Fungal Mycobiome of Mature Strawberry Fruits (Fragaria x ananassa Variety ‘Monterey’) Suggests a Potential Market Site Contamination with Harmful Yeasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fruit Sampling and Processing
2.2. Library Construction, Sequencing, Data Processing, and Analysis
2.3. Rarefaction Curves
2.4. Analysis of Diversity
2.5. Statistical Significance Tests
3. Results and Discussion
3.1. Fungal Communities in Ripe Strawberry Fruits: Alpha-Diversity Analysis
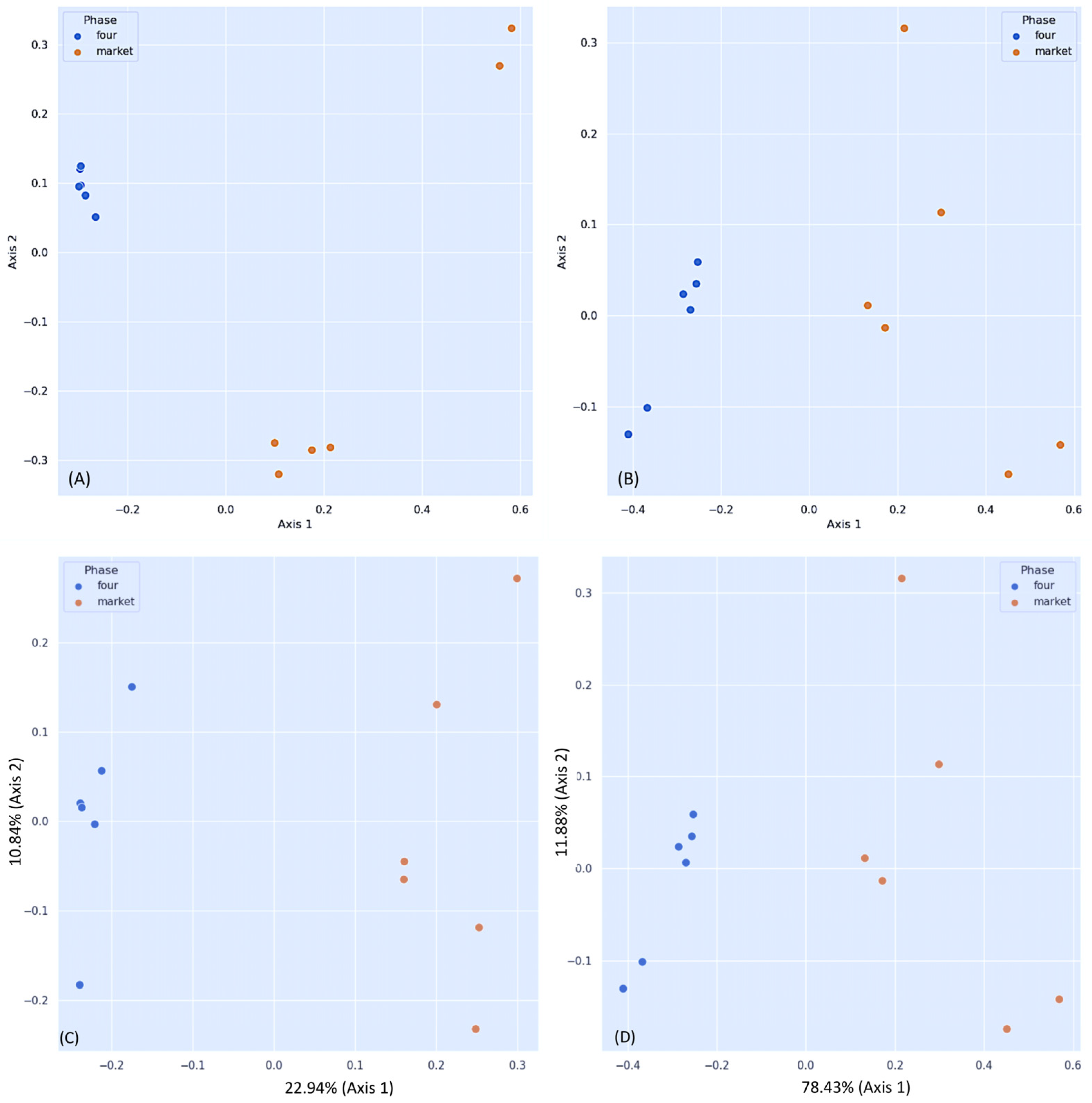
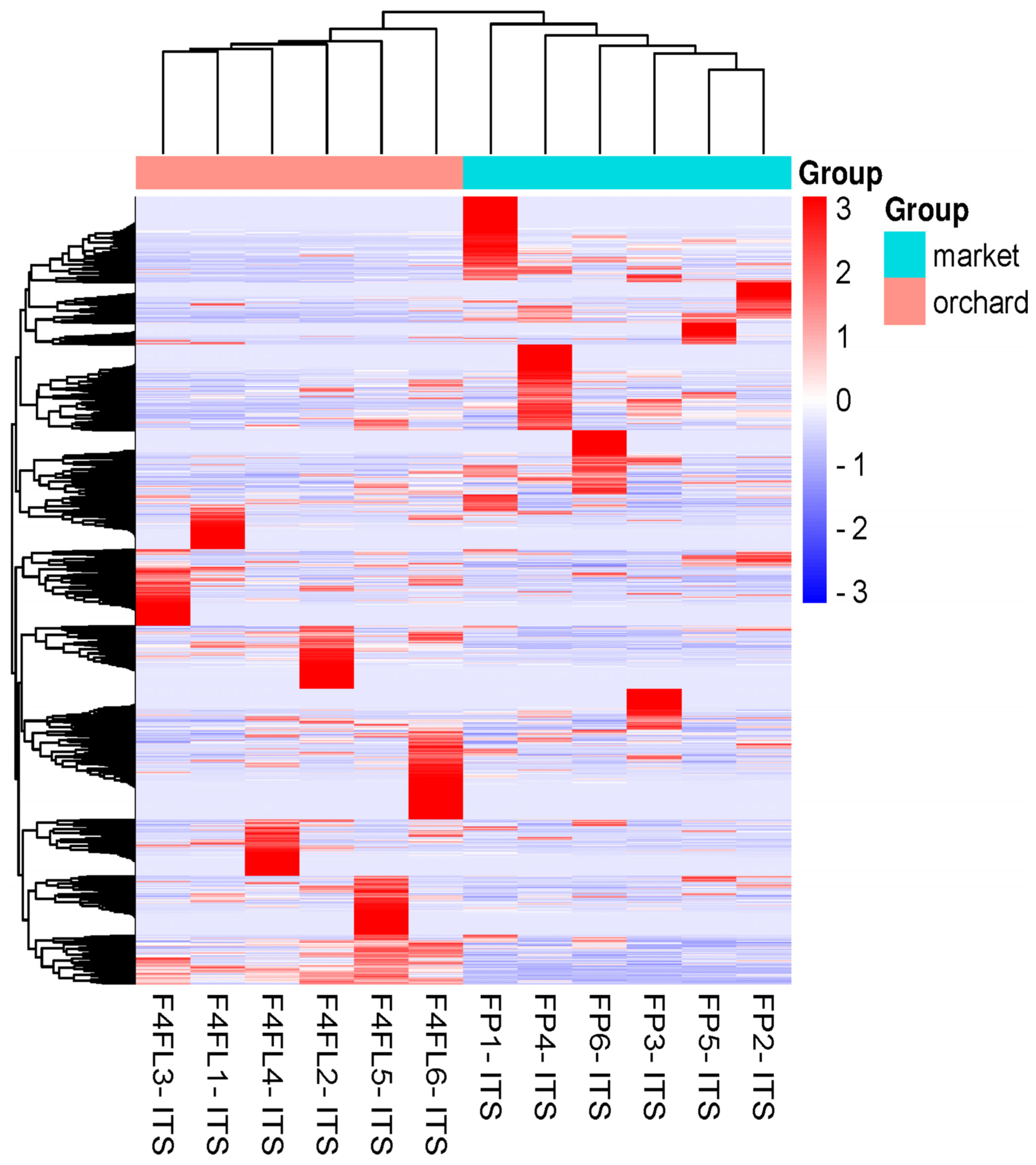
3.2. Differential Fungal Community of Ripe Strawberry Fruits: Beta-Diversity Analysis
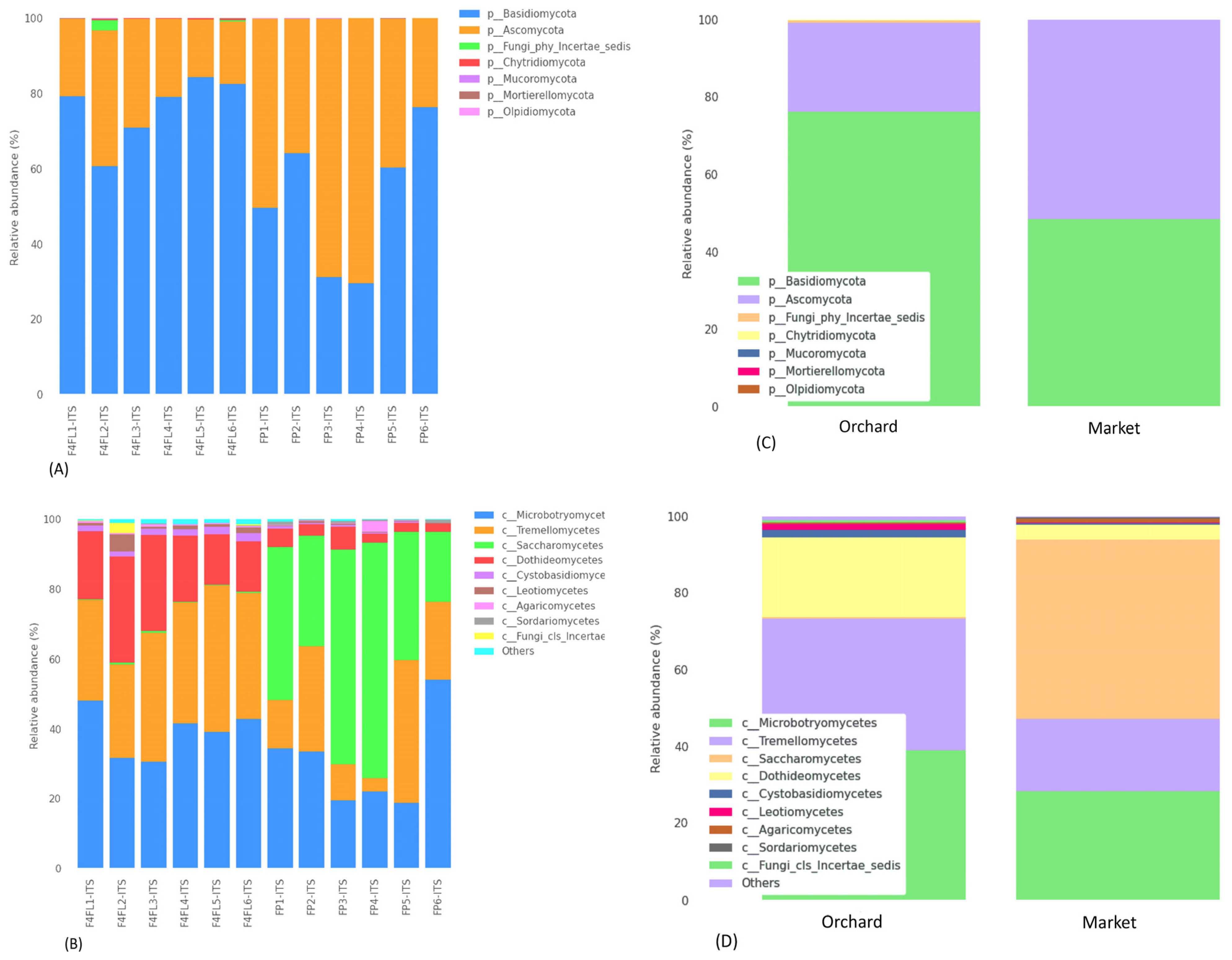
3.3. Taxonomic Assignment of Fungi
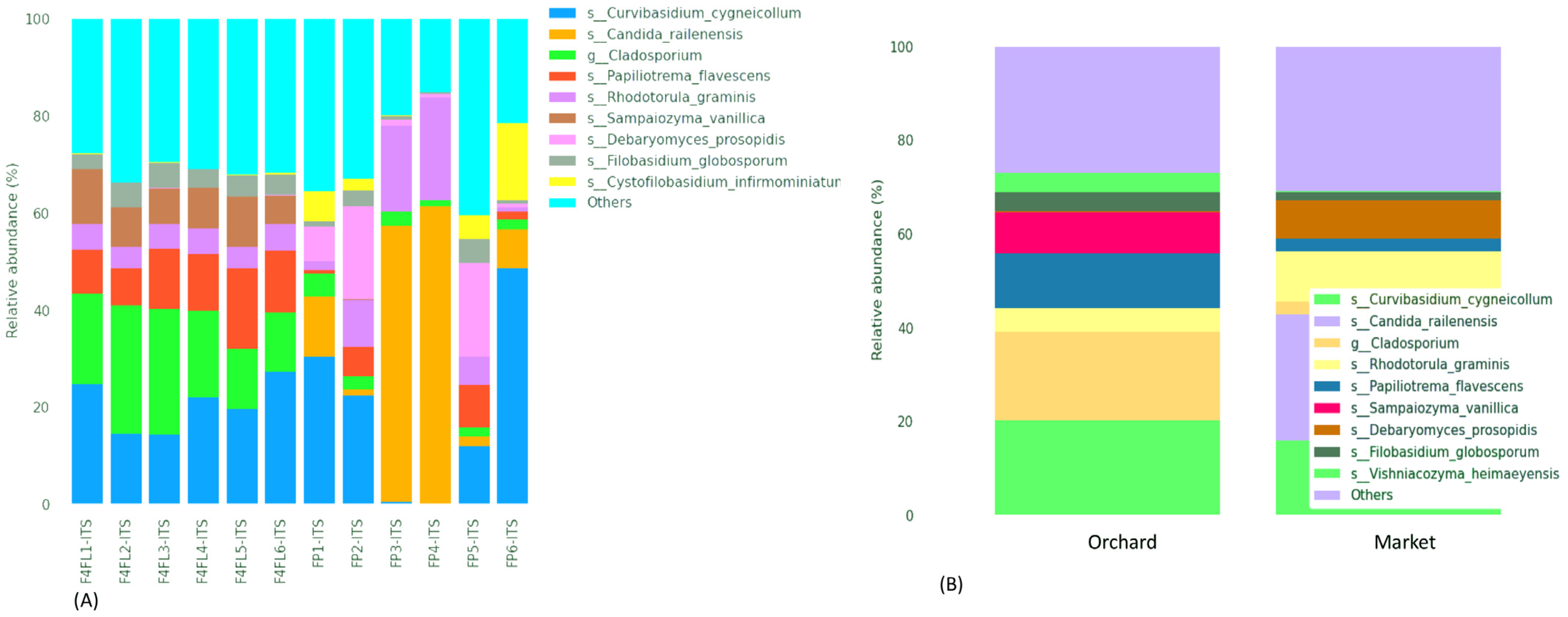
3.4. Taxonomic Assignment Suggests Both Beneficial and Harmful Yeasts in Strawberry Fruits
3.5. Clinically Related Yeasts Were Detected in Strawberries Fruits from Market
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kahramanoğlu, İ.; Panfilova, O.; Kesimci, T.G.; Bozhüyük, A.U.; Gürbüz, R.; Alptekin, H. Control of postharvest gray mold at strawberry fruits caused by botrytis cinerea and improving fruit storability through Origanum onites L. and Ziziphora clinopodioides L. volatile essential oils. Agronomy 2022, 12, 389. [Google Scholar] [CrossRef]
- Satitmunnaithum, J.; Kitazawa, H.; Arofatullah, N.A.; Widiastuti, A.; Kharisma, A.D.; Yamane, K.; Tanabata, S.; Sato, T. Microbial population size and strawberry fruit firmness after drop shock-induced mechanical damage. Postharvest Biol. Technol. 2022, 192, 112008. [Google Scholar] [CrossRef]
