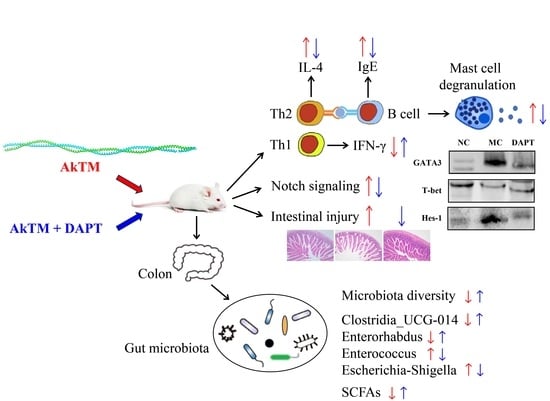
Notch Signaling Inhibition Alleviates Allergies Caused by Antarctic Krill Tropomyosin through Improving Th1/Th2 Imbalance and Modulating Gut Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Antarctic Krill TM
2.2. Protocols of Animal Experiment
2.3. Spleen Cells Culture and Cytokine Assay
2.4. Measurement of Serum Antibodies, Histamine and mMCP-1
2.5. Quantitative Real-Time PCR (qPCR)
2.6. Western Blotting
2.7. Histologic Staining
2.8. 16S rRNA Gene Sequencing and Data Analysis
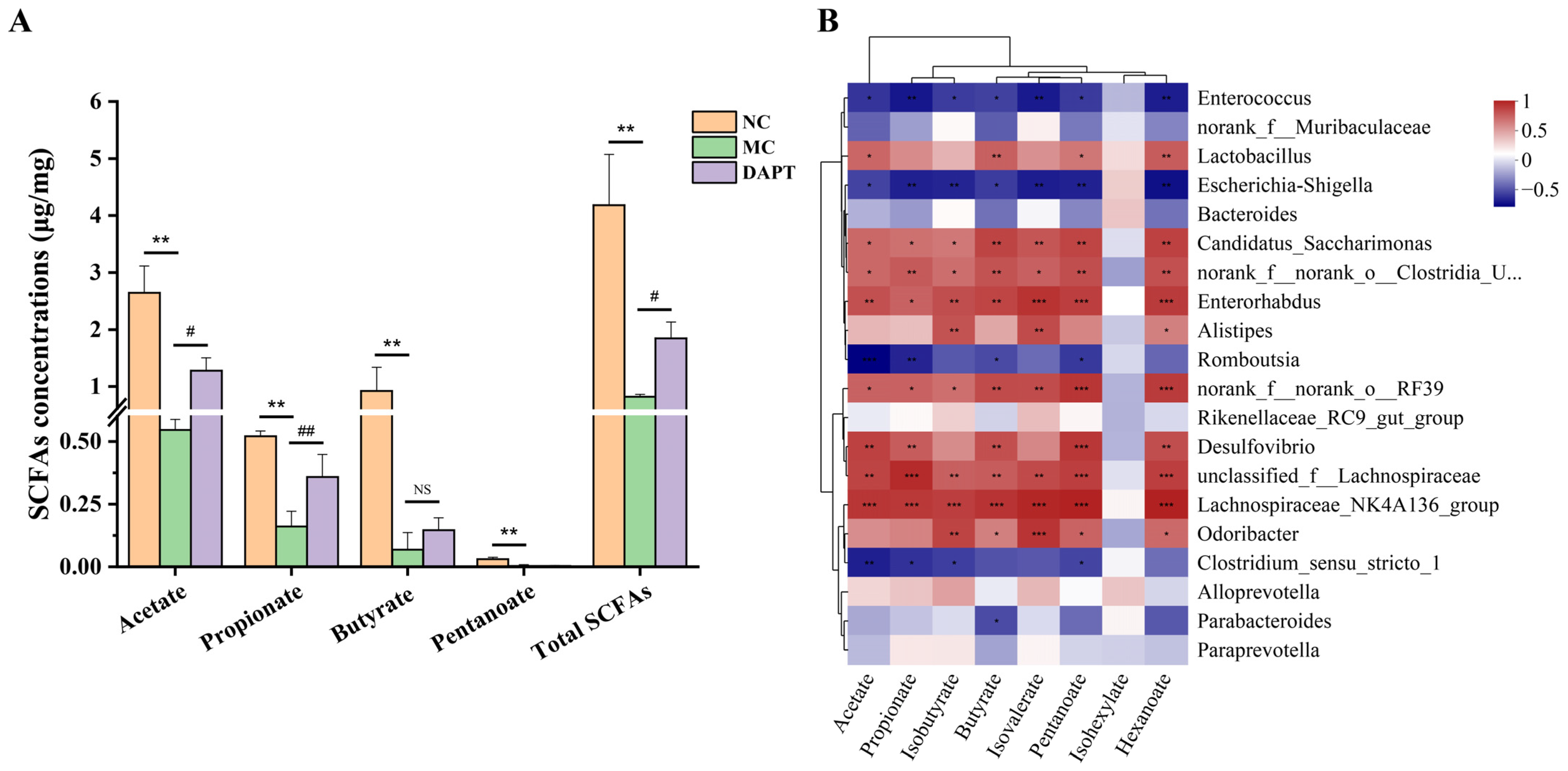
2.9. SCFAs Extraction and Analysis
2.10. Statistical Analysis
3. Results
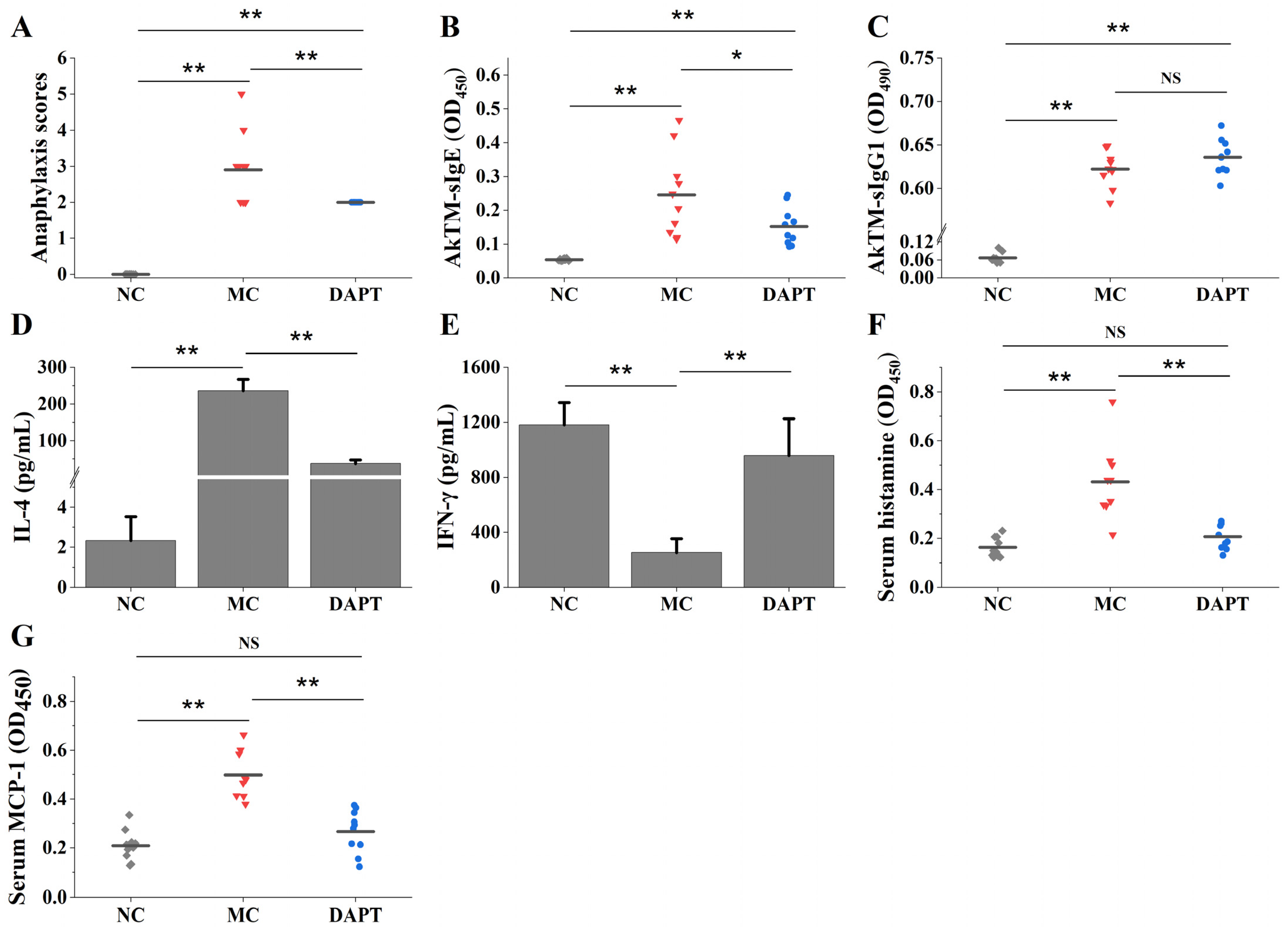
3.1. DAPT Treatment Alleviated Allergic Symptoms, Reduced the Levels of Specific IgE, and Affected the Secretion of Cytokines
3.2. DAPT Treatment Reduced Levels of Serum Histamine and mMCP-1
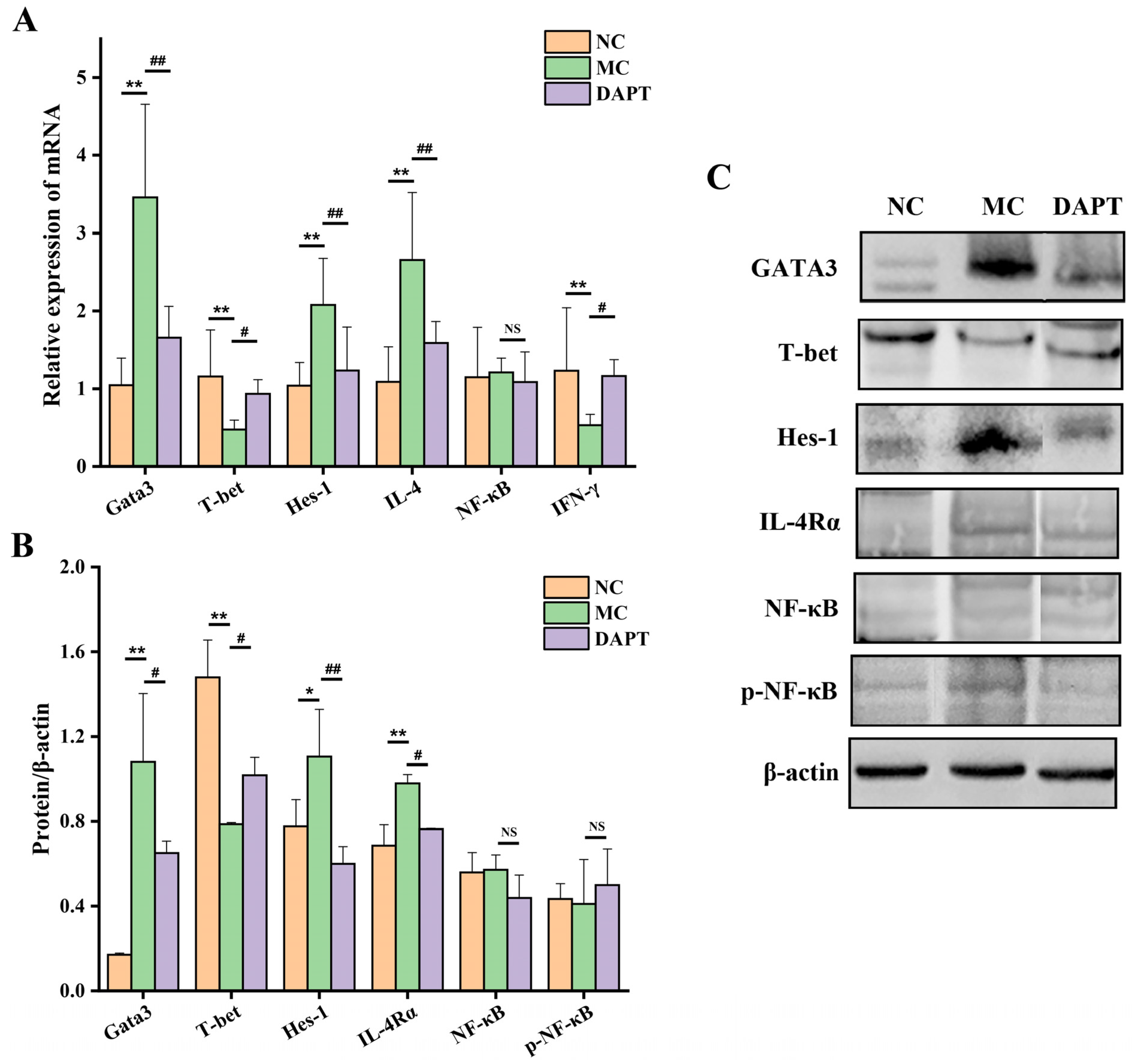
3.3. Effects of DAPT Treatment on the Expression of Hes-1, Gata3 and T-Bet
3.4. Effects of DAPT Treatment on the mRNA Levels of IL-4 and IFN-γ
3.5. Effects of DAPT Treatment on the Expression of IL-4Rα, NF-κB and p-NF-κB
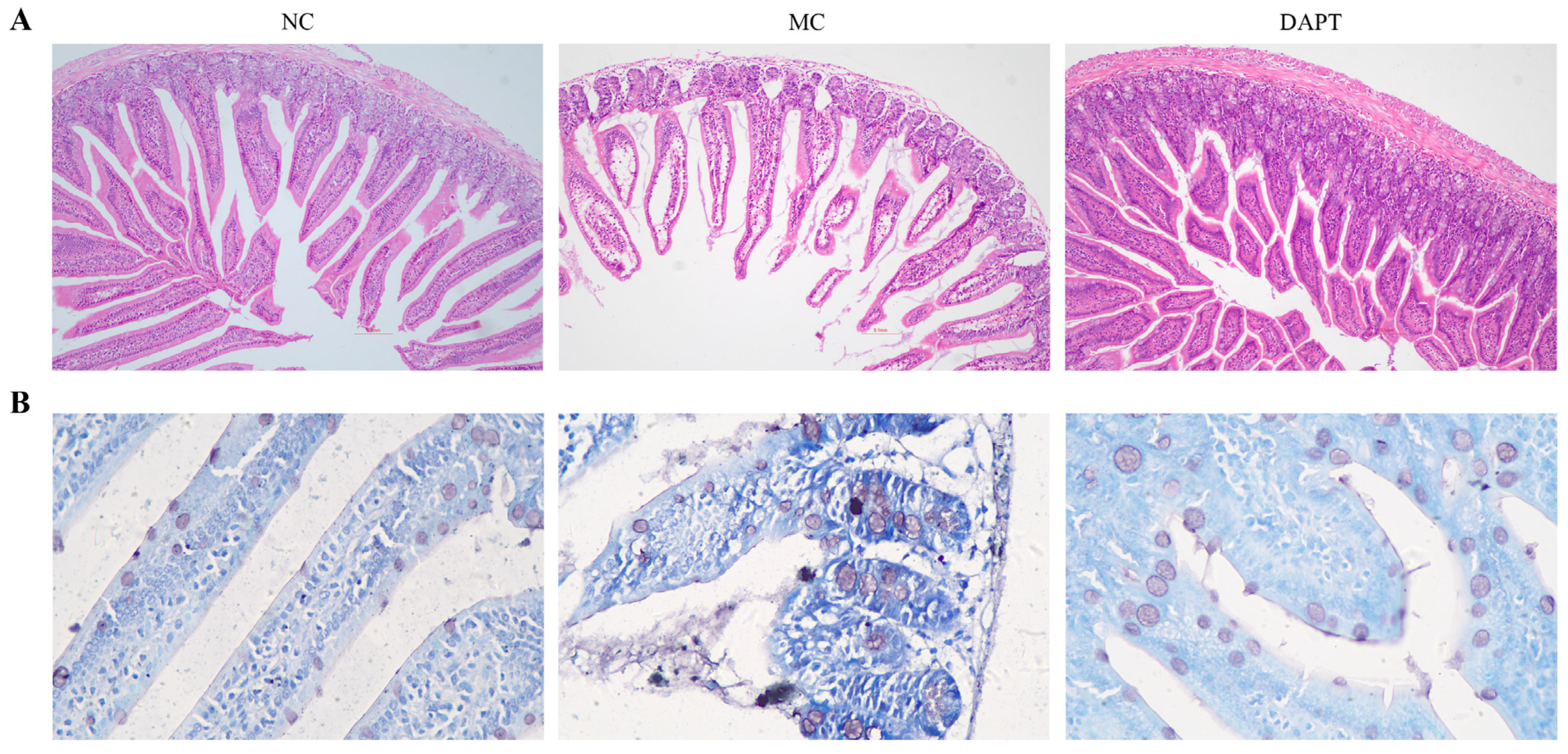
3.6. Histopathological Examination of Small Intestine
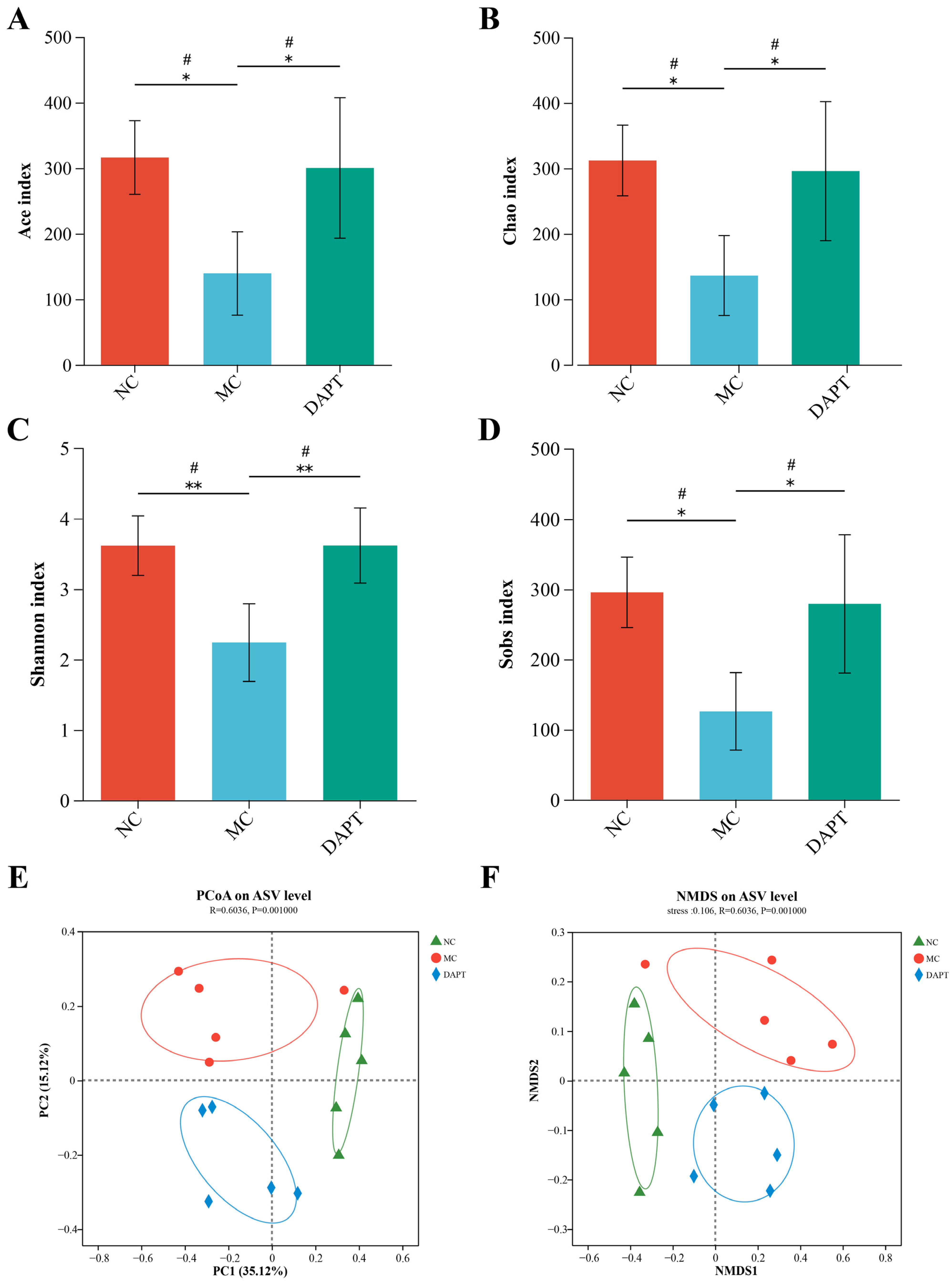
3.7. Gut microbiota Diversity Analysis
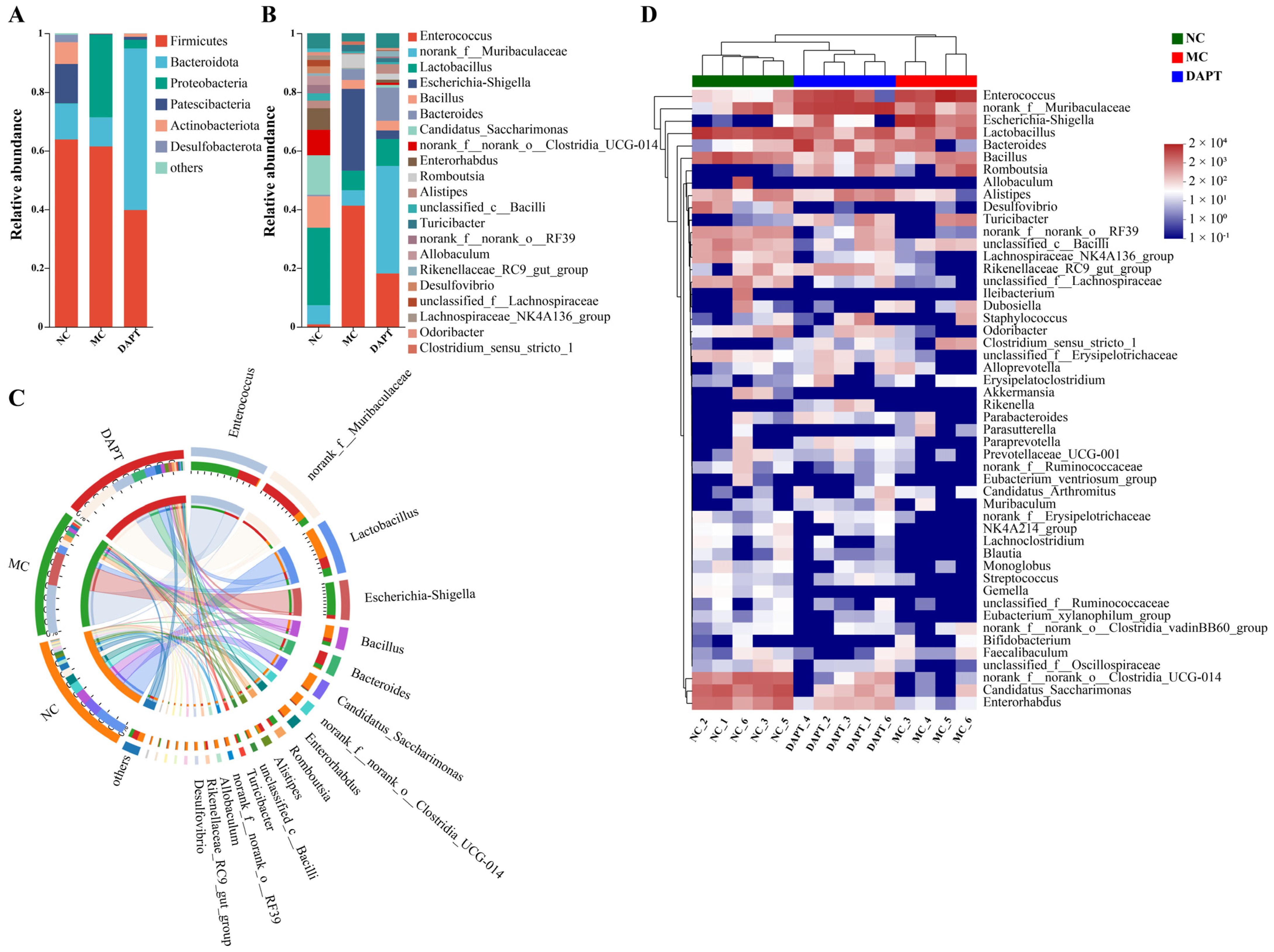
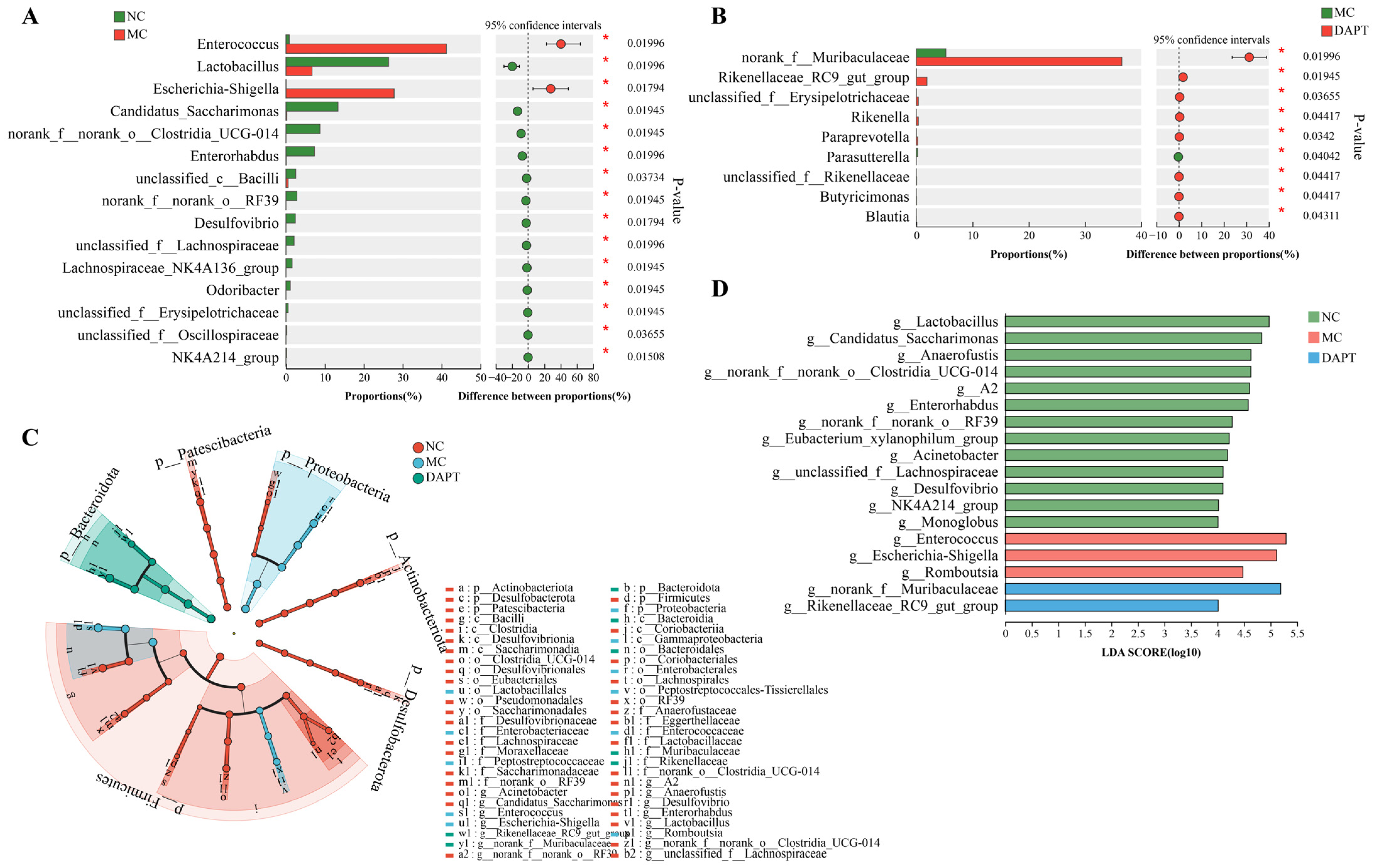
3.8. Changes in the Gut Microbiota Composition at Different Taxonomic Levels
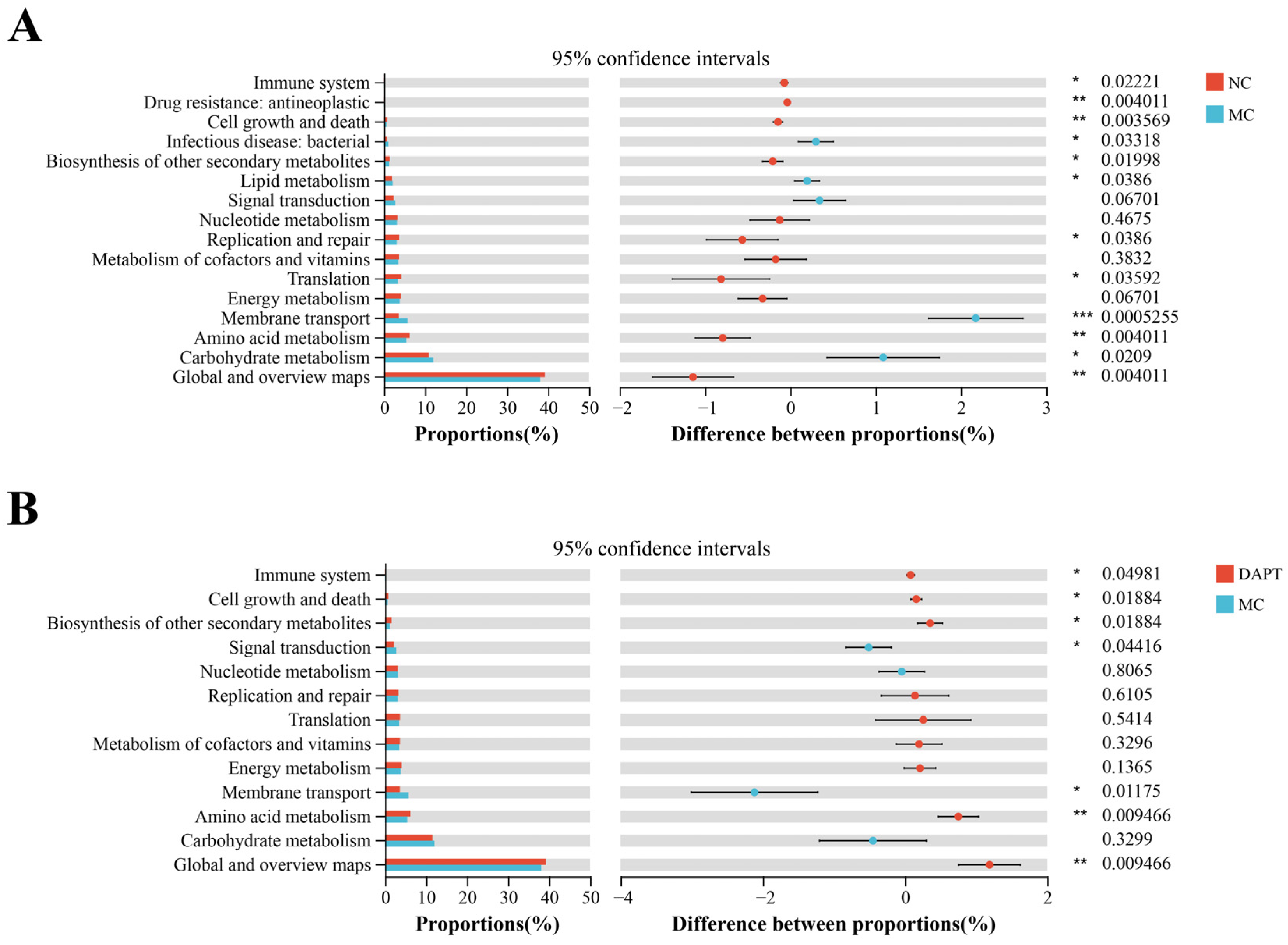
3.9. Functional Prediction Analysis of Gut Microbiota
3.10. Correlation Analysis between Gut Microbiota and SCFAs Concentrations
3.11. Correlation Analysis between Gut Microbiota and Allergic Indices
