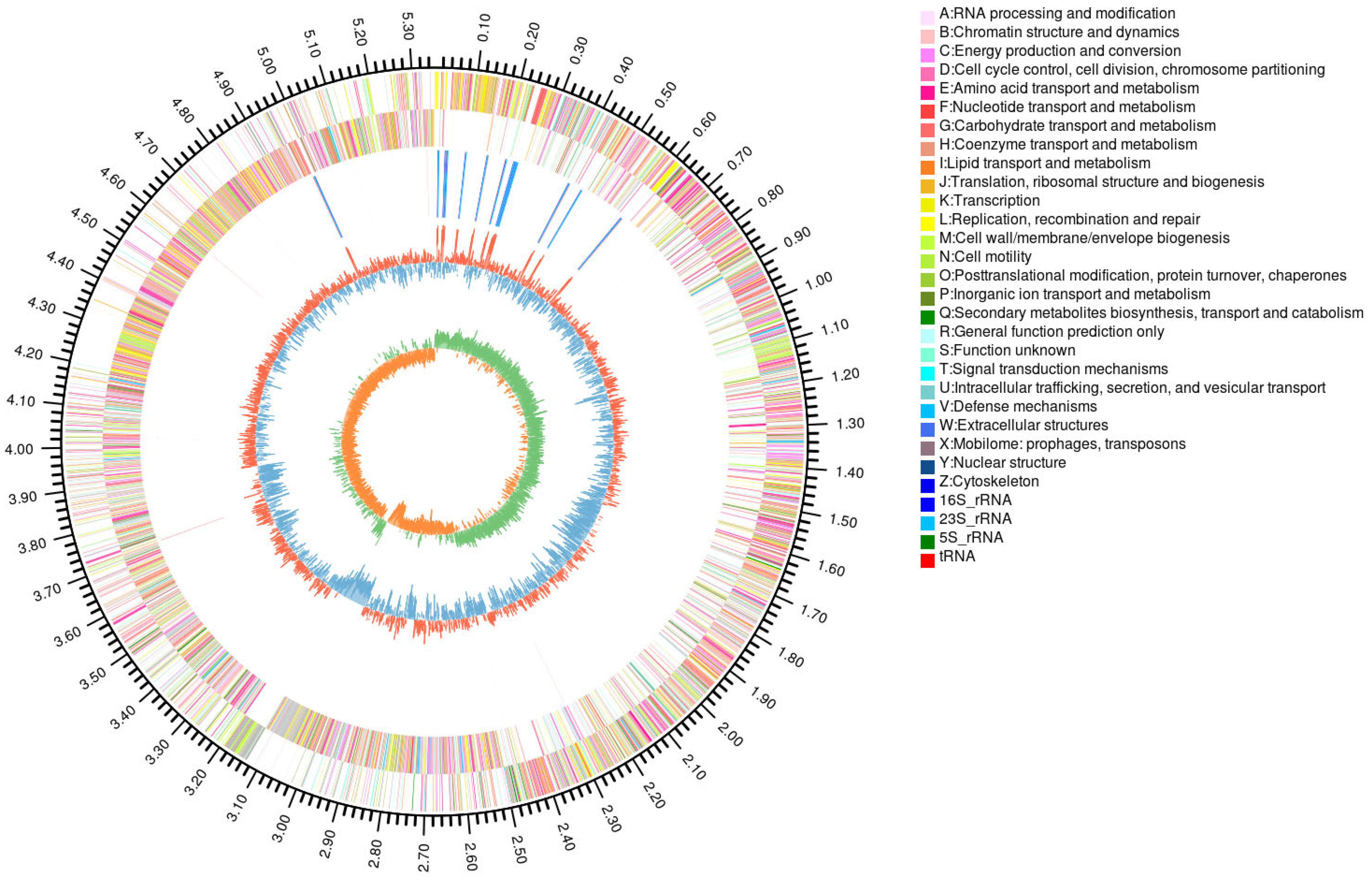
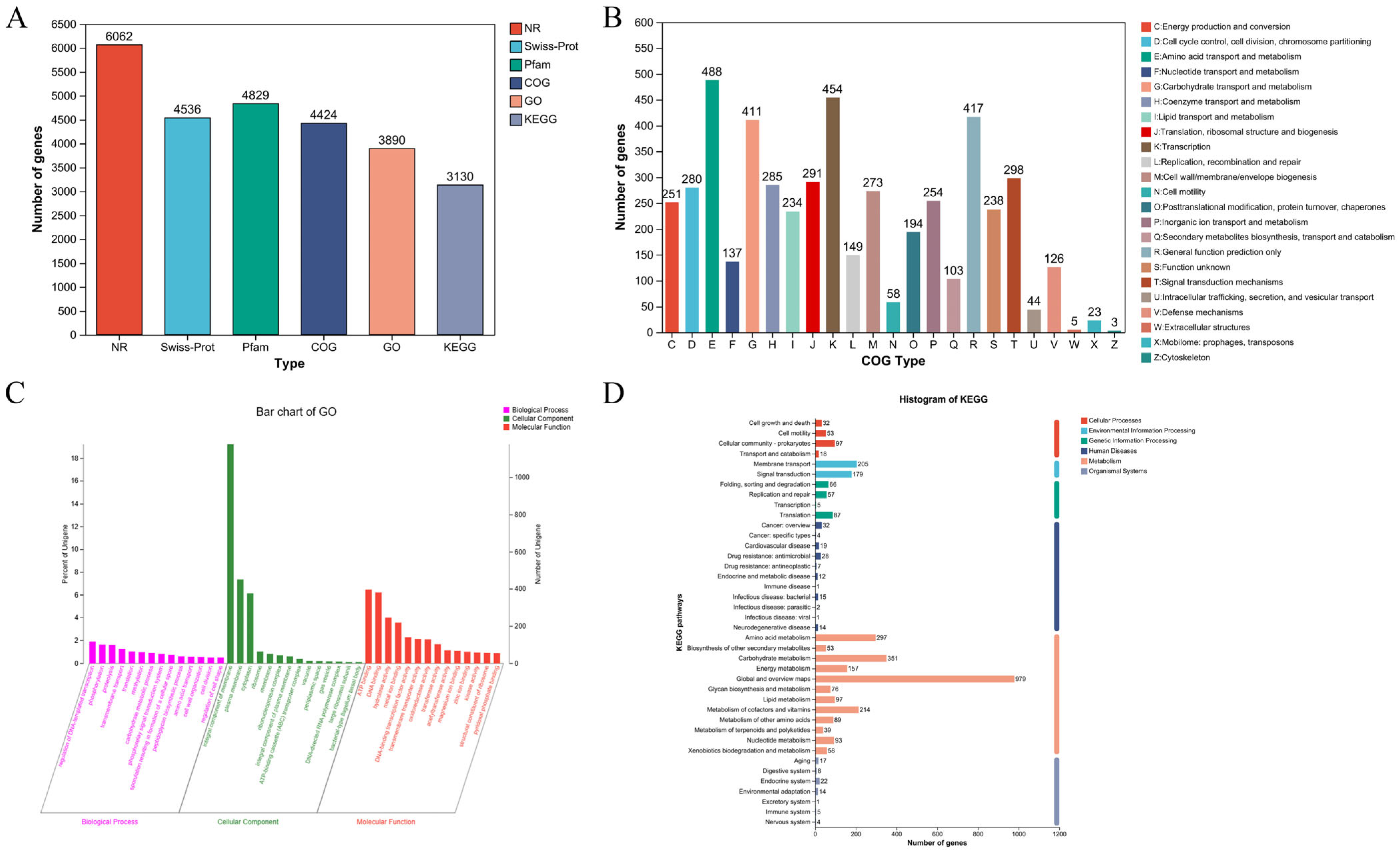
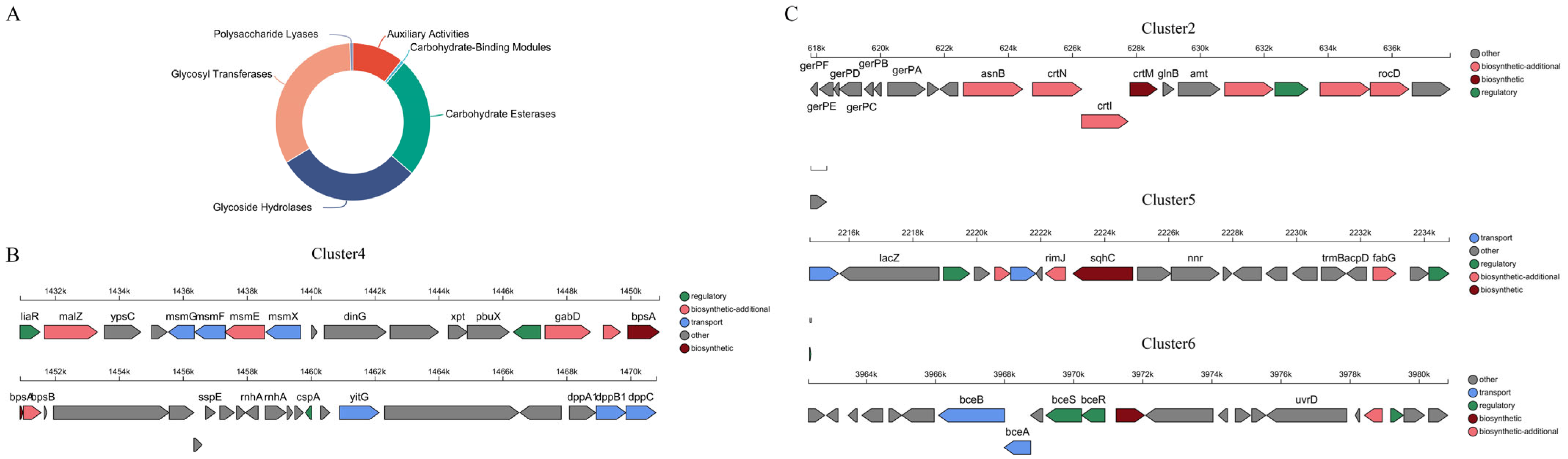

| Parameter | Value |
|---|---|
| Total Bases (bp) | 4,318,106,144 |
| Largest (bp) | 317,278 |
| Chromosome No. | 1 |
| Plasmid No. | 7 |
| Genome Size (bp) | 5,960,365 |
| G + C (%) | 37.62 |
| Reads N50 (bp) | 11,391 |
| Type | Parameter | Value |
|---|---|---|
| Gene | Gene Total Len (bp) | 4,877,301 |
| GC Content in Gene Region (%) | 38.59 | |
| Gene/Genome (%) | 81.83 | |
| Gene Average Len (bp) | 795.39 | |
| tRNAs | tRNAs | 140 |
| rRNAs | 16S rRNA | 15 |
| 23S rRNA | 15 | |
| 5S rRNA | 17 | |
| sRNA | sRNA No | 115 |
| Total Len (bp) | 17,949 | |
| In Genome (%) | 0.3011 | |
| Methylation modification | M4C; modified_base; m4C; m6A | 4 |
| Cluster ID | Location | Type | Gene No. | MIBiG Accession |
|---|---|---|---|---|
| Cluster1 | PlasmidB | lanthipeptide-class-i | 25 | - |
| Cluster2 | Chromosome | terpene | 20 | carotenoid |
| Cluster3 | Chromosome | phosphonate | 35 | - |