- Jensen, B.; Knudsen, I.M.B.; Andersen, B.; Nielsen, K.F.; Thrane, U.; Jensen, D.F.; Larsen, J. Characterization of microbial communities and fungal metabolites on field grown strawberries from organic and conventional production. Int. J. Food Microbiol. 2013, 160, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Zain, M.E. Impact of mycotoxins on humans and animals. J. Saudi Chem. Soc. 2011, 15, 129–144. [Google Scholar] [CrossRef]
- Koch, S.; Epp, A.; Lohmann, M.; Böl, G.F. Pesticide residues in food: Attitudes, beliefs, and misconceptions among conventional and organic consumers. J. Food Prot. 2017, 80, 2083–2089. [Google Scholar] [CrossRef] [PubMed]
- Ruxton, G.D.; Wilkinson, D.M.; Schaefer, H.M.; Sherratt, T.N. Why fruit rots: Theoretical support for Janzen’s theory of microbe–macrobe competition. Proc. R. Soc. B Biol. Sci. 2014, 281, 20133320. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef] [PubMed]
- Wassermann, B.; Müller, H.; Berg, G. An apple a day: Which bacteria do we eat with organic and conventional apples? Front. Microbiol. 2019, 10, 1629. [Google Scholar] [CrossRef] [PubMed]
- Vermote, L.; Verce, M.; Mozzi, F.; De Vuyst, L.; Weckx, S. microbiomes associated with the surfaces of northern argentinian fruits show a wide species diversity. Front. Microbiol. 2022, 13, 872281. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Orikasa, T.; Nakamura, N.; Hayashi, K.; Yasaka, Y.; Makino, N.; Shobatake, K.; Koide, S.; Shiina, T. Dataset for life cycle assessment of strawberry-package supply chain with considering food loss during transportation. Data Brief 2021, 39, 107473. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Wisniewski, M.; Li Destri Nicosia, M.G.; Cacciola, S.O.; Schena, L. Metagenomic analysis of fungal diversity on strawberry plants and the effect of management practices on the fungal community structure of aerial organs. PLoS ONE 2016, 11, e0160470. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Freilich, S.; Bartuv, R.; Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Burchard, E.; Dardick, C.; et al. Global analysis of the apple fruit microbiome: Are all apples the same? Environ. Microbiol. 2021, 23, 6038–6055. [Google Scholar] [CrossRef] [PubMed]
- De Tender, C.; Haegeman, A.; Vandecasteele, B.; Clement, L.; Cremelie, P.; Dawyndt, P.; Maes, M.; Debode, J. Dynamics in the strawberry rhizosphere microbiome in response to biochar and Botrytis cinerea Leaf Infection. Front. Microbiol. 2016, 7, 2062. [Google Scholar] [CrossRef] [PubMed]
- Kusstatscher, P.; Cernava, T.; Abdelfattah, A.; Gokul, J.; Korsten, L.; Berg, G. Microbiome approaches provide the key to biologically control postharvest pathogens and storability of fruits and vegetables. FEMS Microbiol Ecol. 2020, 96, fiaa119. [Google Scholar] [CrossRef] [PubMed]
- Tournas, V.H.; Katsoudas, E. Mould and yeast flora in fresh berries, grapes and citrus fruits. Int. J. Food Microbiol. 2005, 105, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Asociación de Municipalidades, Ecuador. 2021. Available online: https://ame.gob.ec/2021/05/14/tisaleo-principal-productor-de-mora-y-fresa/ (accessed on 8 December 2023).
