Specificity of the AMP-6000 Method for Enumerating Clostridium Endospores in Milk
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Isolates from Raw Milk and Teat Skin Samples
2.2. Strain Selection for Inclusivity and Exclusivity Testing
2.3. Inclusivity Testing with Pure Cultures of Spore-Forming Bacteria
2.3.1. Strain Activation for Inclusivity Testing
2.3.2. Alternative MPN Method: AMP-6000

2.3.3. Reference MPN Method: Bryant and Burkey
2.3.4. Non-Selective Method: Spread Plate on RCA
2.4. Exclusivity Testing with Spore-Forming Bacteria and Non-Spore-Forming Bacteria
2.4.1. Strain Activation and Spore Preparation
2.4.2. Sample Preparation
2.4.3. Alternative Method: AMP-6000
2.4.4. Reference Method: Bryant and Burkey
2.4.5. Non-Selective Spread Plate on RCA
2.5. Statistical Analyses
3. Results and Discussion
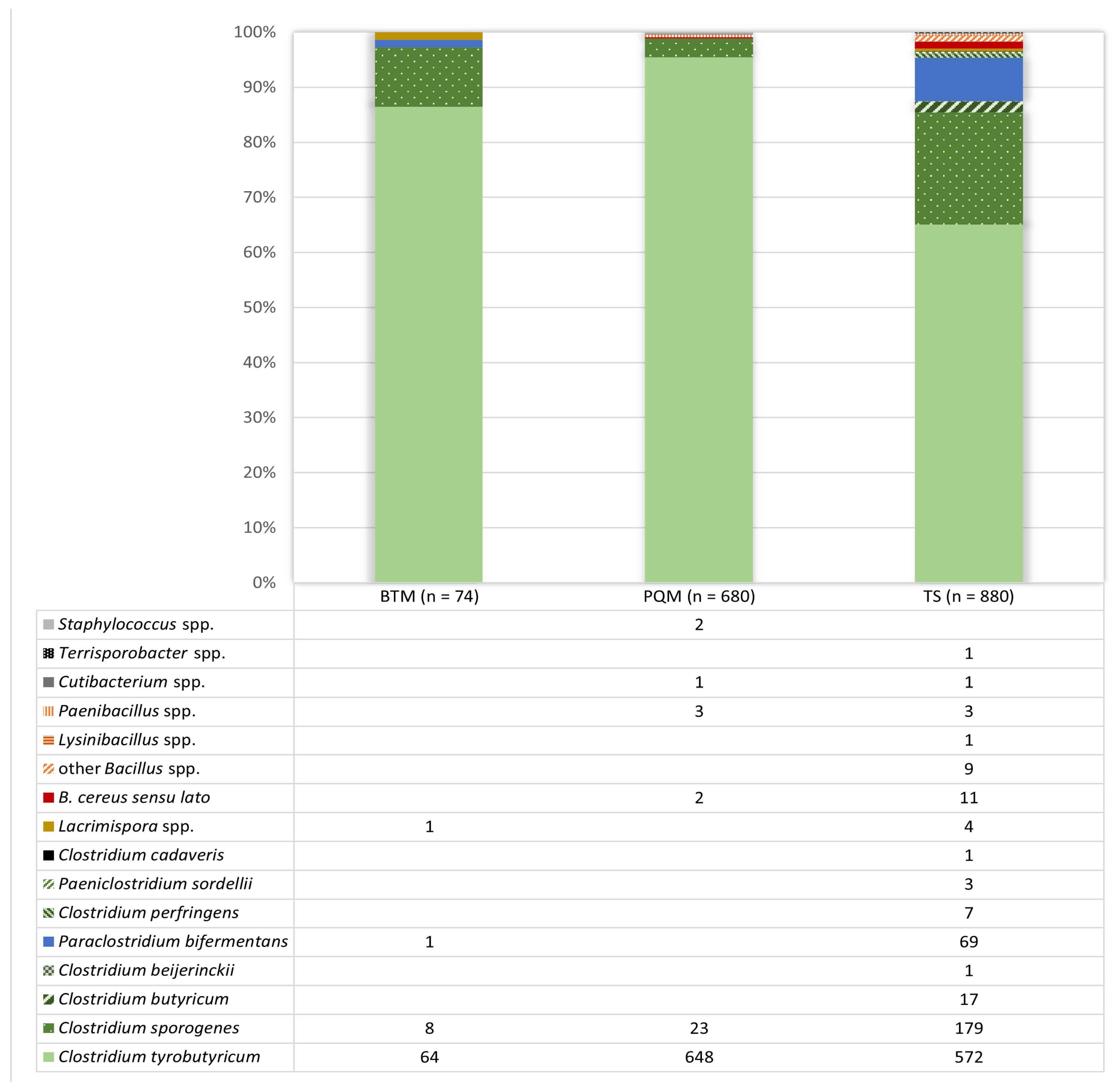
3.1. Isolate Identification from Positive Reactions after the Analysis of Milk and Teat Skin Swab Samples Using the AMP-6000 Method
3.2. Inclusivity of the AMP-6000 Method Based on Pure Culture Testing of C. tyrobutyricum
| Nr. | Species | Internal/International Strain Number | Source | Alternative MPN: AMP-6000 [S/L] | Reference MPN: BB [S/L] | Spread on RCA [S/L] | Inclusive (I)/Exclusive (E) |
|---|---|---|---|---|---|---|---|
| 1 | C. tyrobutyricum | Cl 14/DSM 663 | Emmental cheese | 1550 | 1400 | <1000 | I |
| 2 | C. tyrobutyricum | Cl 82 | milk | 1460 | 1700 | 2000 | I |
| 3 | C. tyrobutyricum | Cl 80 | milk | 7710 | 5400 | 4000 | I |
| 4 | C. tyrobutyricum | Cl 84 | milk | 2320 | 9200 | 3000 | I |
| 5 | C. tyrobutyricum | Cl 188 | cheese-sausage mixture | 8400 | 16,000 | 3500 | I |
| 6 | C. tyrobutyricum | Cl 68 | cheddar cheese | 7490 | >16,000 | 7500 | I |
| 7 | C. tyrobutyricum | Cl 69 | mountain cheese | 2020 | 2400 | 5500 | I |
| 8 | C. tyrobutyricum | Cl 171 | semi-hard cheese | 11,368 | 9200 | 10,500 | I |
| 9 | C. tyrobutyricum | Cl 83 | milk | 1827 | 790 | 1000 | I |
| 10 | C. tyrobutyricum | Cl 102 | milk | 10,027 | 3500 | 2000 | I |
| 11 | C. tyrobutyricum | Cl 168 | semi-hard cheese | 16,611 | 9200 | 10,000 | I |
| 12 | C. tyrobutyricum | Cl 189 | hard cheese | 1639 | 5400 | 4000 | I |
| 13 | C. tyrobutyricum | Cl 15/DSM 664 | raw milk | 1827 | >16,000 | 1000 | I |
