Study of the Microbiome of the Cretan Sour Cream Staka Using Amplicon Sequencing and Shotgun Metagenomics and Isolation of Novel Strains with an Important Antimicrobial Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Physicochemical Analyses
2.3. Microbiological Analysis
2.4. Amplicon and Shotgun Metagenomics Sequencing and Analysis
2.5. Typing and Identification of Isolates with Rep-PCR Fingerprinting and 16S rDNA Sequencing
2.6. Species Discrimination by Biochemical Tests
2.7. Antimicrobial Activity of the Isolated Strains
3. Results and Discussion
3.1. Physicochemical Analyses
3.2. Culture-Based Microbiological Analysis
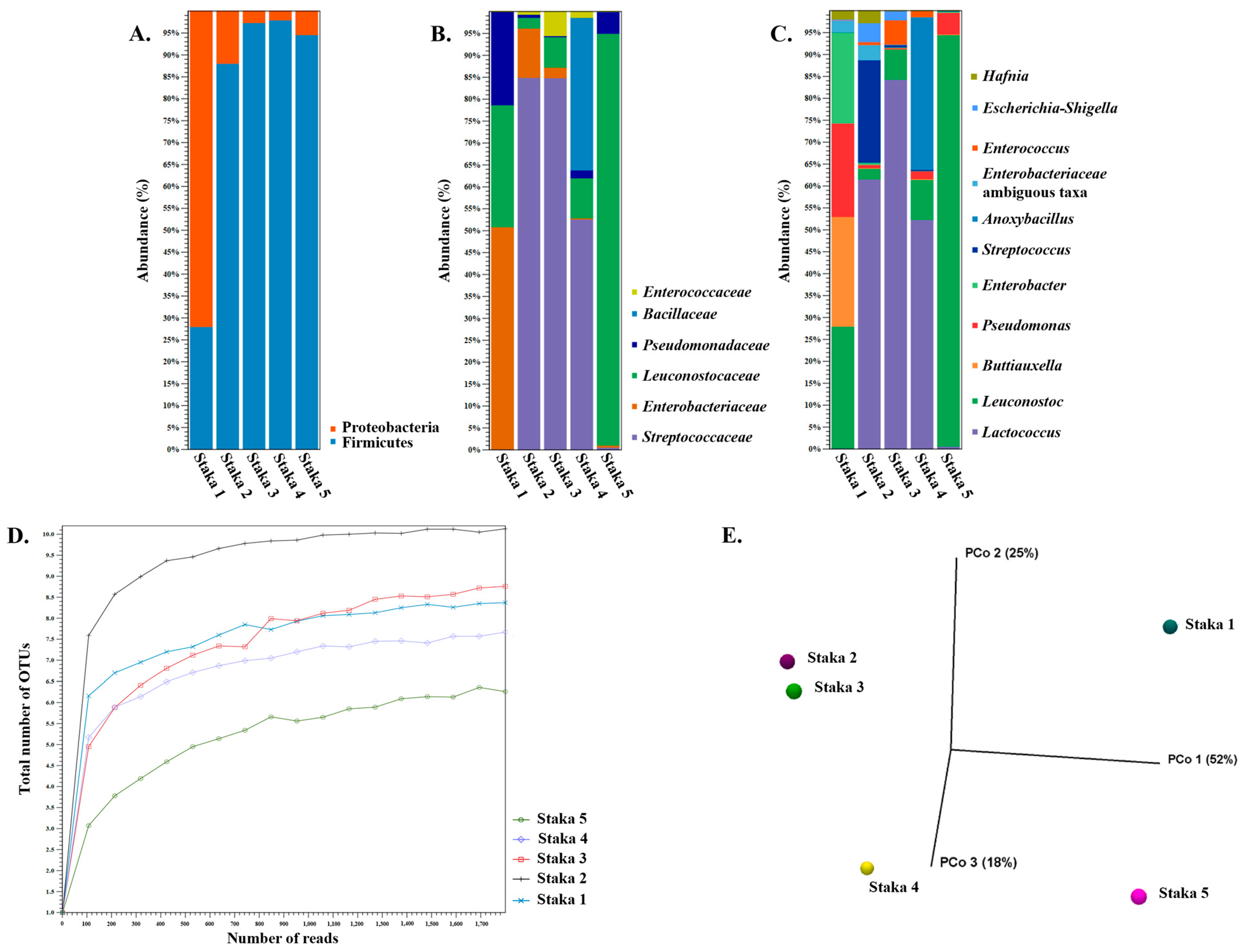
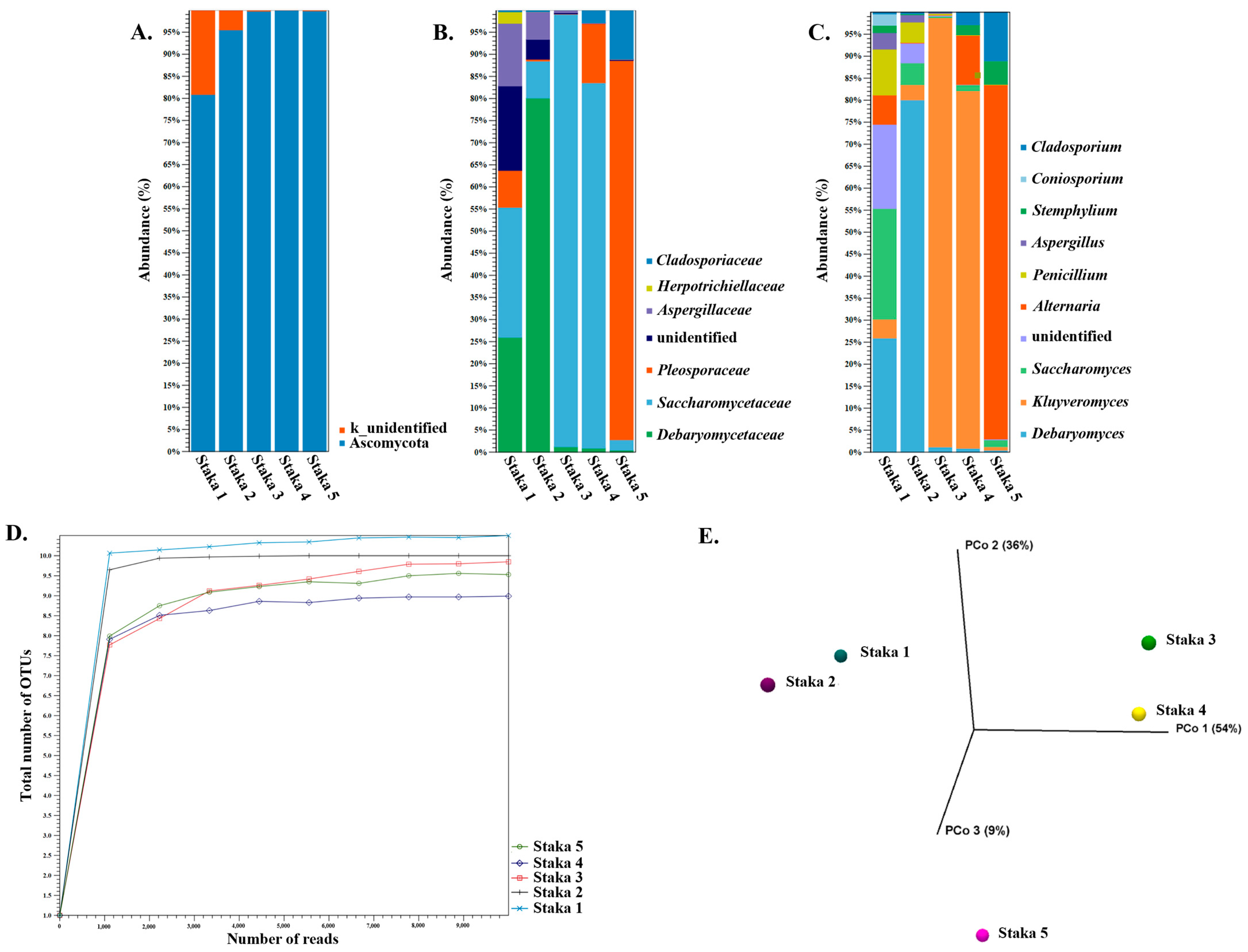
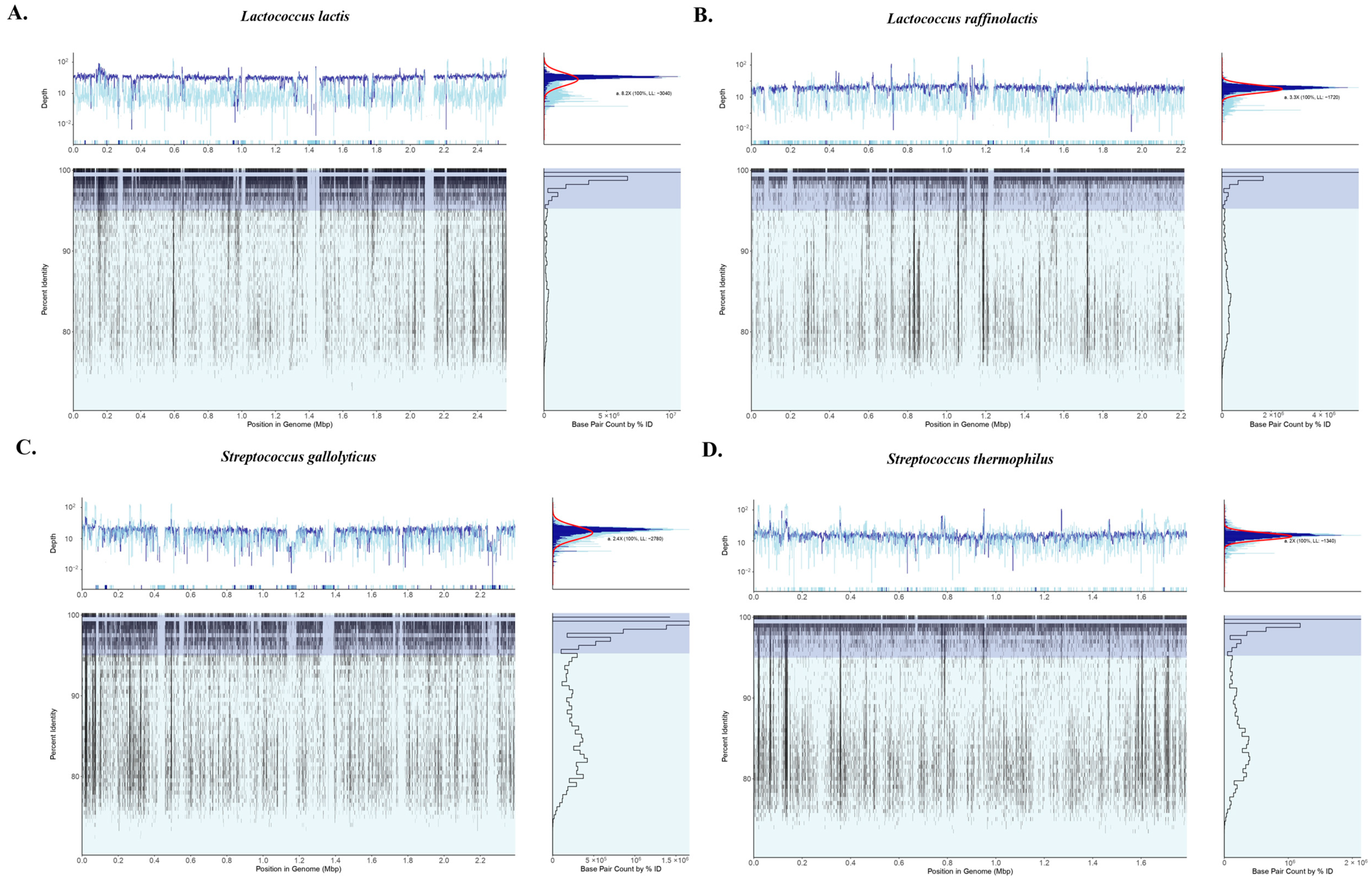
3.3. Amplicon Sequencing and Shotgun Metagenomics
3.4. Isolates Typing and Identification
3.5. Antimicrobial Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Butler, G.; Nielsen, J.H.; Slots, T.; Seal, C.; Eyre, M.D.; Sanderson, R.; Leifert, C. Fatty acid and fat-soluble antioxidant concentrations in milk from high- and low-input conventional and organic systems: Seasonal variation. J. Sci. Food Agric. 2008, 88, 1431–1441. [Google Scholar] [CrossRef]