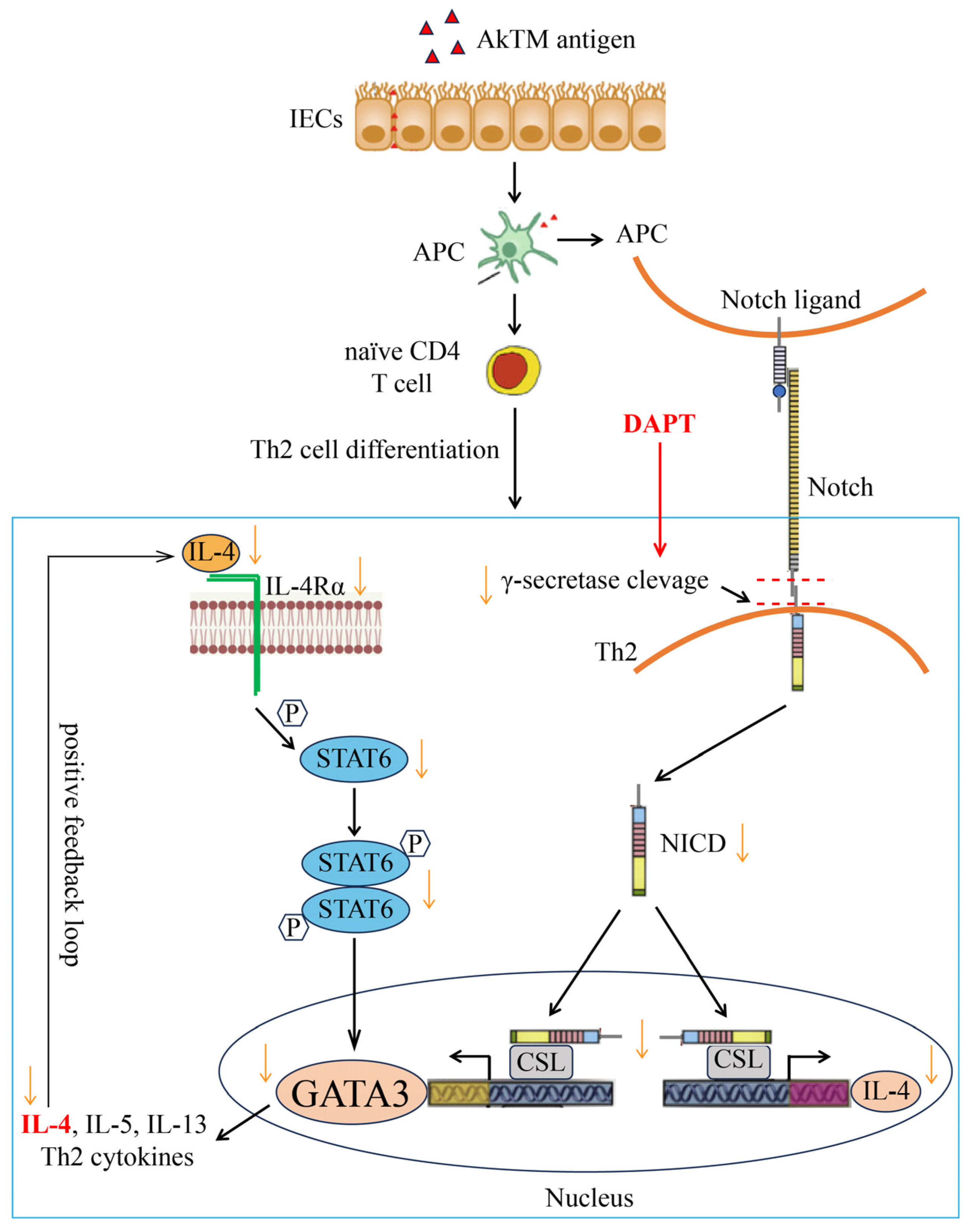
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Suzuki, T.; Shibata, N. The utilization of Antarctic krill for human food. Food Rev. Int. 1990, 6, 119–147. [Google Scholar] [CrossRef]
- Lin, N.; Chi, H.; Ni, L.; Zhang, H.; Liu, Z.D. Study on the Sensitization and Antigenic Epitopes of Tropomyosin from Antarctic Krill (Euphausia superba). J. Agric. Food Chem. 2023, 71, 6445–6457. [Google Scholar] [CrossRef]
- Motoyama, K.; Suma, Y.; Ishizaki, S.; Nagashima, Y.; Lu, Y.; Ushio, H.; Shiomi, K. Identification of Tropomyosins as Major Allergens in Antarctic Krill and Mantis Shrimp and Their Amino Acid Sequence Characteristics. Mar. Biotechnol. 2008, 10, 709–718. [Google Scholar] [CrossRef]
- Sampath, V.; Abrams, E.M.; Adlou, B. Food allergy across the globe. J. Allergy Clin. Immun. 2021, 148, 1347–1364. [Google Scholar] [CrossRef]
- Crespo, J.F.; Cabanillas, B. Recent advances in cellular and molecular mechanisms of IgE-mediated food allergy. Food Chem. 2023, 411, 135500. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Layhadi, J.A.; Kirtland, M.; Shamji, M.H. Mechanisms of Allergy; Academic Press: Oxford, UK, 2022; pp. 435–447. [Google Scholar]
- Amsen, D.; Blander, J.M.; Lee, G.R.; Tanigaki, K.; Honjo, T.; Flavell, R.A. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 2004, 117, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Amsen, D.; Antov, A.; Jankovic, D.; Sher, A.; Radtke, F.; Souabni, A.; Busslinger, M.; McCright, B.; Gridley, T.; Flavell, R.A. Direct regulation of Gata3 expression determines the T helper differentiation potential of notch. Immunity 2007, 27, 89–99. [Google Scholar] [CrossRef]