| 14 | C. tyrobutyricum | Cl 74 | milk | 3536 | 1700 | 4000 | I |
| 15 | C. tyrobutyricum | Cl 86 | milk | 5012 | 2800 | 2500 | I |
| 16 | C. tyrobutyricum | Cl 87 | milk | 1827 | 9200 | <1000 | I |
| 17 | C. tyrobutyricum | Cl 77 | milk | 3182 | 1300 | <1000 | I |
| 18 | C. tyrobutyricum | Cl 78 | milk | 3068 | 2800 | <1000 | I |
| 19 | C. tyrobutyricum | Cl 105 | milk | 1192 | 1100 | 1500 | I |
| 20 | C. tyrobutyricum | Cl 106 | milk | 2420 | 1300 | 1000 | I |
| 21 | C. tyrobutyricum | Cl 107 | milk | 1021 | 450 | 1000 | I |
| 22 | C. tyrobutyricum | Cl 165 | mountain cheese | 8647 | 9200 | 13,000 | I |
| 23 | C. tyrobutyricum | Cl 103 | milk | 5317 | 5400 | 3000 | I |
| 24 | C. tyrobutyricum | Cl 172 | Swiss cheese | 12,574 | 9200 | 5000 | I |
| 25 | C. tyrobutyricum | Cl 174 | cheese | 3658 | 3500 | 1000 | I |
| 26 | C. tyrobutyricum | Cl 175 | mountain cheese | 223 | 2400 | <1000 | I |
| 27 | C. tyrobutyricum | Cl 182 | Gouda cheese | 4864 | 1100 | 2500 | I |
| 28 | C. tyrobutyricum | Cl 169 | mountain cheese | 1457 | 1700 | <1000 | I |
| 29 | C. tyrobutyricum | Cl 75 | milk | 1732 | 2400 | 1500 | I |
| 30 | C. tyrobutyricum | Cl 162 | milk | 1457 | 1300 | 2000 | I |
| 31 | C. tyrobutyricum | Cl 167 | mountain cheese | 8162 | 3500 | 4000 | I |
| 32 | C. tyrobutyricum | Cl 16 | n.a. | 7494 | 16,000 | 4000 | I |
| 33 | C. tyrobutyricum | Cl 149 | milk | 4169 | 3500 | 2000 | I |
| 34 | C. tyrobutyricum | Cl 81 | milk | 4864 | 9200 | 1000 | I |
| 35 | C. tyrobutyricum | Cl 88 | milk | 3068 | 5400 | 3000 | I |
| 36 | C. tyrobutyricum | Cl 110 | milk | 2524 | 1300 | 1000 | I |
| 37 | C. tyrobutyricum | Cl 89 | milk | 2117 | 2400 | 7000 | I |
| 38 | C. tyrobutyricum | Cl 90 | milk | 2317 | 450 | 1500 | I |
| 39 | C. tyrobutyricum | Cl 91 | milk | 3536 | 1300 | 1000 | I |
| 40 | C. tyrobutyricum | Cl 178 | hard cheese | 10,665 | 2400 | 14,500 | I |
| 41 | C. tyrobutyricum T | Cl 20/DSM 2637 | n.a. | 9442 | >16,000 | 11,500 | E * |
| 42 | C. tyrobutyricum | Cl 21/DSM 1460 | contaminated culture of C. kluyveri | 3182 | 1300 | <1000 | E * |
| 43 | C. tyrobutyricum | Cl 33 | n.a. | 13,511 | 3500 | 7500 | E * |
| 44 | C. tyrobutyricum | Cl 180 | semi-hard cheese | 1457 | 2400 | 4500 | E * |
| 45 | C. tyrobutyricum | Cl 85 | milk | 8162 | >16,000 | 16,000 | E * |