- Tenea, G.N.; Reyes, P. Bacterial community changes in strawberry fruits (Fragaria × ananassa variety “Monterey”) from farm field to retail market sites an indicator of postharvest contamination. Front. Microbiol. 2024, 15, 1348316. [Google Scholar] [CrossRef] [PubMed]
- Angamarca, E.; Castillejo, P.; Tenea, G.N. Microbiota and its antibiotic resistance profile in avocado Guatemalan fruits (Persea nubigena var. guatemalensis) sold at retail markets of Ibarra city, northern Ecuador. Front. Microbiol. 2023, 14, 1228079. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Reeder, J.; Knight, R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 2010, 7, 668–669. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2020, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Alishum, A. DADA2 formatted 16S rRNA gene sequences for both bacteria & archaea (Version 4.2) [Data set]. Zenodo 2021. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Kandlikar, G.S.; Gold, Z.J.; Cowen, M.C.; Meyer, R.S.; Freise, A.C.; Kraft, N.J.B.; Moberg-Parker, J.; Sprague, J.; Kushner, D.J.; Curd, E.E. Ranacapa: An R package and shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000Research 2018, 7, 1734. [Google Scholar] [CrossRef]
- Krebs, C.J. Species diversity measures. In Ecological Methodology; University of British Columbia: Vancouver, BC, Canada, 2014; pp. 532–593. [Google Scholar]
- Lozupone, C.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Yinglin, X.; Jun, S.; Ding-Geng, C. Statistical Analysis of Microbiome Data with R; Springer Nature: Singapore, 2018; pp. 191–241. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Tenea, G.N.; Reyes, P.; Molina, D.; Ortega, C. Pathogenic microorganisms linked to fresh fruits and juices purchased at low-cost markets in Ecuador, potential carriers of antibiotic resistance. Antibiotics 2023, 12, 236. [Google Scholar] [CrossRef]