| Cluster4 | Chromosome | T3PKS | 37 | - |
| Cluster5 | Chromosome | terpene | 20 | - |
| Cluster6 | Chromosome | terpene | 22 | surfactin |
| Cluster7 | Chromosome | siderophore | 15 | - |
| Parameters | Gene Information | |
|---|---|---|
| Description Info | Gene Name | lsa |
| Location | Plasmid E: complement (26,583–28,061) | |
| Description | Lsa family ABC-F type ribosomal protection protein | |
| Name | Drug Content (μg) | Diameter of Judgement Criteria (mm) | Diameter (mm) | Grade | ||
|---|---|---|---|---|---|---|
| R | I | S | ||||
| Penicillin (PEN) | 10 | ≤28 | - | ≥29 | 0 | R |
| Chloramphenicol (CLM) | 10 | ≤16 | 18~24 | ≥25 | 20.67 ± 0.31 | I |
| Erythromycin (ERM) | 30 | ≤13 | 14~20 | ≥21 | 4.07 ± 0.01 | R |
| Tetracycline (TET) | 30 | ≤14 | 15~16 | ≥17 | 7.47 ± 0.13 | R |
| Gentamicin (GEN) | 15 | ≤13 | 14~22 | ≥23 | 6.67 ± 0.12 | R |
| Lincomycin (LIM) | 5 | ≤15 | 16~20 | ≥17 | 0 | R |
| Ciprofloxacin (CFX) | 10 | ≤12 | 13~14 | ≥15 | 10.67 ± 0.12 | R |
| Ceftriaxone (CTR) | 23.75 | ≤10 | 11~15 | ≥16 | 12.67 ± 0.12 | I |