- Banasiewicz, T.; Domagalska, D.; Borycka-Kiciak, K.; Rydzewska, G. Determination of butyric acid dosage based on clinical and experimental studies—A literature review. Gastroenterol. Rev. 2020, 15, 119–125. [Google Scholar] [CrossRef] [PubMed]
- den Hartigh, L.J. Conjugated linoleic acid effects on cancer, obesity, and atherosclerosis: A review of pre-clinical and human trials with current perspectives. Nutrients 2019, 11, 370. [Google Scholar] [CrossRef] [PubMed]
- Santa-María, C.; López-Enríquez, S.; Montserrat-de la Paz, S.; Geniz, I.; Reyes-Quiroz, M.E.; Moreno, M.; Palomares, F.; Sobrino, F.; Alba, G. Update on anti-inflammatory molecular mechanisms induced by oleic acid. Nutrients 2023, 15, 224. [Google Scholar] [CrossRef] [PubMed]
- Nestel, P. Chapter 16—Fermented dairy foods and cardiovascular risk. In Dairy in Human Health and Disease Across the Lifespan; Watson, R.R., Collier, R.J., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 225–229. [Google Scholar] [CrossRef]
- Ledoux, M.; Chardigny, J.-M.; Darbois, M.; Soustre, Y.; Sébédio, J.-L.; Laloux, L. Fatty acid composition of French butters, with special emphasis on conjugated linoleic acid (CLA) isomers. J. Food Compos. Anal. 2005, 18, 409–425. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- 21CFR131.160. Milk and Cream. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=131.160 (accessed on 22 December 2023).