- Olimi, E.; Kusstatscher, P.; Wicaksono, W.A.; Abdelfattah, A.; Cernava, T.; Berg, G. Insights into the microbiome assembly during different growth stages and storage of strawberry plants. Environ. Microbiome 2022, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Lattin, J.M.; Carroll, J.D.; Green, P.E. Analyzing Multivariate Data; Thomson Brooks/Cole: Pacific Grove, CA, USA, 2003. [Google Scholar]
- Jones, R.; Fountain, M.T.; Andreani, N.A.; Günther, C.S.; Goddard, M.R. The relative abundances of yeasts attractive to Drosophila suzukii differ between fruit types and are greatest on raspberries. Sci. Rep. 2022, 12, 10382. [Google Scholar] [CrossRef]
- Artimová, R.; Játiová, M.; Baumgartnerová, J.; Lipková, N.; Petrová, J.; Maková, J.; Javoreková, S.; Hleba, L.; Medová, J.; Medo, J. Microbial communities on samples of commercially available fresh-consumed leafy vegetables and small berries. Horticulturae 2023, 9, 150. [Google Scholar] [CrossRef]
- Nam, M.H.; Park, M.S.; Kim, H.S.; Kim, T.I.; Kim, H.G. Cladosporium cladosporioides and C. tenuissimum cause blossom blight in strawberry in Korea. Mycobiology 2015, 43, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Ayoubi, N.; Soleimani, M.J.; Zare, R. Cladosporium species, a new challenge in strawberry production in Iran. Phytopathol. Mediterr. 2017, 56, 486–493. Available online: http://www.jstor.org/stable/44809359 (accessed on 9 April 2024).
