Mechanistic Study of Novel Dipeptidyl Peptidase IV Inhibitory Peptides from Goat’s Milk Based on Peptidomics and In Silico Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. In Silico-Simulated Hydrolysis of DPP-IV Inhibitory Peptides
2.3. Enzymatic Hydrolysis of Goat Milk Protein
2.4. Peptidomic Profiling Using LC-MS/MS
2.5. Screening of DPP-IV Inhibitory Peptides
2.5.1. Virtual Screening of Potential DPP-IV Inhibitory Peptides
2.5.2. In Silico Analysis of Pharmacokinetic Properties and Physicochemical Properties
2.6. Solid-Phase Synthesis of Peptides
2.7. DPP-IV Inhibitory Assay
2.8. In Situ Cell-Based DPP-IV Activity Assay
2.8.1. Cell Culture
2.8.2. Cell Viability Assay
2.8.3. In Situ Inhibition of DPP-IV
2.9. Molecular Docking Analysis
2.10. Statistical Analysis
3. Results and Discussion
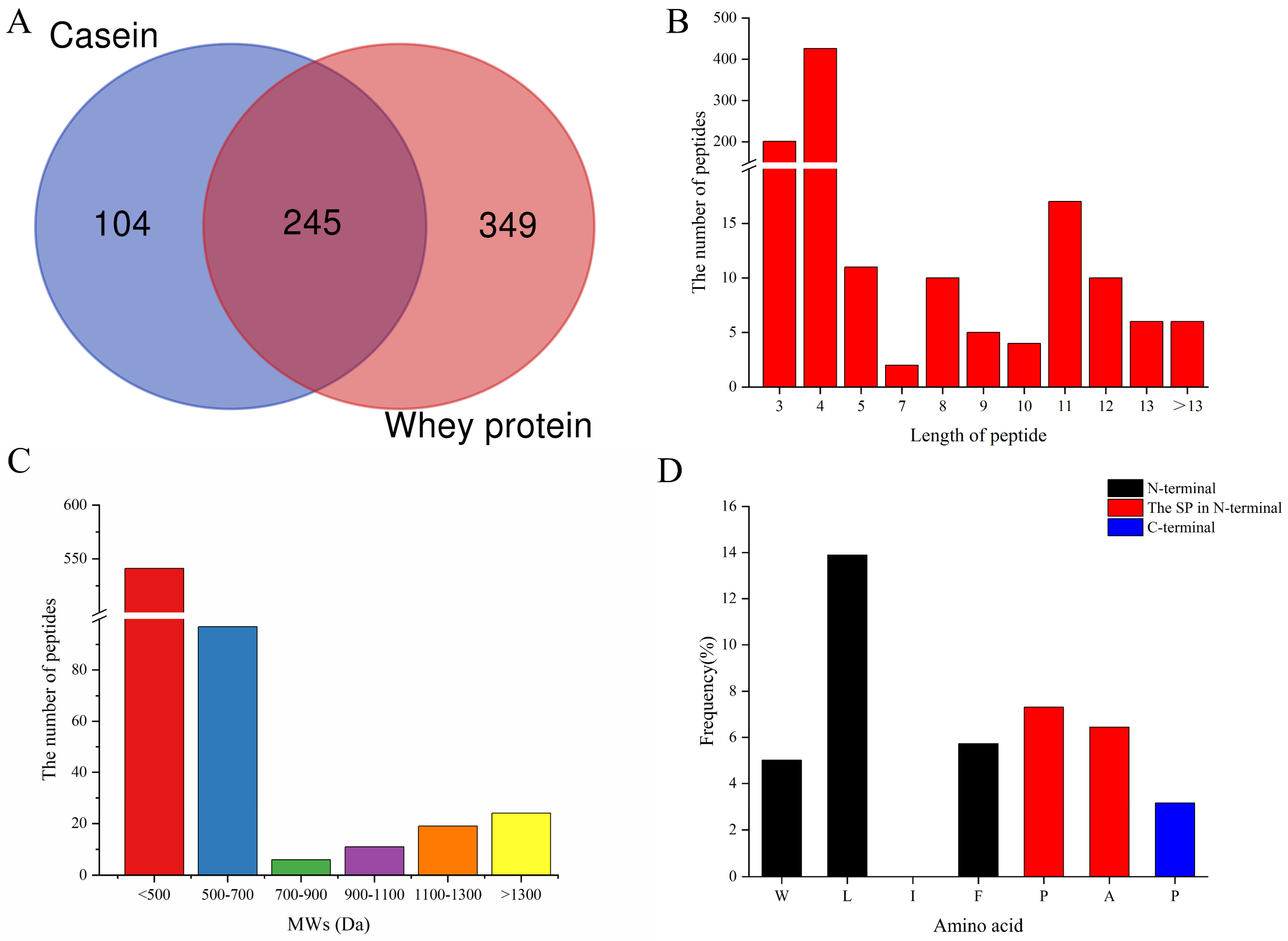
3.1. Analysis of Goat Milk Protein Hydrolysate Enriched in DPP-IV Inhibitory Peptides
3.2. The Amino Acid Profiles of Identified Peptides
3.3. Virtual Screening of Potential DPP-IV Inhibitory Peptides
3.4. Prediction of Pharmacokinetic Properties and Physicochemical Properties
3.5. Validation and Interactive Mechanisms of Screened DPP-IV Inhibitory Peptides
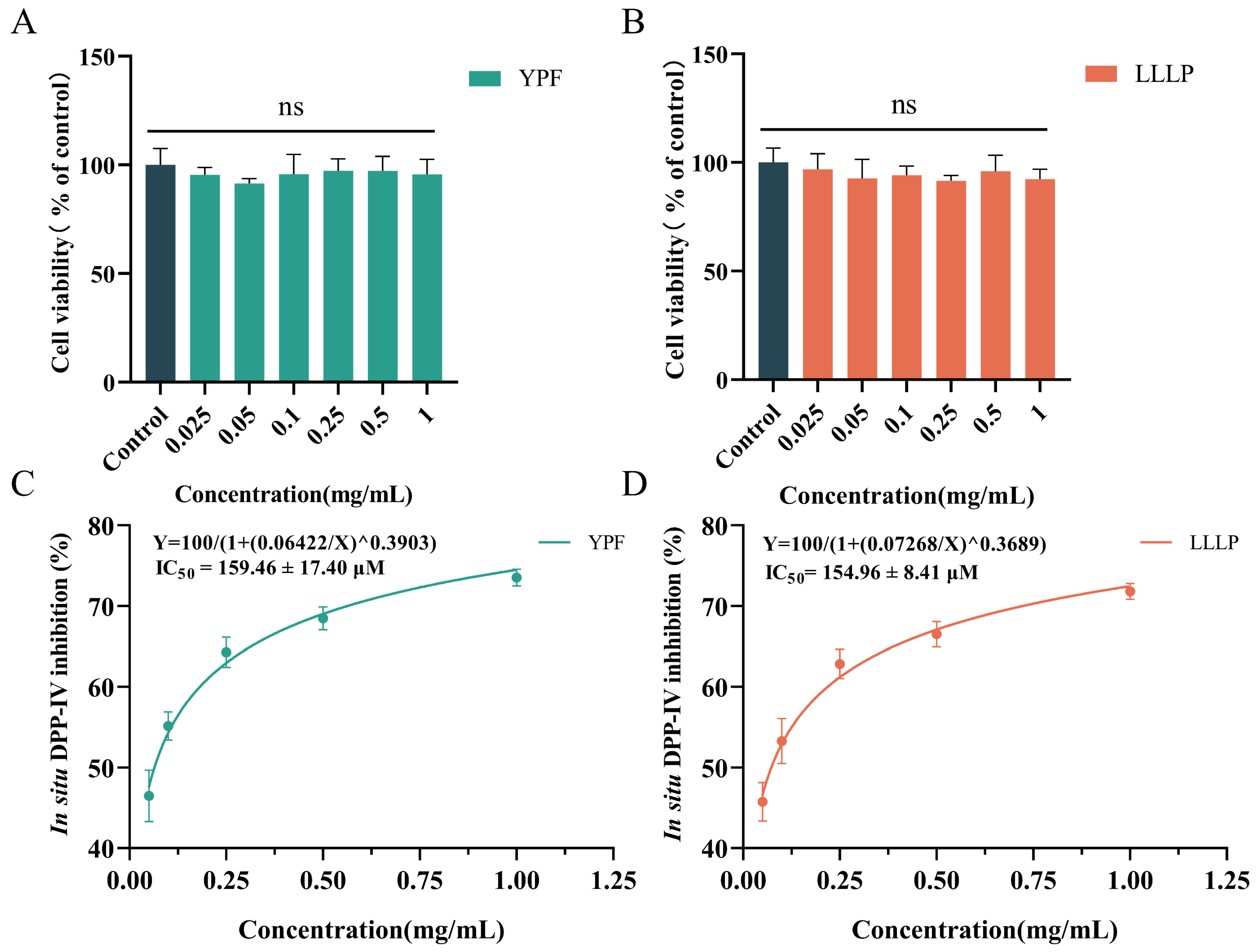
3.6. Cell Viability and In Situ DPP-IV Inhibition of Screening Peptides on Caco-2 Cells
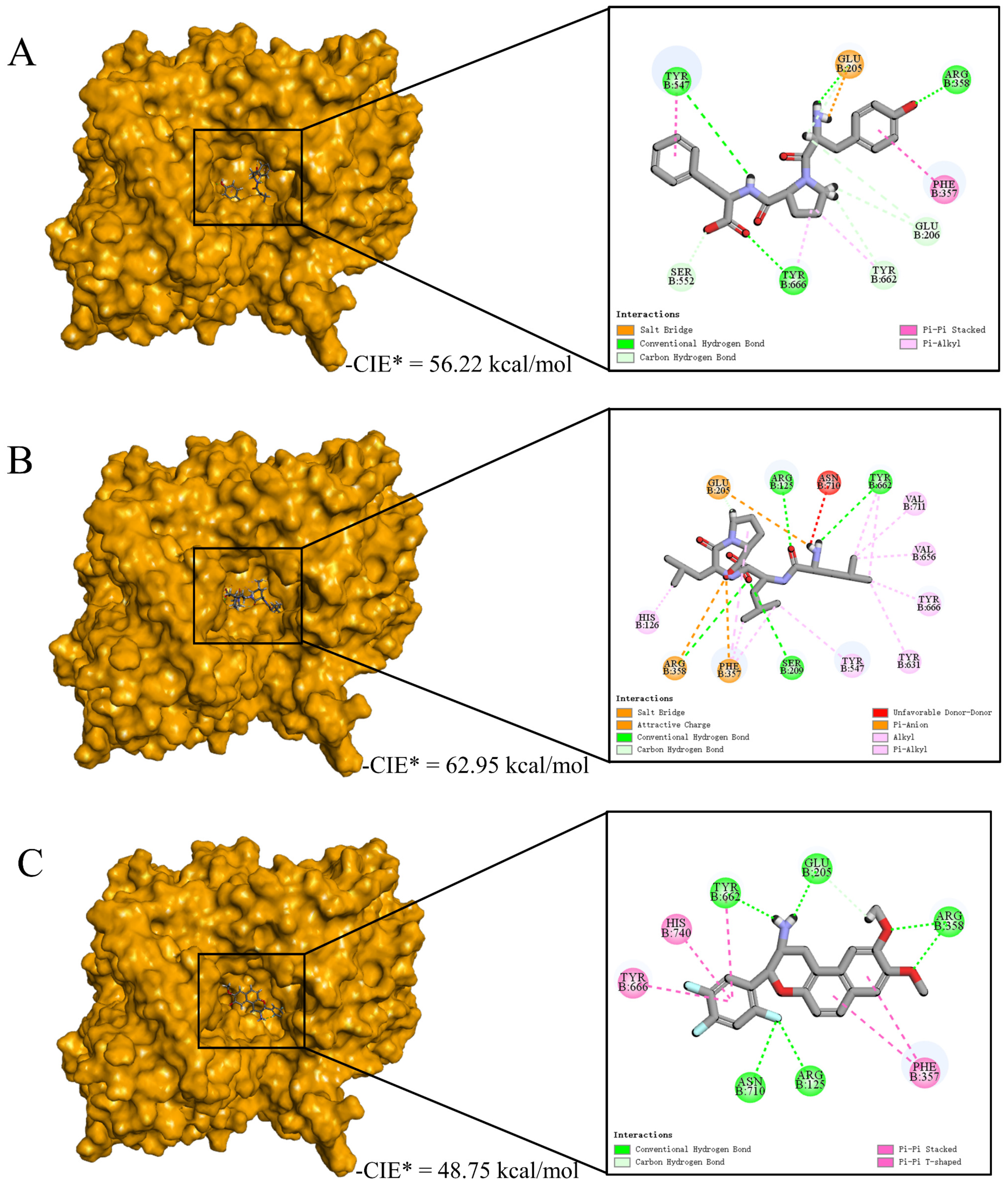
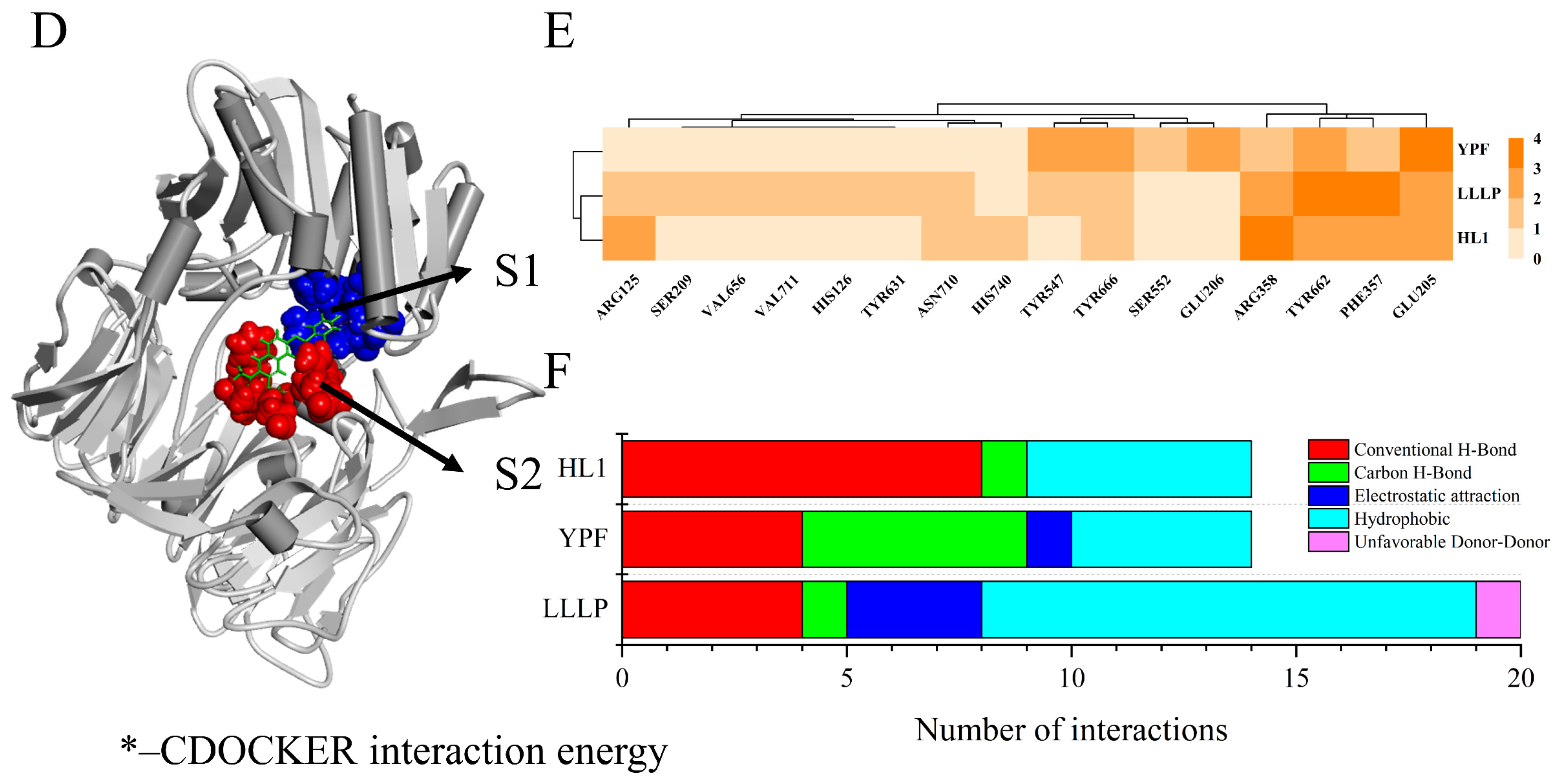
3.7. Interactive Mechanism of Screening Peptides against DPP-IV
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and Regional Diabetes Prevalence Estimates for 2019 and Projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th Edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.H.Y.; Shin, J.Y. Treating type 2 diabetes: Moving towards precision medicine. Lancet Digit. Health 2022, 4, e851–e852. [Google Scholar] [CrossRef]
- Deacon, C.F. Dipeptidyl Peptidase 4 Inhibitors in the Treatment of Type 2 Diabetes Mellitus. Nat. Rev. Endocrinol. 2020, 16, 642–653. [Google Scholar] [CrossRef]
- Andersen, E.S.; Deacon, C.F.; Holst, J.J. Do We Know the True Mechanism of Action of the DPP-4 Inhibitors? Diabetes Obes. Metab. 2018, 20, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Bouchi, R.; Sugiyama, T.; Goto, A.; Imai, K.; Ihana-Sugiyama, N.; Ohsugi, M.; Yamauchi, T.; Kadowaki, T.; Ueki, K. Retrospective Nationwide Study on the Trends in First-Line Antidiabetic Medication for Patients with Type 2 Diabetes in Japan. J. Diabetes Investig. 2022, 13, 280–291. [Google Scholar] [CrossRef]
- Suzuki, L.; Kanazawa, A.; Uzawa, H.; Osonoi, Y.; Masuyama, A.; Azuma, K.; Takeno, K.; Takayanagi, N.; Sato, J.; Someya, Y.; et al. Safety and Efficacy of Metformin Up-Titration in Japanese Patients with Type 2 Diabetes Mellitus Treated with Vildagliptin and Low-Dose Metformin. Expert. Opin. Pharmacother. 2017, 18, 1921–1928. [Google Scholar] [CrossRef]