- ISO 27205-IDF 149:2010; Fermented Milk Products—Bacterial Starter Cultures—Standard of Identity. ISO: London, UK, 2010.
- Meunier-Goddik, L. Sour Cream and Crème Fraîche; CRC Press: Boca Raton, FL, USA, 2004; pp. 235–246. [Google Scholar] [CrossRef]
- Niamsiri, N.; Batt, C.A. Dairy Products. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 34–44. [Google Scholar]
- 21CFR131.162. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/cfrsearch.cfm?fr=131.162 (accessed on 22 December 2023).
- Aryana, K.J.; Olson, D.W. A 100-Year Review: Yogurt and other cultured dairy products. J. Dairy Sci. 2017, 100, 9987–10013. [Google Scholar] [CrossRef] [PubMed]
- Monnet, V.C.S.; Cogan, T.M.; Gripon, K.C. Metabolism of starter cultures. In Dairy Starter Cultures; Cogan, T.M., Accolas, J.P., Eds.; VCH Publishers: New York, NY, USA, 1995; pp. 47–100. [Google Scholar]
- Page, R.A.; Lavalie, V.G. Sour Cream Dairy Product. US Patent No. 2,719,793, 4 October 1955. [Google Scholar]
- Cakmakci, S.; Hayaloglu, A.A. Evaluation of the chemical, microbiological and volatile aroma characteristics of Ispir Kaymak, a traditional Turkish dairy product. Int. J. Dairy Technol. 2011, 64, 444–450. [Google Scholar] [CrossRef]
- Dairy, A.M.-G.t.P. A Mini-Guide to Polish Dairy. Available online: https://culture.pl/en/article/a-mini-guide-to-polish-dairy (accessed on 10 November 2023).