- Jiang, S.S.; Da, Y.F.; Han, S.W.; He, Y.H.; Che, H.L. Notch ligand Delta-like1 enhances degranulation and cytokine production through a novel Notch/Dok-1/MAPKs pathway in vitro. Immunol. Res. 2018, 66, 87–96. [Google Scholar] [CrossRef]
- Jiang, S.S.; Han, S.W.; Chen, J.Y.; Li, X.J.; Che, H.L. Inhibition effect of blunting Notch signaling on food allergy through improving TH1/TH2 balance in mice. Ann. Allerg. Asthma Immunol. 2017, 118, 94–102. [Google Scholar] [CrossRef]
- Zhao, C.L.; Li, J.; Yang, J.S.; Yang, L.; Chen, P.W.; Dou, J.M.; Zhao, S.Q. Inhibitor of gamma-secretase alleviates middle ear inflammation by regulating Th2 response in OVA-mediated allergic OME in vivo. Immunobiology 2019, 224, 765–773. [Google Scholar] [CrossRef]
- Radtke, F.; Fasnacht, N.; MacDonald, H.R. Notch Signaling in the Immune System. Immunity 2010, 32, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.C.; Yashiro-Ohtani, Y.; Del Bianco, C.; Knoblock, D.M.; Blacklow, S.C.; Pear, W.S. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity 2007, 27, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.L.; Fang, T.C.; Artis, D.; Shestova, O.; Pross, S.E.; Maillard, I.; Pear, W.S. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005, 202, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Kim, B.S.; Uhm, T.G.; Lee, S.H.; Lee, G.R.; Park, C.S.; Chung, I.Y. gamma-Secretase Inhibitor Reduces Allergic Pulmonary Inflammation by Modulating Th1 and Th2 Responses. Am. J. Resp. Crit. Care 2009, 179, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Zhang, X.J.; Wang, Y.J.; Ji, H.; Du, Y.Y.; Liu, H.C. DAPT protects brain against cerebral ischemia