- Willemen, M.J.; Mantel-Teeuwisse, A.K.; Straus, S.M.; Meyboom, R.H.; Egberts, T.C.; Leufkens, H.G. Use of Dipeptidyl Peptidase-4 Inhibitors and the Reporting of Infections: A Disproportionality Analysis in the World Health Organization VigiBase. Diabetes Care 2011, 34, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Jedlowski, P.M.; Jedlowski, M.F.; Fazel, M.T. DPP-4 Inhibitors and Increased Reporting Odds of Bullous Pemphigoid: A Pharmacovigilance Study of the FDA Adverse Event Reporting System (FAERS) from 2006 to 2020. Am. J. Clin. Dermatol. 2021, 22, 891–900. [Google Scholar] [CrossRef]
- Bommer, C.; Sagalova, V.; Heesemann, E.; Manne-Goehler, J.; Atun, R.; Bärnighausen, T.; Davies, J.; Vollmer, S. Global Economic Burden of Diabetes in Adults: Projections from 2015 to 2030. Diabetes Care 2018, 41, 963–970. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Food-Derived Dipeptidyl-Peptidase IV Inhibitors as a Potential Approach for Glycemic Regulation—Current Knowledge and Future Research Considerations. Trends Food Sci. Technol. 2016, 54, 1–16. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Prospects for the Management of Type 2 Diabetes Using Food Protein-Derived Peptides with Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Activity. Curr. Opin. Food Sci. 2016, 8, 19–24. [Google Scholar] [CrossRef]
- Oseguera Toledo, M.E.; Gonzalez de Mejia, E.; Sivaguru, M.; Amaya-Llano, S.L. Common Bean (Phaseolus vulgaris L.) Protein-Derived Peptides Increased Insulin Secretion, Inhibited Lipid Accumulation, Increased Glucose Uptake and Reduced the Phosphatase and Tensin Homologue Activation in Vitro. J. Funct. Foods 2016, 27, 160–177. [Google Scholar] [CrossRef]
- Wang, Y.; Landheer, S.; van Gilst, W.H.; van Amerongen, A.; Hammes, H.P.; Henning, R.H.; Deelman, L.E.; Buikema, H. Attenuation of Renovascular Damage in Zucker Diabetic Fatty Rat by NWT-03, an Egg Protein Hydrolysate with ACE- and DPP4-Inhibitory Activity. PLoS ONE 2012, 7, e46781. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Evaluation of the Potential of Dietary Proteins as Precursors of Dipeptidyl Peptidase (DPP)-IV Inhibitors by an in-Silico Approach. J. Funct. Foods 2012, 4, 403–422. [Google Scholar] [CrossRef]
- Ishikawa, Y.; Hira, T.; Inoue, D.; Harada, Y.; Hashimoto, H.; Fujii, M.; Kadowaki, M.; Hara, H. Rice Protein Hydrolysates Stimulate GLP-1 Secretion, Reduce GLP-1 Degradation, and Lower the Glycemic Response in Rats. Food Funct. 2015, 6, 2525–2534. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, R.; Chen, X.; Zeng, Z.; Ma, H.; Chen, S. Dipeptidyl Peptidase IV-Inhibitory Peptides Derived from Silver Carp (Hypophthalmichthys molitrix Val.) Proteins. J. Agric. Food Chem. 2016, 64, 831–839. [Google Scholar] [CrossRef]