- Garmasheva, I. Isolation and characterization of lactic acid bacteria from Ukrainian traditional dairy products. AIMS Microbiol. 2016, 2, 372–387. [Google Scholar] [CrossRef]
- Motato, K.E.; Milani, C.; Ventura, M.; Valencia, F.E.; Ruas-Madiedo, P.; Delgado, S. Bacterial diversity of the Colombian fermented milk “Suero Costeno” assessed by culturing and high-throughput sequencing and DGGE analysis of 16S rRNA gene amplicons. Food Microbiol. 2017, 68, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Ferrocino, I.; Rantsiou, K.; Cocolin, L. Investigating dairy microbiome: An opportunity to ensure quality, safety and typicity. Curr. Opin. Biotechnol. 2022, 73, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, M.; O’Sullivan, O.; Cotter, P.D.; Sinderen, D.V.; Kenny, J.G. The application of metagenomics to study microbial communities and develop desirable traits in fermented foods. Foods 2022, 11, 3297. [Google Scholar] [CrossRef] [PubMed]
- Budhkar, Y.A.; Bankar, S.B.; Singhal, R.S. Milk and milk products|Microbiology of cream and butter. In Encyclopedia of Food Microbiology; Academic Press: Cambridge, MA, USA, 2014; pp. 728–737. [Google Scholar] [CrossRef]
- Kochetkova, T.V.; Grabarnik, I.P.; Klyukina, A.A.; Zayulina, K.S.; Elizarov, I.M.; Shestakova, O.O.; Gavirova, L.A.; Malysheva, A.D.; Shcherbakova, P.A.; Barkhutova, D.D.; et al. Microbial communities of artisanal fermented milk products from Russia. Microorganisms 2022, 10, 2140. [Google Scholar] [CrossRef] [PubMed]
- Syromyatnikov, M.Y.; Kokina, A.V.; Solodskikh, S.A.; Panevina, A.V.; Popov, E.S.; Popov, V.N. High-Throughput 16S rRNA gene sequencing of butter microbiota reveals a variety of opportunistic pathogens. Foods 2020, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Lappa, I.K.; Gantzias, C.; Manolopoulou, E.; De Brandt, E.; Aerts, M.; Vandamme, P.; Tsakalidou, E.; Georgalaki, M. MALDI-TOF MS insight into the biodiversity of Staka, the artisanal Cretan soured cream. Int. Dairy J. 2021, 116, 104969. [Google Scholar] [CrossRef]
- ISO 2920-IDF 58:2004; Whey Cheese. ISO: London, UK, 2004.
- AOAC. Official Methods of Analysis: Official Method for Ash; Method No. 936.03; AOAC: Rockville, MD, USA, 2000. [Google Scholar]
- Ardö, Y.; Polychroniadou, A. Laboratory Manual for Chemical Analysis of Cheese. Improvement of the Quality of the Production of Raw Milk Cheeses; Publications Office: Luxembourg, 1999. [Google Scholar]
- Angelopoulou, A.; Alexandraki, V.; Georgalaki, M.; Anastasiou, R.; Manolopoulou, E.; Tsakalidou, E.; Papadimitriou, K. Production of probiotic Feta cheese using Propionibacterium freudenreichii subsp. shermanii as adjunct. Int. Dairy J. 2017, 66, 135–139. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Anastasiou, R.; Georgalaki, M.; Bounenni, R.; Paximadaki, A.; Charmpi, C.; Alexandraki, V.; Kazou, M.; Tsakalidou, E. Comparison of the microbiome of artisanal homemade and industrial Feta cheese through amplicon sequencing and shotgun metagenomics. Microorganisms 2022, 10, 1073. [Google Scholar] [CrossRef] [PubMed]
- Papademas, P.; Aspri, M.; Mariou, M.; Dowd, S.E.; Kazou, M.; Tsakalidou, E. Conventional and omics approaches shed light on Halitzia cheese, a long-forgotten white-brined cheese from Cyprus. Int. Dairy J. 2019, 98, 72–83. [Google Scholar] [CrossRef]
- Schmartz, G.P.; Hirsch, P.; Amand, J.; Dastbaz, J.; Fehlmann, T.; Kern, F.; Müller, R.; Keller, A. BusyBee Web: Towards comprehensive and differential composition-based metagenomic binning. Nucleic Acids Res. 2022, 50, W132–W137. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Bagchi, S.; Chaterji, S.; Gerlach, W.; Grama, A.; Harrison, T.; Paczian, T.; Trimble, W.L.; Wilke, A. MG-RAST version 4—Lessons learned from a decade of low-budget ultra-high-throughput metagenome analysis. Brief. Bioinform. 2017, 20, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- De Roos, J.; Vandamme, P.; De Vuyst, L. Wort substrate consumption and metabolite production during Lambic beer fermentation and maturation explain the successive growth of specific bacterial and yeast species. Front. Microbiol. 2018, 9, 2763. [Google Scholar] [CrossRef] [PubMed]