by down-regulating the expression of Notch 1 and Nuclear factor kappa B in rats. Neurol. Sci. 2012, 33, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, C.L.; Zhang, K.; Xue, W.T. The role of gut microbiota and its metabolites short-chain fatty acids in food allergy. Food Sci. Hum. Well. 2023, 12, 702–710. [Google Scholar] [CrossRef]
- Yang, H.; Qu, Y.Z.; Gao, Y.R.; Sun, S.Y.; Wu, R.N.; Wu, J.R. Research Progress on the Correlation between the Intestinal Microbiota and Food Allergy. Foods 2022, 11, 2913. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.R.; Mor, H.; Neriya, D.M.; Magzal, F.; Muller, E.; Appel, M.Y.; Nachshon, L.; Borenstein, E.; Tamir, S.; Louzoun, Y.; et al. Microbial signature in IgE-mediated food allergies. Genome Med. 2020, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, C.H.; Nagler, C.R. The Influence of the Microbiome on Allergic Sensitization to Food. J. Immunol. 2017, 198, 581–589. [Google Scholar] [CrossRef]
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, M. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019, 74, 799–809. [Google Scholar] [CrossRef]
- Cait, A.; Cardenas, E.; Dimitriu, P.A.; Amenyogbe, N.; Dai, D.; Cait, J.; Sbihi, H.; Stiemsma, L.; Subbarao, P.; Mandhane, P.J. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J. Allergy Clin. Immun. 2019, 144, 1638–1647. [Google Scholar] [CrossRef]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Pablos-Tanarro, A.; Lozano-Ojalvo, D.; Martinez-Blanco, M.; Molina, E.; Lopez-Fandino, R. Egg Yolk Provides Th2 Adjuvant Stimuli and Promotes Sensitization to Egg White Allergens in BALB/c Mice. Mol. Nutr. Food Res. 2018, 62, 1800057. [Google Scholar] [CrossRef]