- Huang, S.L.; Hung, C.C.; Jao, C.L.; Tung, Y.S.; Hsu, K.C. Porcine Skin Gelatin Hydrolysate as a Dipeptidyl Peptidase IV Inhibitor Improves Glycemic Control in Streptozotocin-Induced Diabetic Rats. J. Funct. Foods 2014, 11, 235–242. [Google Scholar] [CrossRef]
- Patil, P.; Mandal, S.; Tomar, S.K.; Anand, S. Food Protein-Derived Bioactive Peptides in Management of Type 2 Diabetes. Eur. J. Nutr. 2015, 54, 863–880. [Google Scholar] [CrossRef]
- Jankiewicz, M.; van Lee, L.; Biesheuvel, M.; Brouwer-Brolsma, E.M.; van der Zee, L.; Szajewska, H. The Effect of Goat-Milk-Based Infant Formulas on Growth and Safety Parameters: A Systematic Review and Meta-Analysis. Nutrients 2023, 15, 2110. [Google Scholar] [CrossRef]
- Mohanty, D.P.; Mohapatra, S.; Misra, S.; Sahu, P.S. Milk Derived Bioactive Peptides and Their Impact on Human Health—A Review. Saudi J. Biol. Sci. 2016, 23, 577–583. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, R.; Ma, H.; Chen, S. Isolation and Identification of Dipeptidyl Peptidase IV-Inhibitory Peptides from Trypsin/Chymotrypsin-Treated Goat Milk Casein Hydrolysates by 2D-TLC and LC-MS/MS. J. Agric. Food Chem. 2015, 63, 8819–8828. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Jiang, C.; Wang, S.; Jing, H.; Mo, L.; Ma, C.; Wang, H. Preparation, Identification, and Inhibitory Mechanism of Dipeptidyl Peptidase IV Inhibitory Peptides from Goat Milk Whey Protein. J. Food Sci. 2023, 88, 3577–3593. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, W.M.; Guimarães Gomes, A.C.; de Caldas Nobre, M.S.; de Souza Pereira, Á.M.; dos Santos Pereira, E.V.; dos Santos, K.M.O.; Florentino, E.R.; Alonso Buriti, F.C. Goat Milk as a Natural Source of Bioactive Compounds and Strategies to Enhance the Amount of These Beneficial Components. Int. Dairy. J. 2023, 137, 105515. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, L.; Wu, G.; Liu, T.; Qi, X.; Zhang, H. Rapid Screening of Novel Dipeptidyl Peptidase-4 Inhibitory Peptides from Pea (Pisum sativum L.) Protein Using Peptidomics and Molecular Docking. J. Agric. Food Chem. 2022, 70, 10221–10228. [Google Scholar] [CrossRef] [PubMed]
- Khedr, M.A.; Mohafez, O.M.M.; Al-Haider, I.A. Virtual Screening-Based Discovery of Potent Hypoglycemic Agents: In Silico, Chemical Synthesis and Biological Study. Curr. Comput. Aided Drug Des. 2019, 16, 741–756. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Li, S.; Luo, Q.; Shen, G.; Wu, H.; Li, M.; Liu, X.; Chen, A.; Ye, M.; Zhang, Z. Discovery and Identification of Antimicrobial Peptides in Sichuan Pepper (Zanthoxylum bungeanum Maxim) Seeds by Peptidomics and Bioinformatics. Appl. Microbiol. Biotechnol. 2019, 103, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Pescuma, M.; Hébert, E.M.; Haertlé, T.; Chobert, J.M.; Mozzi, F.; Font De Valdez, G. Lactobacillus Delbrueckii Subsp. Bulgaricus CRL 454 Cleaves Allergenic Peptides of β-Lactoglobulin. Food Chem. 2015, 170, 407–414. [Google Scholar] [CrossRef]