- Verce, M.; De Vuyst, L.; Weckx, S. Shotgun metagenomics of a water kefir fermentation ecosystem reveals a novel Oenococcus species. Front. Microbiol. 2019, 10, 479. [Google Scholar] [CrossRef] [PubMed]
- Georgalaki, M.; Zoumpopoulou, G.; Mavrogonatou, E.; Van Driessche, G.; Alexandraki, V.; Anastasiou, R.; Papadelli, M.; Kazou, M.; Manolopoulou, E.; Kletsas, D.; et al. Evaluation of the antihypertensive angiotensin-converting enzyme inhibitory (ACE-I) activity and other probiotic properties of lactic acid bacteria isolated from traditional Greek dairy products. Int. Dairy J. 2017, 75, 10–21. [Google Scholar] [CrossRef]
- Ntougias, S.; Zervakis, G.I.; Ehaliotis, C.; Kavroulakis, N.; Papadopoulou, K.K. Ecophysiology and molecular phylogeny of bacteria isolated from alkaline two-phase olive mill wastes. Res. Microbiol. 2006, 157, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Quiloan, M.L.G.; Vu, J.; Carvalho, J. Enterococcus faecalis can be distinguished from Enterococcus faecium via differential susceptibility to antibiotics and growth and fermentation characteristics on mannitol salt agar. Front. Biol. 2012, 7, 167–177. [Google Scholar] [CrossRef]
- Manero, A.B.R.A. Identification of Enterococcus spp. with a Biochemical Key. Appl. Environ. Microbiol. 1999, 65, 4425–4430. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Quality control Lactobacillus strains for use with the API 50CH and API ZYM systems at 37 °C. J. Basic Microbiol. 2001, 41, 241–251. [Google Scholar] [CrossRef]
- Garvie, E.I. The growth factor and amino acid requirements of species of the genus Leuconostoc, including Leuconostoc paramesenteroides (sp.nov.) and Leuconostoc Oenos. J. Gen. Microbiol. 1967, 48, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; Cnockaert, M.; Abbott, S.L.; Janda, J.M.; Vandamme, P. Hafnia paralvei sp. nov., formerly known as Hafnia alvei hybridization group 2. Int. J. Syst. Evol. Microbiol. 2010, 60, 1725–1728. [Google Scholar] [CrossRef]
- Zoumpopoulou, G.; Tzouvanou, A.; Mavrogonatou, E.; Alexandraki, V.; Georgalaki, M.; Anastasiou, R.; Papadelli, M.; Manolopoulou, E.; Kazou, M.; Kletsas, D.; et al. Probiotic features of lactic acid bacteria isolated from a diverse pool of traditional Greek dairy products regarding specific strain-host interactions. Probiotics Antimicrob. Proteins 2018, 10, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Shepard, L.; Miracle, R.E.; Leksrisompong, P.; Drake, M.A. Relating sensory and chemical properties of sour cream to consumer acceptance. J. Dairy Sci. 2013, 96, 5435–5454. [Google Scholar] [CrossRef] [PubMed]
- CXS 288-1976; Standard for Cream and Prepared Creams. WHO: Geneva, Switzerland, 1976.
- USDA-AMS. Specifications for Sour Cream and Acidified Sour Cream; Agricultural Marketing Service: Washington, DC, USA, 2005. [Google Scholar]
- Ribar, S.; Karmelić, I.; Mesarić, M. Sphingoid bases in dairy products. Food Res. Int. 2007, 40, 848–854. [Google Scholar] [CrossRef]
- Yu, J.; Wang, H.M.; Zha, M.S.; Qing, Y.T.; Bai, N.; Ren, Y.; Xi, X.X.; Liu, W.J.; Menghe, B.L.; Zhang, H.P. Molecular identification and quantification of lactic acid bacteria in traditional fermented dairy foods of Russia. J. Dairy Sci. 2015, 98, 5143–5154. [Google Scholar] [CrossRef] [PubMed]
- Shelley, A.; Deeth, H.; MacRae, I. Review of methods of enumeration, detection and isolation of lipolytic microorganisms with special reference to dairy applications. J. Microbiol. Methods 1987, 6, 123–137. [Google Scholar] [CrossRef]
- Smacchi, E.; Gobbetti, M. Bioactive peptides in dairy products: Synthesis and interaction with proteolytic enzymes. Food Microbiol. 2000, 17, 129–141. [Google Scholar] [CrossRef]
- Fox, P.F. Proteolysis during cheese manufacture and ripening. J. Dairy Sci. 1989, 72, 1379–1400. [Google Scholar] [CrossRef]
- Seiler, H. Yeasts in milk and dairy products. Encycl. Dairy Sci. 2002, 4, 2761–2769. [Google Scholar]
- Yu, Z.; Peng, C.; Kwok, L.; Zhang, H. The Bacterial diversity of spontaneously fermented dairy products collected in Northeast Asia. Foods 2021, 10, 2321. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H.; Mian, M.A. The occurrence and growth of yeasts in dairy products. Int. J. Food Microbiol. 1987, 4, 145–155. [Google Scholar] [CrossRef]