| Ampicillin (AMP) | 30 | ≤12 | 13~17 | ≥18 | 0 | R |
| Paediatric Compound Sulfamethoxazole Tablets (T/S) | 2 | ≤13 | 14~17 | ≥18 | 4.67 ± 0.12 | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liang, J.; Zhang, D.; Wang, L.; Ye, S. Unraveling Whole-Genome Sequence and Functional Characterization of P. megaterium PH3. Foods 2024, 13, 3555. https://doi.org/10.3390/foods13223555
Zhang X, Liang J, Zhang D, Wang L, Ye S. Unraveling Whole-Genome Sequence and Functional Characterization of P. megaterium PH3. Foods. 2024; 13(22):3555. https://doi.org/10.3390/foods13223555
Chicago/Turabian StyleZhang, Xiaohan, Junbo Liang, Dong Zhang, Liang Wang, and Shuhong Ye. 2024. "Unraveling Whole-Genome Sequence and Functional Characterization of P. megaterium PH3" Foods 13, no. 22: 3555. https://doi.org/10.3390/foods13223555
APA StyleZhang, X., Liang, J., Zhang, D., Wang, L., & Ye, S. (2024). Unraveling Whole-Genome Sequence and Functional Characterization of P. megaterium PH3. Foods, 13(22), 3555. https://doi.org/10.3390/foods13223555