- Zhu, K.; Zheng, Z.; Dai, Z. Identification of Antifreeze Peptides in Shrimp Byproducts Autolysate Using Peptidomics and Bioinformatics. Food Chem. 2022, 383, 132568. [Google Scholar] [CrossRef]
- Foroutan, B.; Abbasian Najafabadi, A.R. Capabilities of Bioinformatics Tools for Optimizing Physicochemical Features of Proteins Used in Nano Biosensors: A Short Overview of the Tools Related to Bioinformatics. Biochem. Biophys. Rep. 2021, 27, 101094. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y.; Tang, H.; Li, H.; Da, S.; Ciren, D.; Peng, X.; Zhao, K. Identification, Characterization, and Insights into the Mechanism of Novel Dipeptidyl Peptidase-IV Inhibitory Peptides from Yak Hemoglobin by in Silico Exploration, Molecular Docking, and in Vitro Assessment. Int. J. Biol. Macromol. 2024, 259, 129191. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, D.; Yu, Z.; Ding, L.; Liu, J. Novel Membrane Peptidase Inhibitory Peptides with Activity against Angiotensin Converting Enzyme and Dipeptidyl Peptidase IV Identified from Hen Eggs. J. Funct. Foods. 2020, 64, 103649. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Mazzocchi, C.; Paolella, S.; FitzGerald, R.J. Release of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides from Milk Protein Isolate (MPI) during Enzymatic Hydrolysis. Food Res. Int. 2017, 94, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Kęska, P.; Stadnik, J. Potential Dpp Iv Inhibitory Peptides from Dry-Cured Pork Loins after Hydrolysis: An in Vitro and in Silico Study. Curr. Issues Mol. Biol. 2021, 43, 1335–1349. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, M.; Pan, F.; Li, J.; Dou, R.; Wang, X.; Wang, Y.; He, Y.; Wang, S.; Cai, S. In Silico Analysis of Novel Dipeptidyl Peptidase-IV Inhibitory Peptides Released from Macadamia Integrifolia Antimicrobial Protein 2 (MiAMP2) and the Possible Pathways Involved in Diabetes Protection. Curr. Res. Food Sci. 2021, 4, 603–611. [Google Scholar] [CrossRef]
- Rahfeld, J.; Schierhorn, M.; Hartrodt, B.; Neubert, K.; Heins, J. Are Diprotin A (Lie-Pro-Lie) and Diprotin B (Val-Pro-Leu) Inhibitors or Substrates of Dipeptidyl Peptidase IV? Biochim. Biophys. Acta 1991, 1076, 314–316. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Wu, T.; Wang, W.; Li, Y.; Liu, X.; Ding, L. Preparation and Identification of Dipeptidyl Peptidase IV Inhibitory Peptides from Quinoa Protein. Food Res. Int. 2022, 156, 111176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, M.; Zhang, G.; Tian, Y.; Kong, F.; Xiong, S.; Zhao, S.; Jia, D.; Manyande, A.; Du, H. Identification of Novel Antioxidant Peptides from Snakehead (Channa argus) Soup Generated during Gastrointestinal Digestion and Insights into the Anti-Oxidation Mechanisms. Food Chem. 2021, 337, 127921. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. Features of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides from Dietary Proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; Fitzgerald, R.J. Structure Activity Relationship Modelling of Milk Protein-Derived Peptides with Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Activity. Peptides 2016, 79, 1–7. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Cadamuro, C.; Le Gouic, A.; Mudgil, P.; Maqsood, S.; FitzGerald, R.J. Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Properties of a Camel Whey Protein Enriched Hydrolysate Preparation. Food Chem. 2019, 279, 70–79. [Google Scholar] [CrossRef]
- Li-Chan, E.C.Y.; Hunag, S.L.; Jao, C.L.; Ho, K.P.; Hsu, K.C. Peptides Derived from Atlantic Salmon Skin Gelatin as Dipeptidyl-Peptidase IV Inhibitors. J. Agric. Food Chem. 2012, 60, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, J.; Mu, T.; Zhang, H.; Cao, J.; Li, H.; Tang, H.; Chen, L.; Liu, H.; Xu, X.; et al. Selection of Goat β-Casein Derived ACE-Inhibitory Peptide SQPK and Insights into Its Effect and Regulatory Mechanism on the Function of Endothelial Cells. Int. J. Biol. Macromol. 2023, 253, 127312. [Google Scholar] [CrossRef]
- Power, O.; Nongonierma, A.B.; Jakeman, P.; Fitzgerald, R.J. Food Protein Hydrolysates as a Source of Dipeptidyl Peptidase IV Inhibitory Peptides for the Management of Type 2 Diabetes. Proc. Nutr. Soc. 2014, 73, 34–46. [Google Scholar] [CrossRef]
- Mu, X.; Wang, R.; Cheng, C.; Ma, Y.; Zhang, Y.; Lu, W. Preparation, Structural Properties, and in Vitro and in Vivo Activities of Peptides against Dipeptidyl Peptidase IV (DPP-IV) and α-Glucosidase: A General Review. Crit. Rev. Food Sci. Nutr. 2023. Published online. [Google Scholar] [CrossRef]
- Lambeir, A.-M.; Durinx, C.; Scharpé, S.; De Meester, I. Dipeptidyl-Peptidase IV from Bench to Bedside: An Update on Structural Properties. Functions, and Clinical Aspects of the Enzyme DPP IV. Crit. Rev. Clin. Lab. Sci. 2003, 40, 209–294. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Isolation and Characterization of Peptides with Dipeptidyl Peptidase-IV Inhibitory Activity from Pepsin-Treated Bovine Whey Proteins. Peptides 2014, 54, 39–48. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Shoombuatong, W.; Charoenkwan, P.; Kanthawong, S.; Nantasenamat, C.; Hasan, M.M. IDPPIV-SCM: A Sequence-Based Predictor for Identifying and Analyzing Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides Using a Scoring Card Method. J. Proteome Res. 2020, 19, 4125–4136. [Google Scholar] [CrossRef]
- Wang, X.; Deng, Y.; Xie, P.; Liu, L.; Zhang, C.; Cheng, J.; Zhang, Y.; Liu, Y.; Huang, L.; Jiang, J. Novel Bioactive Peptides from Ginkgo Biloba Seed Protein and Evaluation of Their α-Glucosidase Inhibition Activity. Food Chem. 2023, 404, 134481. [Google Scholar] [CrossRef]