- Kosikowski, F.V.; Brown, D.P. Influence of carbon dioxide and nitrogen on microbial populations and shelf life of cottage cheese and sour cream. J. Dairy Sci. 1973, 56, 12–18. [Google Scholar] [CrossRef]
- Lu, M.; Wang, N.S. Chapter 7—Spoilage of milk and dairy products. In The Microbiological Quality of Food; Bevilacqua, A., Corbo, M.R., Sinigaglia, M., Eds.; Woodhead Publishing: Sawston, UK, 2017; pp. 151–178. [Google Scholar] [CrossRef]
- Geronikou, A.; Srimahaeak, T.; Rantsiou, K.; Triantafillidis, G.; Larsen, N.; Jespersen, L. Occurrence of yeasts in white-brined cheeses: Methodologies for identification, spoilage potential and good manufacturing practices. Front. Microbiol. 2020, 11, 582778. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.; Narvhus, J. Yeasts and their possible beneficial and negative effects on the quality of dairy products. Int. Dairy J. 1996, 6, 755–768. [Google Scholar] [CrossRef]
- Kure, C.F.; Skaar, I. The fungal problem in cheese industry. Curr. Opin. Food Sci. 2019, 29, 14–19. [Google Scholar] [CrossRef]
- Zheng, X.; Shi, X.; Wang, B. A Review on the general cheese processing technology, flavor biochemical pathways and the influence of yeasts in cheese. Front. Microbiol. 2021, 12, 703284. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.M.R.; Tanzina, A.Y.; Foysal, M.J.; Hoque, M.N.; Rumi, M.H.; Siddiki, A.M.A.M.Z.; Tay, A.C.-Y.; Hossain, M.J.; Bakar, M.A.; Mostafa, M.; et al. Insights into the nutritional properties and microbiome diversity in sweet and sour yogurt manufactured in Bangladesh. Sci. Rep. 2021, 11, 22667. [Google Scholar] [CrossRef] [PubMed]
- Kamilari, E.; Tsaltas, D.; Stanton, C.; Ross, R.P. Metataxonomic mapping of the microbial diversity of Irish and eastern mediterranean cheeses. Foods 2022, 11, 2483. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, K.; Touchette, M.; St-Gelais, D.; Labrie, S. Characterization of the fungal microflora in raw milk and specialty cheeses of the province of Quebec. Dairy Sci. Technol. 2012, 92, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.M.; Dawyndt, P.; Hoste, B.; Gevers, D.; Vandemeulebroecke, K.; Cleenwerck, I.; Vancanneyt, M.; Swings, J. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 2007, 57, 2777–2789. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.; Thompson, F.L.; Hoste, B.; Gevers, D.; Vandemeulebroecke, K.; Cleenwerck, I.; Thompson, C.C.; Vancanneyt, M.; Swings, J. Phylogeny and identification of enterococci by atpA gene sequence analysis. J. Clin. Microbiol. 2005, 43, 2224–2230. [Google Scholar] [CrossRef] [PubMed]
- Chun, B.H.; Kim, K.H.; Jeon, H.H.; Lee, S.H.; Jeon, C.O. Pan-genomic and transcriptomic analyses of Leuconostoc mesenteroides provide insights into its genomic and metabolic features and roles in kimchi fermentation. Sci. Rep. 2017, 7, 11504. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Russo, F.; Blaiotta, G.; Pepe, O.; Mauriello, G.; Villani, F. Simultaneous detection of Pseudomonas fragi, P. lundensis, and P. putida from meat by use of a multiplex PCR assay targeting the carA gene. Appl. Environ. Microbiol. 2007, 73, 2354–2359. [Google Scholar] [CrossRef] [PubMed]
- Ammoun, I.; Kothe, C.I.; Mohellibi, N.; Beal, C.; Yaacoub, R.; Renault, P. Lebanese fermented goat milk products: From tradition to meta-omics. Food Res. Int. 2023, 168, 112762. [Google Scholar] [CrossRef] [PubMed]
- Barzideh, Z.; Siddiqi, M.; Mohamed, H.M.; LaPointe, G. Dynamics of starter and non-starter lactic acid bacteria populations in long-ripened cheddar cheese using propidium monoazide (PMA) treatment. Microorganisms 2022, 10, 1669. [Google Scholar] [CrossRef] [PubMed]
- Ruggirello, M.; Giordano, M.; Bertolino, M.; Ferrocino, I.; Cocolin, L.; Dolci, P. Study of Lactococcus lactis during advanced ripening stages of model cheeses characterized by GC-MS. Food Microbiol. 2018, 74, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hao, S.; Ren, Q. Analysis of bacterial diversity in fermented grains of Baijiu based on culturomics and amplicon sequencing. Fermentation 2023, 9, 260. [Google Scholar] [CrossRef]