- Xu, H.; Quan, Q.; Chang, X.; Ge, S.; Xu, S.; Wang, R.; Xu, Y.; Luo, Z.; Shan, Y.; Ding, S. Maintaining the balance of fungal community through active packaging film makes strawberry fruit pose pleasant flavor during storage. Postharvest Biol. Technol. 2024, 211, 112815. [Google Scholar] [CrossRef]
- Sangiorgio, D.; Cellini, A.; Donati, I.; Ferrari, E.; Tanunchai, B.; Mohamed Wahdan, S.F.; Sadubsarn, D.; Farneti, B.; Checcucci, A.; Buscot, F.; et al. Taxonomical and functional composition of strawberry microbiome is genotype-dependent. J. Adv. Res. 2022, 42, 189–204. [Google Scholar] [CrossRef]
- Fernández-Ortuño, D.; Li, X.P.; Wang, F.; Schnabel, G. First report of gray mold of strawberry caused by Botrytis caroliniana in North Carolina. Plant Dis. 2012, 96, 914. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhao, Z.; Virag, A.; Becker, T.; Zhao, L.; Liu, W.; Xia, Y. Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries. Bio-Protocol 2023, 13, e4859. [Google Scholar] [CrossRef]
- Lachance, M.A.; Starmer, W.T. Kurtzmaniella gen. nov. and description of the heterothallic, haplontic yeast species Kurtzmaniella cleridarum sp. nov.; the teleomorph of Candida cleridarum. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 2, 520–524. [Google Scholar] [CrossRef]
- Li, A.H.; Yuan, F.X.; Groenewald, M.; Bensch, K.; Yurkov, A.M.; Li, K.; Han, P.J.; Guo, L.D.; Aime, M.C.; Sampaio, J.P.; et al. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: Proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud. Mycol. 2020, 96, 17–140. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, X.; Zheng, X.; Apaliya, M.T.; Yang, Q.; Zhao, L.; Gu, X.; Zhang, H. Control of postharvest blue mold decay in pears by Meyerozyma guilliermondii and it’s effects on the protein expression profile of pears. Postharvest Biol. Technol. 2018, 136, 124–131. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, L.; Li, Z.; Li, C.; Li, B.; Gu, X.; Zhang, X.; Zhang, H. Screening and identification of an antagonistic yeast controlling postharvest blue mold decay of pears and the possible mechanisms involved. Biol. Control 2019, 133, 26–33. [Google Scholar] [CrossRef]
- Apaliya, M.T.; Zhang, H.; Yang, Q.; Zheng, X.; Zhao, L.; Kwaw, E.; Mahunu, G.K. Hanseniaspora uvarum enhanced with trehalose induced defense-related enzyme activities and relative genes expression levels against Aspergillus tubingensis in table grapes. Postharvest Biol. Technol. 2017, 132, 162–170. [Google Scholar] [CrossRef]