- Du, Z.; Comer, J.; Li, Y. Bioinformatics Approaches to Discovering Food-Derived Bioactive Peptides: Reviews and Perspectives. TrAC Trends Anal. Chem. 2023, 162, 117051. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Paolella, S.; Mudgil, P.; Maqsood, S.; FitzGerald, R.J. Identification of Novel Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides in Camel Milk Protein Hydrolysates. Food Chem. 2018, 244, 340–348. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Inhibition of Dipeptidyl Peptidase (DPP)-IV and α-Glucosidase Activities by Pepsin-Treated Whey Proteins. J. Agric. Food Chem. 2013, 61, 7500–7506. [Google Scholar] [CrossRef]
- Akan, E. An Evaluation of the in Vitro Antioxidant and Antidiabetic Potentials of Camel and Donkey Milk Peptides Released from Casein and Whey Proteins. J. Food Sci. Technol. 2021, 58, 3743–3751. [Google Scholar] [CrossRef]
- Parastouei, K.; Jabbari, M.; Javanmardi, F.; Barati, M.; Mahmoudi, Y.; Khalili-Moghadam, S.; Ahmadi, H.; Davoodi, S.H.; Mousavi Khaneghah, A. Estimation of Bioactive Peptide Content of Milk from Different Species Using an in-Silico Method. Amino Acids. 2023, 55, 1261–1278. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Guan, C.; Luo, J.; Li, S.; Tan, Z.L.; Wang, Y.; Chen, H.; Yamamoto, N.; Zhang, C.; Lu, Y.; Chen, J.; et al. Exploration of DPP-IV Inhibitory Peptide Design Rules Assisted by the Deep Learning Pipeline That Identifies the Restriction Enzyme Cutting Site. ACS Omega 2023, 8, 39662–39672. [Google Scholar] [CrossRef]
- Yeung, J.H.K.; Or, P.M.Y. Polysaccharide Peptides from Coriolus Versicolor Competitively Inhibit Model Cytochrome P450 Enzyme Probe Substrates Metabolism in Human Liver Microsomes. Phytomedicine 2012, 19, 457–463. [Google Scholar] [CrossRef]
- Silveira, S.T.; Martínez-Maqueda, D.; Recio, I.; Hernández-Ledesma, B. Dipeptidyl Peptidase-IV Inhibitory Peptides Generated by Tryptic Hydrolysis of a Whey Protein Concentrate Rich in β-Lactoglobulin. Food Chem. 2013, 141, 1072–1077. [Google Scholar] [CrossRef]
- Magnin, D.R.; Robl, J.A.; Sulsky, R.B.; Augeri, D.J.; Huang, Y.; Simpkins, L.M.; Taunk, P.C.; Betebenner, D.A.; Robertson, J.G.; Abboa-Offei, B.E.; et al. Synthesis of Novel Potent Dipeptidyl Peptidase IV Inhibitors with Enhanced Chemical Stability: Interplay between the N-Terminal Amino Acid Alkyl Side Chain and the Cyclopropyl Group of α -Aminoacyl-L-Cis-4,5-Methanoprolinenitrile-Based Inhibitors. J. Med. Chem. 2004, 47, 2587–2598. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. Inhibition of Dipeptidyl Peptidase IV (DPP-IV) by Proline Containing Casein-Derived Peptides. J. Funct. Foods 2013, 5, 1909–1917. [Google Scholar] [CrossRef]
- Harnedy, P.A.; O’Keeffe, M.B.; Fitzgerald, R.J. Purification and Identification of Dipeptidyl Peptidase (DPP) IV Inhibitory Peptides from the Macroalga Palmaria Palmata. Food Chem. 2015, 172, 400–406. [Google Scholar] [CrossRef]