- Ganesan, B.; Stuart, M.R.; Weimer, B.C. Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl. Environ. Microbiol. 2007, 73, 2498–2512. [Google Scholar] [CrossRef] [PubMed]
- Ghodhbane, H.; Elaidi, S.; Sabatier, J.M.; Achour, S.; Benhmida, J.; Regaya, I. Bacteriocins active against multi-resistant gram negative bacteria implicated in nosocomial infections. Infect. Disord. Drug Targets 2015, 15, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Line, J.E.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S.; Siragusa, G.R.; et al. Isolation and purification of enterocin E-760 with Broad Antimicrob. Act. Against gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 2008, 52, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Svetoch, E.A.; Eruslanov, B.V.; Levchuk, V.P.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Stepanshin, J.; Dyatlov, I.; Seal, B.S.; Stern, N.J. Isolation of Lactobacillus salivarius 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum. Appl. Environ. Microbiol. 2011, 77, 2749–2754. [Google Scholar] [CrossRef] [PubMed]
- Xiraphi, N.; Georgalaki, M.; Driessche, G.V.; Devreese, B.; Beeumen, J.V.; Tsakalidou, E.; Metaxopoulos, J.; Drosinos, E.H. Purification and characterization of curvaticin L442, a bacteriocin produced by Lactobacillus curvatus L442. Antonie Van Leeuwenhoek 2006, 89, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Ono, H.; Kitagawa, H.; Ito, H.; Mu, F.; Sawa, N.; Zendo, T.; Sonomoto, K. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl. Environ. Microbiol. 2011, 77, 8164–8170. [Google Scholar] [CrossRef]
- Martin, R.; Olivares, M.; Marin, M.L.; Xaus, J.; Fernandez, L.; Rodriguez, J.M. Characterization of a reuterin-producing Lactobacillus coryniformis strain isolated from a goat’s milk cheese. Int. J. Food Microbiol. 2005, 104, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Harris, H.M.; McCann, A.; Guo, C.; Argimon, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef] [PubMed]
- Matthijs, S.; Vander Wauven, C.; Cornu, B.; Ye, L.; Cornelis, P.; Thomas, C.M.; Ongena, M. Antimicrobial properties of Pseudomonas strains producing the antibiotic mupirocin. Res. Microbiol. 2014, 165, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Ghequire, M.G.; Kemland, L.; Anoz-Carbonell, E.; Buchanan, S.K.; De Mot, R. A natural chimeric Pseudomonas bacteriocin with novel pore-forming activity parasitizes the ferrichrome transporter. mBio 2017, 8, e01961-16. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.A.; Jabeen, N.; Sohail, M.; Rasool, S.A. Biophysicochemical characterization of Pyocin SA189 produced by Pseudomonas aeruginosa SA189. Braz. J. Microbiol. 2015, 46, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.A.; Elshawadfy, A.M.; Amin, G.; Askora, A. Characterization of R-pyocin activity against Gram-positive pathogens for the first time with special focus on Staphylococcus aureus. J. Appl. Microbiol. 2021, 131, 2780–2792. [Google Scholar] [CrossRef] [PubMed]

| Concentration (% w/w) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Staka Sample | Production | Geographical Origin c | Milk Type | pH | TA f | Moisture | Dry Matter | Ash | Fat | Protein |
| 1 | C a | Sfakia, Chania | S d | 5.2 ± 0.2 | 0.3 ± 0.1 | 34.52 ± 1.5 | 65.48 ± 1.5 | 0.27 ± 0.07 | 41.87 ± 0.90 | 2.31 ± 0.40 |
| 2 | C | Keramia, Chania | S | 4.6 ± 0.3 | 0.4 ± 0.1 | 29.54 ± 1.9 | 70.46 ± 0.9 | 0.21 ± 0.04 | 46.47 ± 0.74 | 2.62 ± 0.15 |
| 3 | C | Varypetro, Chania | S & G e | 4.7 ± 0.4 | 0.5 ± 0.1 | 36.43 ± 0.8 | 63.57 ± 2.0 | 0.22 ± 0.03 | 40.57 ± 0.88 | 3.00 ± 0.25 |
| 4 | H-M b | Palaiokastro, Sitia | S & G | 5.2 ± 0.2 | 1.2 ± 0.3 | 27.05 ± 1.1 | 72.95 ± 1.7 | 3.42 ± 0.49 | 32.68 ± 0.66 | 16.00 ± 0.78 |
| 5 | C | Chamezi, Sitia | S & G | 5.3 ± 0.1 | 0.2 ± 0.1 | 48.25 ± 0.7 | 51.75 ± 1.2 | 2.15 ± 0.31 | 30.31 ± 1.70 | 6.40 ± 0.11 |
| Concentration (g/kg) | |||||||
|---|---|---|---|---|---|---|---|
| Staka Sample | Lactose | Lactic Acid | Acetic Acid | Succinic Acid | Butyric Acid | Propionic Acid | Ethanol |
| 1 | 24.7 ± 0.4 | 8.1 ± 0.5 | 1.4 ± 0.4 | 0.3 ± 0.1 | nd | nd | 0.4 ± 0.2 |