| 46 | C. butyricum T | Cl 19/DSM 10702 | pig intestine | 531 | 2400 | <1000 | I |
| 47 | C. butyricum | Cl 65 | milk | 2117 | >16,000 | 1000 | I |
| 48 | C. butyricum | Cl 17/DSM 2477 4P1 | cotton tree | 2524 | >16,000 | 2000 | I |
| 49 | C. butyricum | Cl 18/DSM 2478 MMP3 | lake sediment | 9728 | >16,000 | 12,000 | I |
| 50 | C. sporogenes | Cl 2 | n.a. | 2737 | 3500 | 4500 | I |
| 51 | C. sporogenes T | Cl 9/DSM 795 | soil | 148 | 690 | <1000 | I |
| 52 | C. sporogenes | Cl 60 | milk | 10,665 | >16,000 | 19,000 | I |
| 53 | C. sporogenes | Cl 12/ATCC 19404 | gas gangrene | 1732 | 5400 | 2500 | I |
| 54 | C. sporogenes | Cl 76 | milk | 1827 | 1100 | <1000 | I |
| 55 | C. sporogenes | Cl 137 | milk | >32,031 | >16,000 | >900,000 | I |
| 56 | C. sporogenes | Cl 140 | milk | >32,031 | >16,000 | 171,000 | I |
| 57 | C. beijerinckii | Cl 49 | grass silage | 937 | 9200 | 16,500 | I |
| 58 | C. beijerinckii | Cl 22 | strain R 1357 | 3182 | >16,000 | <1000 | I |
| 59 | C. beijerinckii | Cl 24 | strain P 1320 | 73 | >16,000 | 71,500 | E |
| 60 | C. beijerinckii | Cl 46 | grass silage | <73 | >16,000 | 11,500 | E |
| 61 | C. beijerinckii T | Cl 36/DSM 791 | soil | <73 | > 16,000 | 9500 | E |
| 62 | C. beijerinckii | Cl 48 | grass silage | <73 | >16,000 | 32,000 | E |
| 63 | C. beijerinckii | Cl 40/ATCC 6015 | n.a. | <73 | >16,000 | 3500 | E |
| 64 | C. beijerinckii | Cl 34/DSM 1820 | soil | <73 | 5400 | 2000 | E |
| 65 | C. beijerinckii | Cl 44 | draff silage | <73 | >16,000 | 2000 | E |
| 66 | C. cadaveris | Cl 67 | milk | 24,321 | 1300 | 15,500 | I |
| 67 | C. acetobutylicum T | Cl 37/DSM 792 | cornmeal | 2019 | >16,000 | <1000 | I |
| 68 | C. acetobutylicum | Cl 1/NCDO 1712 | n.a. | 4864 | 2400 | 4500 | E* |
| 69 | C. pasteurianum T | Cl 192/LMG 3285 | n.a. | <73 | 3500 | 14,000 | E |
| 70 | P. bifermentans | Cl 59 | milk | 6321 | 180 | 12,500 | I |
| 71 | P. bifermentans | Cl 164 | milk | 5317 | >16,000 | 74,000 | I |
| 72 | P. bifermentans | Cl 133 | milk | 1732 | 2400 | 38,500 | I |
| 73 | P. bifermentans | Cl 126 | milk | 4169 | 200 | 4000 | I |
| 74 | P. bifermentans | Cl 142 | milk | 12,574 | <180 | 60,000 | I |
| 75 | Pa. sordellii T | Cl 191/LMG 15708 | spore appendages | 5801 | <180 | 5000 | I |