- Tulipano, G.; Sibilia, V.; Caroli, A.M.; Cocchi, D. Whey Proteins as Source of Dipeptidyl Dipeptidase IV (Dipeptidyl Peptidase-4) Inhibitors. Peptides 2011, 32, 835–838. [Google Scholar] [CrossRef]
- Bella, A.M.; Erickson, R.H.; Kim’, Y.S. Rat Intestinal Brush Border Membrane Dipeptidyl-Aminopeptidase IV: Kinetic Properties and Substrate Specificities of the Purified Enzyme. Arch. Biochem. Biophys. 1982, 218, 156–162. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Mooney, C.; Shields, D.C.; Fitzgerald, R.J. In Silico Approaches to Predict the Potential of Milk Protein-Derived Peptides as Dipeptidyl Peptidase IV (DPP-IV) Inhibitors. Peptides 2014, 57, 43–51. [Google Scholar] [CrossRef]
- Harnedy-Rothwell, P.A.; McLaughlin, C.M.; O’Keeffe, M.B.; Le Gouic, A.V.; Allsopp, P.J.; McSorley, E.M.; Sharkey, S.; Whooley, J.; McGovern, B.; O’Harte, F.P.M.; et al. Identification and Characterisation of Peptides from a Boarfish (Capros aper) Protein Hydrolysate Displaying In vitro Dipeptidyl Peptidase-IV (DPP-IV) Inhibitory and Insulinotropic Activity. Food Res. Int. 2020, 131, 108989. [Google Scholar] [CrossRef]
- Liu, R.; Zhou, L.; Zhang, Y.; Sheng, N.J.; Wang, Z.K.; Wu, T.Z.; Wang, X.Z.; Wu, H. Rapid Identification of Dipeptidyl Peptidase-IV (Dpp-IV) Inhibitory Peptides from Ruditapes Philippinarum Hydrolysate. Molecules 2017, 22, 1714. [Google Scholar] [CrossRef]
- Juillerat-Jeanneret, L. Dipeptidyl Peptidase IV and Its Inhibitors: Therapeutics for Type 2 Diabetes and What Else? J. Med. Chem. 2014, 57, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.; Hoffmann, T.; Wagner, L.; Wermann, M.; Heiser, U.; Kiefersauer, R.; Huber, R.; Bode, W.; Demuth, H.-U.; Brandstetter, H. The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals Its Functional Regulation and Enzymatic Mechanism. Proc. Natl. Acad. Sci. USA 2003, 100, 5063–5068. [Google Scholar] [CrossRef] [PubMed]


| Peptide | Sequence | Water Solubility (ESOL) | Water Solubility (Ali) | Water Solubility (Silicos-IT) | GI Absorption a | BBB Permeant b | P-gp Substrate c | CYP1A2 Inhibitor d | CYP2C19 Inhibitor e | CYP2C9 Inhibitor f | CYP2D6 Inhibitor g | CYP3A4 Inhibitor h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | PLPP | VS | VS | S | High | No | Yes | No | No | No | No | No |
| P2 | VPPF | VS | VS | S | High | No | Yes | No | No | No | No | No |
| P3 | LPPL | VS | VS | S | High | No | Yes | No | No | No | No | No |
| P4 | YPF | VS | VS | MS | High | No | Yes | No | No | No | No | No |
| P5 | PALF | VS | VS | MS | High | No | Yes | No | No | No | No | No |
| P6 | FVY | VS | VS | MS | High | No | Yes | No | No | No | No | No |
| P7 | LYL | VS | VS | S | High | No | Yes | No | No | No | No | No |
| P8 | LLPL | VS | VS | S | High | No | Yes | No | No | No | No | No |