- Zhou, M.S.; Zhang, B.; Gao, Z.L.; Zheng, R.P.; Marcellin, D.; Saro, A.; Pan, J.; Chu, L.; Wang, T.S.; Huang, J.F. Altered diversity and composition of gut microbiota in patients with allergic rhinitis. Microb. Pathog. 2021, 161, 105272. [Google Scholar] [CrossRef]
- Vaali, K.; Puumalainen, T.; Wolff, H.; Alenius, H.; Palosuo, T. Mucosal mast cell protease-1 (MMCP-1) as a marker of intestinal immunopathology in food allergy model. J. Allergy Clin. Immun. 2005, 115, S240. [Google Scholar] [CrossRef]
- Artavanis-Tsakonas, S. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef]
- Zheng, W.P.; Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. J. Immunol. 2016, 196, 4426–4435. [Google Scholar] [CrossRef]
- Rengarajan, J.; Szabo, S.J.; Glimcher, L.H. Transcriptional regulation of Th1/Th2 polarization. Immunol. Today 2000, 21, 479–483. [Google Scholar] [CrossRef]
- Ouyang, W.J.; Löhning, M.; Gao, Z.G.; Assenmacher, M.; Ranganath, S.; Radbruch, A.; Murphy, K.M. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 2000, 12, 27–37. [Google Scholar] [CrossRef]
- Li-Weber, M.; Giaisi, M.; Baumann, S.; Pálfi, K.; Krammer, P.H. NF-κB synergizes with NF-AT and NF-IL6 in activation of the IL-4 gene in T cells. Eur. J. Immunol. 2004, 34, 1111–1118. [Google Scholar] [CrossRef]
- KleinJan, A.; de Bruijn, M.; van Nimwegen, M.; Bergen, I.; Hoogsteden, H.C.; Amsen, D.; Hendriks, R.W. The Notch Mediator RBP-J in CD4 T Cells Plays a Crucial Role in the Induction of Allergic Asthma in Mice. J. Allergy Clin. Immun. 2013, 131, AB147. [Google Scholar] [CrossRef]
- Ulla, M.F.; Arias, A.M. Cell and molecular biology of Notch. J. Endocrinol. 2007, 194, 459–474. [Google Scholar]