| 76 | L. celerecrescens T | Cl 194/DSM 5628 | culture | <73 | 200 | 1500 | E |
3.3. Inclusivity of the AMP-6000 Method Based on Pure Culture Testing of Clostridium spp.
3.4. Inclusivity of the AMP-6000 Method Based on Pure Culture Testing of Former Clostridium spp.
3.5. Exclusivity of the AMP-6000 Method Based on Pure Culture Testing
| Nr. | Species | Internal/ International Strain Number | Alternative MPN: AMP-6000 [S/L] | Reference MPN: BB [S/L] | Spread on RCA (37 °C, Anaerobic) [S/L] | Spread on RCA (30 °C, Aerobic) [S/L] | Inclusive (I)/Exclusive (E) |
|---|---|---|---|---|---|---|---|
| 1 | Bacillus cereus T | DSM 31 | <73 | <4100 3 | 3.0 × 106 | 2.0 × 106 | E |
| 2 | Bacillus cereus | LMG 8221 | <73 | <4100 3 | 1.4 × 106 | 1.6 × 105 | E |
| 3 | Bacillus cereus | LMG 12,334 | <73 | <4100 3 | 5.0 × 105 | 7.3 × 105 | E |
| 4 | Bacillus cereus | ATCC 7004 | <73 | <4100 | 1.9 × 109 | 1.4 × 109 | E |
| 5 | Bacillus cereus s.l. | I-Bc 1 (milk) | <73 | <4100 | 1.4 × 109 | 1.9 × 109 | E |
| 6 | Bacillus cytotoxicus T | LMG 26,718 | <73 | <4100 3 | 3.6 × 105 | 7.6 × 105 | E |
| 7 | Bacillus thuringiensis T | LMG 7138 | <73 | <4100 | 6.0 × 105 | 6.3 × 105 | E |
| 8 | Bacillus mycoides T | LMG 7128 | <73 | <4100 | <1.0 × 105 | >3.0 × 106 | E |
| 9 | Bacillus pseudomycoides | LMG 18,993 | <73 | <4100 3 | 1.2 × 105 | 1.4 × 104 | E |
| 10 | Bacillus wiedmannii T | LMG 29,269 | <73 | <4100 3 | 8.7 × 105 | 9.7 × 105 | E |
| 11 | Bacillus mycoides T,2 | DSM 11,821 | <73 | <4100 3 | <1.0 × 104 | 1.4 × 106 | E |
| 12 | Bacillus licheniformis T | DSM 13 | <73 | <4100 3 | 2.4 × 105 | 3.3 × 105 | E |
| 13 | Bacillus sonorensis T | LMG 21,636 | <73 | <4100 3 | 1.2 × 105 | 9.4 × 105 | E |
| 14 | Bacillus sonorensis | I-Bc 3 (milk) | <73 | <4100 | >3.0 × 101 | >3.0 × 1010 | E |
| 15 | Bacillus oleronius T | LMG 17,952 | <73 | <4100 3 | <1.0 × 104 | 3.5 × 104 | E |
| 16 | Bacillus oleronius | I-Bc 2 (teat skin) | <73 | <4100 | <1.0 × 105 | 1.1 × 1010 | E |
| 17 | Bacillus galactosidilyticus T | LMG 17,892 | <73 | <4100 | <1.0 × 105 | 2.8 × 107 | E |
| 18 | Bacillus galactosidilyticus | I-Bc 4 (milk) | <73 | <4100 | <1.0 × 105 | 5.6 × 108 | E |
| 19 | Bacillus pumilus T | LMG 18,928 | <73 | <4100 | <1.0 × 105 | 1.1 × 107 | E |
| 20 | Bacillus pumilus | I-Bc 5 (milk) | <73 | <4100 3 | <1.0 × 104 | 4.3 × 105 | E |
| 21 | Bacillus subtilis ssp. subtilis T | LMG 7135 | <73 | <4100 | >3.0 × 101 | >3.0 × 1010 | E |
| 22 | Bacillus subtilis ssp. spizizenii T | DSM 15,029 | <73 | <4100 3 | <1.0 × 104 | 5.7 × 105 | E |
| 23 | Lysinibacillus sphaericus T | LMG 7134 | <73 | <4100 | <1.0 × 105 | >3.0 × 1010 | E |
| 24 | Lysinibacillus sphaericus | I-LBc 1 (teat skin) | <73 | <4100 | <1.0 × 105 | >3.0 × 1010 | E |
| 25 | Paenibacillus lactis T | LMG 21,910 | <73 | <4100 3 | <1.0 × 104 | 7.7 × 104 | E |