| P9 | LLLP | VS | VS | S | High | No | Yes | No | No | No | No | No |
| P10 | WVL | VS | VS | MS | High | No | Yes | No | No | No | No | No |
| P11 | LVW | VS | VS | MS | High | No | Yes | No | No | No | No | No |
| P12 | LWV | VS | VS | MS | High | No | Yes | No | No | No | No | No |
| P13 | FWV | S | S | PS | High | No | Yes | No | No | No | Yes | No |
| P14 | LLW | VS | S | MS | High | No | Yes | No | No | No | No | No |
| P15 | LFF | VS | VS | PS | High | No | Yes | No | No | No | Yes | Yes |
| P16 | FLF | VS | VS | PS | High | No | Yes | No | No | No | Yes | Yes |
| P17 | LWF | S | S | PS | High | No | Yes | No | No | No | Yes | No |
| P18 | LFW | S | S | PS | High | No | Yes | No | No | No | Yes | No |
| P19 | IPI | VS | VS | S | High | No | Yes | No | No | No | No | No |
| Peptide | Sequence | Mass (Da) | Retention Time (min) | Identification Score | DPP-IV Inhibitory Activity (IC50, μM) | Source |
|---|---|---|---|---|---|---|
| P1 | PLPP | 422.25 | 13.884 | 8.0791 | ND | Whey protein |
| P2 | VPPF | 458.25 | 50.601 | 44.845 | 1573.85 ± 256.05 | Casein and whey protein |
| P3 | LPPL | 438.28 | 27.735 | 8.7137 | 2558.90 ± 5.70 | Casein and whey protein |
| P4 | YPF | 425.19 | 24.940 | 55.918 | 368.54 ± 12.97 | Casein and whey protein |
| P5 | PALF | 446.25 | 38.521 | 9.4209 | 1746.7 ± 29.90 | Whey protein |
| P6 | FVY | 427.21 | 21.367 | 43.876 | ND | Casein and whey protein |
| P7 | LYL | 407.24 | 28.303 | 36.833 | ND | Whey protein |
| P8 | LLPL | 454.31 | 35.932 | 38.717 | ND | Casein and whey protein |
| P9 | LLLP | 454.31 | 32.377 | 28.962 | 213.99 ± 0.64 | Casein and whey protein |
| P11 | LVW | 416.24 | 30.462 | 55.918 | ND | Whey protein |
| P12 | LWV | 416.24 | 29.518 | 36.05 | ND | Casein and whey protein |
| P14 | LLW | 430.26 | 36.161 | 48.874 | ND | Casein and whey protein |
| P19 | IPI | 341.23 | - | - | 9.28 ± 0.52 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Zhang, J.; Zhu, R.; Zhang, H.; Li, D.; Li, H.; Tang, H.; Chen, L.; Peng, X.; Xu, X.; et al. Mechanistic Study of Novel Dipeptidyl Peptidase IV Inhibitory Peptides from Goat’s Milk Based on Peptidomics and In Silico Analysis. Foods 2024, 13, 1194. https://doi.org/10.3390/foods13081194
Wu Y, Zhang J, Zhu R, Zhang H, Li D, Li H, Tang H, Chen L, Peng X, Xu X, et al. Mechanistic Study of Novel Dipeptidyl Peptidase IV Inhibitory Peptides from Goat’s Milk Based on Peptidomics and In Silico Analysis. Foods. 2024; 13(8):1194. https://doi.org/10.3390/foods13081194
Chicago/Turabian StyleWu, Yulong, Jin Zhang, Ruikai Zhu, Hong Zhang, Dapeng Li, Huanhuan Li, Honggang Tang, Lihong Chen, Xinyan Peng, Xianrong Xu, and et al. 2024. "Mechanistic Study of Novel Dipeptidyl Peptidase IV Inhibitory Peptides from Goat’s Milk Based on Peptidomics and In Silico Analysis" Foods 13, no. 8: 1194. https://doi.org/10.3390/foods13081194