| 2 | 17.2 ± 0.3 | 6.7 ± 0.5 | 1.0 ± 0.2 | 0.3 ± 0.2 | 0.1 ± 0 | nd | nd |
| 3 | 18.5 ± 0.3 | 10.0 ± 0.6 | 0.8 ± 0.2 | nd | nd | nd | 0.3 ± 0.2 |
| 4 | 40.0 ± 1.1 | 16.3 ± 0.5 | nd | nd | 1.3 ± 0.2 | 1.7 ± 0.3 | nd |
| 5 | 13.6 ± 0.6 | 2.0 ± 0.3 | 1.6 ± 0.3 | 0.1 ± 0.1 | nd | nd | 0.2 ± 0.1 |
| Staka Sample | |||||
|---|---|---|---|---|---|
| Presumptive Microbial Group | 1 | 2 | 3 | 4 | 5 |
| Total mesophilic bacteria (PCA agar, 30 °C) | 8.09 ± 0.09 | 7.10 ± 0.18 | 7.88 ± 0.26 | 0.00 | 0.00 |
| Thermophilic lactobacilli (MRS agar pH 5.4, 42 °C) | 4.51 ± 0.35 | 6.13 ± 0.17 | 7.45 ± 0.21 | 0.00 | 0.00 |
| Mesophilic lactobacilli (MRS agar pH 5.4, 22 °C) | 7.99 ± 0.11 | 6.88 ± 0.39 | 7.33 ± 0.22 | 0.00 | 0.00 |
| Thermophilic cocci (M17 agar, 42 °C) | 4.86 ± 0.19 | 6.18 ± 0.28 | 7.67 ± 0.42 | 3.80 ± 0.22 | 0.00 |
| Mesophilic cocci (M17 agar, 22 °C) | 8.14 ± 0.17 | 6.44 ± 0.81 | 7.60 ± 0.19 | 0.00 | 0.00 |
| NSLAB (Rogosa agar, 30 °C) | 7.84 ± 0.41 | 6.92 ± 0.16 | 7.52 ± 0.31 | 2.80 ± 0.23 | 0.00 |
| Enterococci (KAA agar, 37 °C) | 4.70 ± 0.19 | 6.03 ± 0.22 | 7.29 ± 0.26 | 0.00 | 0.00 |
| Lipolytic bacteria (PCA-tributyrin, 30 °C) | 8.05 ± 0.08 | 6.97 ± 0.10 | 7.91 ± 0.04 | 2.60 ± 0.08 | 0.00 |
| Proteolytic bacteria (PCA-milk, 30 °C) | 7.78 ± 0.35 | 6.70 ± 0.17 | 7.95 ± 0.59 | 0.00 | 0.00 |
| Psychrotrophic bacteria (PCA, 7 °C) | 7.35 ± 0.46 | 4.13 ± 0.09 | 5.92 ± 0.16 | 0.00 | 0.00 |
| Pseudomonas spp. (Pseudomonas agar base, 30 °C) | 7.72 ± 0.25 | 0.00 | 5.85 ± 0.30 | 0.00 | 0.00 |
| Coliforms (VRBL agar, 37 °C) | 7.54 ± 0.34 | 0.00 | 5.60 ± 0.26 | 0.00 | 0.00 |
| Yeasts and molds (YGC agar, 25 °C) | 4.11 ± 0.26 | 3.41 ± 0.40 | 5.50 ± 0.35 | 0.00 | 0.00 |
| Producer Strain ACA-DC Number | Target Strain (Growth Medium of the Producer Strain—mm Inhibition Zone) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| S. mutans LMG 14558T | S. oralis LMG 14532T | S. pneumoniae LMG 14545T | S. agalactiae LMG 14694T | S. salivarius LMG 11489T | S. sanguinis DSM 20068 | S. sobrinus LMG 14641T | S. gordonii LMG 14518T | S. anginosus LMG 14502T | |
| L. curvatus 1135 | MRS-10 | mye-7t | MRS-10 mye -9 | nd | nd | MRS-15 | nd | MRS-15 | MRS-13 |
| L. coryniformis 1251 | nd | nd | MRS-9 mye-7 | nd | nd | nd | nd | nd | |
| L. paracasei 1260 | nd | nd | MRS-11 | nd | nd | mye-9t | nd | nd | MRS-10 mye-bl |
| C. versmoldensis 1262 | MRS-10 | nd | MRS-13 | nd | nd | nd | nd | nd | MRS-10 |
| L. pseudomesenteroides 1130 | mye-10 | mye15 | MRS-20 mye-15 | MRS-8t mye-6t | mye-7t | mye-20 | mye-11 | mye-20 | MRS-14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadimitriou, K.; Georgalaki, M.; Anastasiou, R.; Alexandropoulou, A.-M.; Manolopoulou, E.; Zoumpopoulou, G.; Tsakalidou, E. Study of the Microbiome of the Cretan Sour Cream Staka Using Amplicon Sequencing and Shotgun Metagenomics and Isolation of Novel Strains with an Important Antimicrobial Potential. Foods 2024, 13, 1129. https://doi.org/10.3390/foods13071129
Papadimitriou K, Georgalaki M, Anastasiou R, Alexandropoulou A-M, Manolopoulou E, Zoumpopoulou G, Tsakalidou E. Study of the Microbiome of the Cretan Sour Cream Staka Using Amplicon Sequencing and Shotgun Metagenomics and Isolation of Novel Strains with an Important Antimicrobial Potential. Foods. 2024; 13(7):1129. https://doi.org/10.3390/foods13071129
Chicago/Turabian StylePapadimitriou, Konstantinos, Marina Georgalaki, Rania Anastasiou, Athanasia-Maria Alexandropoulou, Eugenia Manolopoulou, Georgia Zoumpopoulou, and Effie Tsakalidou. 2024. "Study of the Microbiome of the Cretan Sour Cream Staka Using Amplicon Sequencing and Shotgun Metagenomics and Isolation of Novel Strains with an Important Antimicrobial Potential" Foods 13, no. 7: 1129. https://doi.org/10.3390/foods13071129
APA StylePapadimitriou, K., Georgalaki, M., Anastasiou, R., Alexandropoulou, A.-M., Manolopoulou, E., Zoumpopoulou, G., & Tsakalidou, E. (2024). Study of the Microbiome of the Cretan Sour Cream Staka Using Amplicon Sequencing and Shotgun Metagenomics and Isolation of Novel Strains with an Important Antimicrobial Potential. Foods, 13(7), 1129. https://doi.org/10.3390/foods13071129