| 26 | Paenibacillus lactis | I-Pa 1 (milk) | <73 | <4100 | 8.0 × 109 | 1.3 × 1010 | E |
| 27 | Paenibacillus macerans T | LMG 6324 | <73 | >11,000 | 2.4 × 106 | 1.7 × 106 | E |
| 28 | Paenibacillus macerans | DSM 24,746 | <73 | >11,000 | 8.8 × 104 | 3.0 × 105 | E |
| 29 | Paenibacillus odorifer T | LMG 19,079 | <73 | <4100 | 9.9 × 109 | 1.4 × 1010 | E |
| 30 | Paenibacillus peoriae T | LMG 14,832 | <73 | <180 3 | 4.2 × 105 | <1.0 × 105 | E |
| 31 | Paenibacillus barengoltzii T | DSM 22,255 | <73 | <180 3 | <1.0 × 105 | 7.7 × 106 | E |
| 32 | Paenibacillus pabuli T | LMG 15,970 | <73 | <4100 | <1.0 × 105 | 8.5 × 107 | E |
| 33 | Paenibacillus polymyxa T | LMG 13,294 | <73 | 1300 | 1.9 × 103 | 5.2 × 103 | E |
| 34 | Paenibacillus glucanolyticus T | LMG 12,239 | <73 | <4100 3 | 5.0 × 104 | 6.4 × 104 | E |
| 35 | Paenibacillus amylolyticus T | LMG 14,012 | <73 | <4100 | 1.6 × 108 | 2.2 × 109 | E |
| 36 | Paenibacillus lautus T | LMG 11,157 | >32,000 | <4100 | 8 × 109 | 9.7 × 108 | I |
| 37 | Paenibacillus lautus | LMG 14,669 | >1732 | <4100 | 6.2 × 105 | 7.3 × 105 | I |
3.6. Exclusivity of the AMP-6000 Method Based on Pure Culture Testing of Bacillus spp.
3.7. Exclusivity of the AMP-6000 Method Based on Pure Culture Testing of Lysinibacillus and Paenibacillus
3.8. Exclusivity of the AMP-6000 Method Based on Pure Culture Testing of Lactic Acid Bacteria
| Nr. | Species | Internal/ International Strain Number | Alternative MPN: AMP-6000 [S/L] | Reference MPN: BB [S/L] | Spread on RCA (37 °C) [CFU/L] (Pasteurized) | Spread on MRS [CFU/L] (Optimum Growth Conditions; Unpasteurized) | Inclusive (I)/Exclusive (E) |
|---|---|---|---|---|---|---|---|
| 1 | Lactobacillus amylovorus T | DSM 20531 | <73 | <3700 | <1.0 × 104 | 4.2 × 1010 | E |
| 2 | Lacticaseibacillus rhamnosus T | DSM 20021 | <73 | <3700 | <1.0 × 104 | 1.1 × 1010 | E |
| 3 | Lactobacillus delbrueckii ssp. bulgaricus T | DSM 20081 | <73 | <3700 | <1.0 × 104 | 8.1 × 109 | E |
| 4 | Lentilactobacillus buchneri T | DSM 20057 | <73 | <3700 | <1.0 × 104 | 1.3 × 1010 | E |
| 5 | Limosilactobacillus fermentum T | DSM 20052 | <73 | <3700 | <1.0 × 104 | 4.0 × 1010 | E |
| 6 | Lactiplantibacillus plantarum T | DSM 20174 | <73 | <3700 | <1.0 × 104 | 7.8 × 1010 | E |
| 7 | Lactobacillus acidophilus T | DSM 20079 | <73 | <3700 | <1.0 × 104 | 5.7 × 109 | E |
| 8 | Lentilactobacillus kefiri T | DSM 20587 | <73 | <3700 | <1.0 × 104 | 1.3 × 1010 | E |
| 9 | Enterococcus faecalis T | DSM 20478 | <73 | <3700 | <1.0 × 104 | 6.1 × 109 | E |
| 10 | Enterococcus durans T | DSM 20633 | <73 | <3700 | <1.0 × 104 | 1.9 × 109 | E |
| 11 | Lactococcus lactis T | DSM 20481 | <73 | <3700 | 1.1 × 104 | 6.8 × 1010 | |
| 12 | Leuconostoc lactis T | DSM 20202 | <73 | <3700 | <1.0 × 104 | 1.5 × 1010 | E |