- Kachalkin, A.V.; Glushakova, A.M.; Venzhik, A.S. Presence of clinically significant endophytic yeasts in agricultural crops: Monitoring and ecological safety assessment. IOP Conf. Ser. Earth Environ. Sci. 2021, 723, 042005. [Google Scholar] [CrossRef]
- Li, J.; Hong, M. Impact of Candida railenensis during fermentation on the aromatic profile of Vidal blanc icewine. Front. Microbiol. 2023, 14, 1192006. [Google Scholar] [CrossRef] [PubMed]
- Breierová, E.; Čertík, M.; Márová, I.; Vadkertiová, R. The Effect of Zn(II) Ions and Reactive Oxygen on the Uptake of Zinc and Production of Carotenoids by Selected Red Yeasts. Chem. Biodivers. 2018, 15, e1800069. [Google Scholar] [CrossRef] [PubMed]
- Baroncelli, R.; Zapparata, A.; Sarrocco, S.; Sukno, S.A.; Lane, C.R.; Thon, M.R.; Vannacci, G.; Holub, E.; Sreenivasaprasad, S. Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum species. PLoS ONE 2015, 10, e0129140. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Li, X.; Gao, Q.-H.; Geng, C.; Duan, K. Colletotrichum species pathogenic to strawberry: Discovery history, global diversity, prevalence in China, and the host range of top two species. Phytopathol. Res. 2022, 4, 42. [Google Scholar] [CrossRef]
- de Hoog, G.S.; Guarro, J.; Gené, J.; Ahmed, S.; Al-Hatmi, A.M.S.; Figueras, M.J.; Vitale, R.G. Atlas of Clinical Fungi, 4th ed.; Foundation Atlas of Clinical Fungi: Hilversum, The Netherlands, 2020. [Google Scholar]
- Xing, H.; Wang, J.; Sun, Y.; Wang, H. Recent advances in the allergic cross-reactivity between fungi and foods. J. Immunol. Res. 2022, 2022, 7583400. [Google Scholar] [CrossRef] [PubMed]
- de Garcia, V.; Zalar, P.; Brizzio, S.; Gunde-Cimerman, N.; van Broock, M. Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiol. Ecol. 2012, 82, 523–539. [Google Scholar] [CrossRef]
- Yadav, V.; Heitman, J. On fruits and fungi: A risk of antifungal usage in food storage and distribution in driving drug resistance in Candida auris. mBio 2022, 13, e00739-22. [Google Scholar] [CrossRef] [PubMed]
- Kohler, J.R.; Hube, B.; Puccia, R.; Casadevall, A.; Perfect, J.R. Fungi that infect humans. Microbiol. Spectr. 2017, 5, 10–1128. [Google Scholar] [CrossRef]
- Lo, H.J.; Tsai, S.H.; Chu, W.L.; Chen, Y.Z.; Zhou, Z.L.; Chen, H.F.; Lee, C.F.; Yang, Y.L. Fruits as the vehicle of drug resistant pathogenic yeasts. J. Infect. 2017, 75, 254–262. [Google Scholar] [CrossRef] [PubMed]
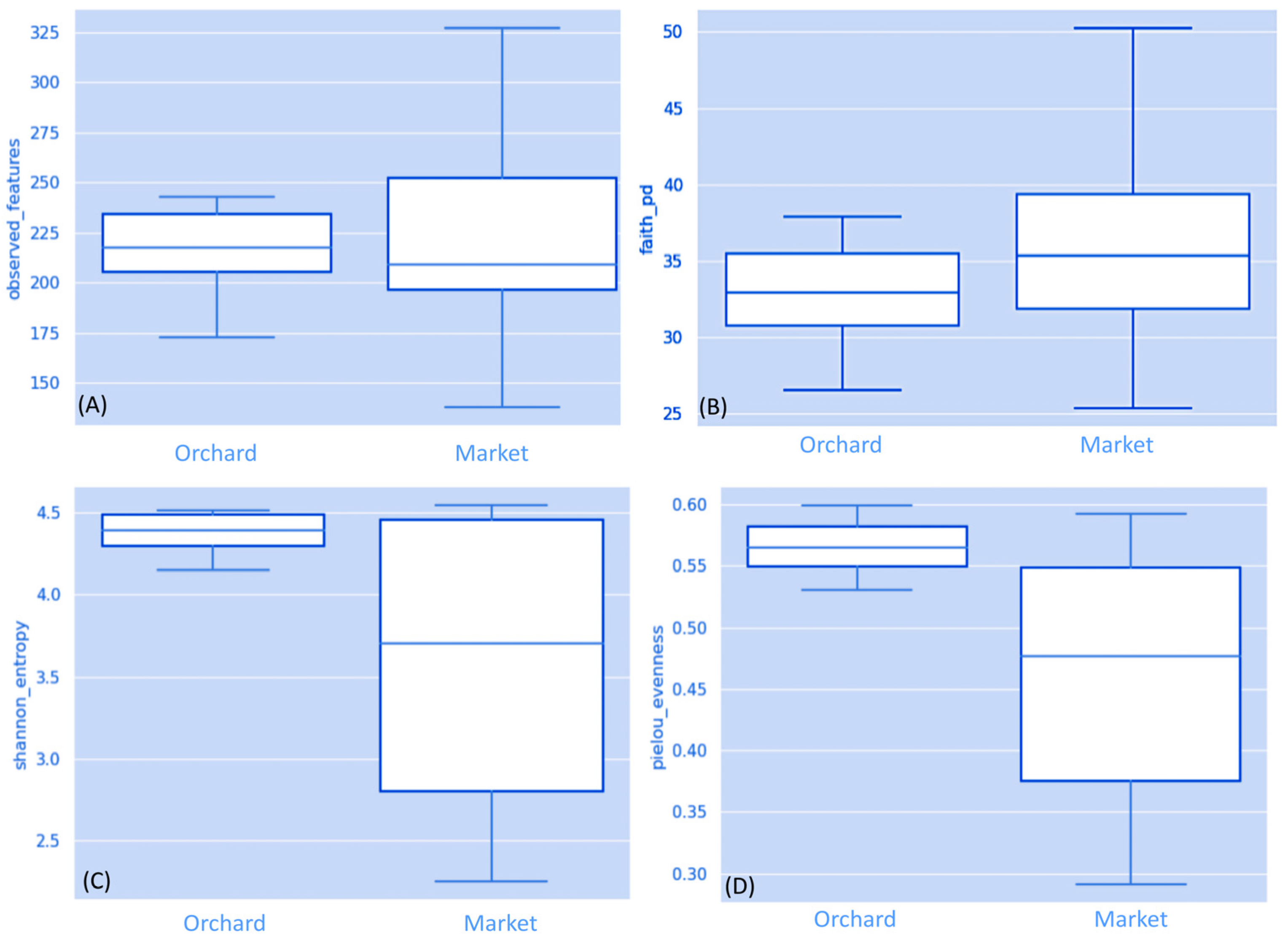


| Sample Origin | Sample ID | Number Reads Passing Quality Filtering | % Reads Classified to Genus | Shannon Species Diversity | Number of Species Identified |