- Eiwegger, T.; Hung, L.; Diego, K.E.S.; O’Mahony, L.; Upton, J. Recent developments and highlights in food allergy. Allergy 2019, 74, 2355–2367. [Google Scholar] [CrossRef]
- Honjo, A.; Nakano, N.; Yamazaki, S.; Hara, M.; Uchida, K.; Kitaura, J.; Nishiyama, C.; Yagita, H.; Ohtsuka, Y.; Ogawa, H.; et al. Pharmacologic inhibition of Notch signaling suppresses food antigen-induced mucosal mast cell hyperplasia. J. Allergy Clin. Immun. 2017, 139, 987–996. [Google Scholar] [CrossRef]
- Amsen, D.; Helbig, C.; Backer, R.A. Notch in T Cell Differentiation: All Things Considered. Trends Immunol. 2015, 36, 802–814. [Google Scholar] [CrossRef]
- Belizário, J.E.; Napolitano, M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 2015, 6, 1050. [Google Scholar] [CrossRef]
- Akagawa, S.; Kaneko, K. Gut microbiota and allergic diseases in children. Allergol. Int. 2022, 71, 301–309. [Google Scholar] [CrossRef]
- Sjödin, K.S.; Hammarström, M.L.; Rydén, P.; Sjödin, A.; Hernell, O.; Engstrand, L.; West, C.E. Temporal and long-term gut microbiota variation in allergic disease: A prospective study from infancy to school age. Allergy 2019, 74, 176–185. [Google Scholar] [CrossRef]
- Gu, S.M.; Xie, Q.; Chen, C.; Liu, C.L.; Xue, W.T. Gut Microbial Signatures Associated with Peanut Allergy in a BALB/c Mouse Model. Foods 2022, 11, 1395. [Google Scholar] [CrossRef]
- Chen, C.C.; Chen, K.J.; Kong, M.S.; Chang, H.J.; Huang, J.L. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr. Allergy Immunol. 2016, 27, 254–262. [Google Scholar] [CrossRef]
- Noval Rivas, M.; Burton, O.T.; Wise, P.; Zhang, Y.Q.; Hobson, S.A.; Lloret, M.G.; Chehoud, C.; Kuczynski, J.; DeSantis, T.; Warrington, J. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immun. 2013, 131, 201–212. [Google Scholar] [CrossRef]