|---|---|---|---|---|---|
| Orchard | F4FL1-ITS | 402,166 | 88.15% | 2.47 | 582 |
| F4FL2-ITS | 413,205 | 83.63% | 2.47 | 563 | |
| F4FL3-ITS | 362,685 | 85.13% | 2.38 | 567 | |
| F4FL4-ITS | 374,315 | 86.46% | 2.56 | 598 | |
| F4FL5-ITS | 473,020 | 82.99% | 2.61 | 628 | |
| F4FL6-ITS | 330,732 | 84.94% | 2.60 | 783 | |
| Market | FP1-ITS | 536,563 | 96.52% | 3.05 | 848 |
| FP2-ITS | 457,487 | 92.27% | 2.78 | 699 | |
| FP3-ITS | 479,798 | 97.05% | 2.54 | 644 | |
| FP4-ITS | 546,063 | 96.72% | 2.26 | 737 | |
| FP5-ITS | 395,658 | 89.55% | 2.85 | 587 | |
| FP6-ITS | 241,369 | 92.60% | 2.45 | 684 |
| Metrics | Pseudo-F * | p-Value | q-Value |
|---|---|---|---|
| Bray–Curtis dissimilarity | 8.19245731 | 0.006 | 0.006 |
| Jaccard distance | 3.11991142 | 0.003 | 0.003 |
| Unweighted_unifrac distance | 3.94475851 | 0.003 | 0.003 |
| Weighted_unifrac distance | 22.2981783 | 0.004 | 0.004 |
| Taxon | W | Reject Null Hypothesis | % of Abundance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 25 | 50 | 75 | 100 | 0 | 25 | 50 | 75 | 100 | |||
| Orchard | Market | |||||||||||
| Phyllozyma spp. | 369 | TRUE | 264 | 533.25 | 671 | 732.25 | 1057 | 1 | 1 | 1 | 1 | 1 |
| Kurtzmaniella spp. | 367 | TRUE | 1 | 1 | 1 | 1 | 1 | 219 | 261.75 | 1128 | 2154.75 | 7074 |
| Hanseniaspora spp. | 348 | TRUE | 1 | 1 | 1 | 1 | 1 | 119 | 134 | 387.5 | 652.25 | 2136 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tenea, G.N.; Reyes, P.; Molina, D. Fungal Mycobiome of Mature Strawberry Fruits (Fragaria x ananassa Variety ‘Monterey’) Suggests a Potential Market Site Contamination with Harmful Yeasts. Foods 2024, 13, 1175. https://doi.org/10.3390/foods13081175
Tenea GN, Reyes P, Molina D. Fungal Mycobiome of Mature Strawberry Fruits (Fragaria x ananassa Variety ‘Monterey’) Suggests a Potential Market Site Contamination with Harmful Yeasts. Foods. 2024; 13(8):1175. https://doi.org/10.3390/foods13081175
Chicago/Turabian StyleTenea, Gabriela N., Pamela Reyes, and Diana Molina. 2024. "Fungal Mycobiome of Mature Strawberry Fruits (Fragaria x ananassa Variety ‘Monterey’) Suggests a Potential Market Site Contamination with Harmful Yeasts" Foods 13, no. 8: 1175. https://doi.org/10.3390/foods13081175
APA StyleTenea, G. N., Reyes, P., & Molina, D. (2024). Fungal Mycobiome of Mature Strawberry Fruits (Fragaria x ananassa Variety ‘Monterey’) Suggests a Potential Market Site Contamination with Harmful Yeasts. Foods, 13(8), 1175. https://doi.org/10.3390/foods13081175