- Zhang, N.; Gou, Y.S.; Liang, S.; Chen, N.; Liu, Y.L.; He, Q.S.; Zhang, J. Dysbiosis of Gut Microbiota Promotes Hepatocellular Carcinoma Progression by Regulating the Immune Response. J. Immunol. Res. 2021, 2021, 4973589. [Google Scholar] [CrossRef]
- Yoo, W.; Zieba, J.K.; Foegeding, N.J.; Torres, T.P.; Shelton, C.D.; Shealy, N.G.; Byndloss, A.J.; Cevallos, S.A.; Gertz, E.; Tiffany, C.R. High-fat diet–induced colonocyte dysfunction escalates microbiota-derived trimethylamine N-oxide. Science 2021, 373, 813–818. [Google Scholar] [CrossRef]
- Zeng, Q.H.; Zeng, R.H.; Ye, J.B. Alteration of the oral and gut microbiota in patients with Kawasaki disease. PEERJ 2023, 11, e15662. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Vuillermin, P.J.; Goverse, G.; Vinuesa, C.G.; Mebius, R.E.; Macia, L.; Mackay, C.R. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep. 2016, 15, 2809–2824. [Google Scholar] [CrossRef]
- Ge, H.F.; Cai, Z.A.Z.; Chai, J.L.; Liu, J.Y.; Liu, B.Q.; Yu, Y.D.; Liu, J.B.; Zhang, T. Egg white peptides ameliorate dextran sulfate sodium-induced acute colitis symptoms by inhibiting the production of pro-inflammatory cytokines and modulation of gut microbiota composition. Food Chem. 2021, 360, 129981. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Honda, K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2014, 500, 232–236. [Google Scholar] [CrossRef]
- Pang, B.; Jin, H.; Liao, N.; Li, J.J.; Jiang, C.M.; Shi, J.L. Vitamin A supplementation ameliorates ulcerative colitis in gut microbiota-dependent manner. Food Res. Int. 2021, 148, 110568. [Google Scholar] [CrossRef]
- Kourosh, A.; Luna, R.A.; Balderas, M.; Nance, C.; Anagnostou, A.; Devaraj, S.; Davis, C.M. Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatr. Allergy Immunol. 2018, 29, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.D.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.J.; Chen, S.Y.; Hsu, Y.H.; Yen, G.C. Protective effect of fermented okara on the regulation of inflammation, the gut microbiota, and SCFAs production in rats with TNBS-induced colitis. Food Res. Int. 2022, 157, 111390. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, N.; Chi, H.; Guo, Q.; Liu, Z.; Ni, L. Notch Signaling Inhibition Alleviates Allergies Caused by Antarctic Krill Tropomyosin through Improving Th1/Th2 Imbalance and Modulating Gut Microbiota. Foods 2024, 13, 1144. https://doi.org/10.3390/foods13081144
Lin N, Chi H, Guo Q, Liu Z, Ni L. Notch Signaling Inhibition Alleviates Allergies Caused by Antarctic Krill Tropomyosin through Improving Th1/Th2 Imbalance and Modulating Gut Microbiota. Foods. 2024; 13(8):1144. https://doi.org/10.3390/foods13081144
Chicago/Turabian StyleLin, Na, Hai Chi, Quanyou Guo, Zhidong Liu, and Ling Ni. 2024. "Notch Signaling Inhibition Alleviates Allergies Caused by Antarctic Krill Tropomyosin through Improving Th1/Th2 Imbalance and Modulating Gut Microbiota" Foods 13, no. 8: 1144. https://doi.org/10.3390/foods13081144
APA StyleLin, N., Chi, H., Guo, Q., Liu, Z., & Ni, L. (2024). Notch Signaling Inhibition Alleviates Allergies Caused by Antarctic Krill Tropomyosin through Improving Th1/Th2 Imbalance and Modulating Gut Microbiota. Foods, 13(8), 1144. https://doi.org/10.3390/foods13081144