| 13 | Leuconostoc mesenteroides ssp. mesenteroides T | LMG 6893 | <73 | <3700 | <1.0 × 104 | 7.5 × 1010 | E |
| 14 | Streptococcus salivarius ssp. thermophilus T | LMG 6896 | <73 | <3700 | 7.9 × 105 | 3.5 × 109 | E |
3.9. Discrepancies between Quantitative Results of the Three Test Methods—General Considerations
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brändle, J.; Domig, K.J.; Kneifel, W. Relevance and analysis of butyric acid producing clostridia in milk and cheese. Food Control. 2016, 67, 96–113. [Google Scholar] [CrossRef]
- Le Bourhis, A.-G.; Doré, J.; Carlier, J.-P.; Chamba, J.-F.; Popoff, M.-R.; Tholozan, J.-L. Contribution of C. beijerinckii and C. sporogenes in association with C. tyrobutyricum to the butyric fermentation in Emmental type cheese. Int. J. Food Microbiol. 2007, 113, 154–163. [Google Scholar] [CrossRef] [PubMed]
- IDF. Enumeration of Butyric Acid Forming (Cheese Spoiling) Clostridia—Methodical Considerations. October, 2022. Available online: https://cdn.shopify.com/s/files/1/0603/5167/6609/files/IDF_Factsheet_2022_Enumeration_of_butyric_acid_forming_cheese_spoiling_clostridia_methodical_considerations_5f7d862b-10d8-4232-acde-4abd29f10f78.pdf?v=1665437378 (accessed on 19 December 2023).
- Stadhouders, J. Prevention of Butyric Acid Fermentation by the Use of Nitrate. In Methods of Detection and Prevention of Anaerobic Spore Formers in Relation to the Quality of Cheese; Bulletin of the International Dairy Federation: Brussels, Belgium, 1990; pp. 40–46. [Google Scholar]
- Brändle, J.; Fraberger, V.; Schuller, K.; Zitz, U.; Kneifel, W.; Domig, K.J. A critical assessment of four most probable number procedures for routine enumeration of cheese-damaging clostridia in milk. Int. Dairy J. 2017, 73, 109–115. [Google Scholar] [CrossRef]
- Brändle, J.; Heinzle, L.; Fraberger, V.; Berta, J.; Zitz, U.; Schinkinger, M.; Stocker, W.; Kneifel, W.; Domig, K.J. Novel approach to enumerate clostridial endospores in milk. Food Control. 2018, 85, 318–326. [Google Scholar] [CrossRef]
- ISO 16140-1:2016; Microbiology of the Food Chain—Method Validation—Part 1: Vocabulary. ISO: Geneva, Switzerland, 2016.
- Burtscher, J.; Rudavsky, T.; Zitz, U.; Neubauer, V.; Domig, K.J. Importance of Pre-Milking Udder Hygiene to Reduce Transfer of Clostridial Spores from Teat Skin to Raw Milk. Microorganisms 2023, 11, 1337. [Google Scholar] [CrossRef]
- ISO 16140-2:2016; Microbiology of the Food Chain—Method Validation—Part 2: Protocol for the Validation of Alternative (Proprietary) Methods Against a Reference Method. ISO: Geneva, Switzerland, 2016.
- Jakob, E.; Glauser, D.L. Vergleich von Methoden zur Bestimmung der Buttersäurebakterien in Milch. Agrar. Schweiz 2019, 10, 388–395. [Google Scholar]
- Burtscher, J.; Küller, F.; Dreier, M.; Arias-Roth, E.; Drissner, D.; Domig, K.J. Characterization of Clostridium tyrobutyricum Strains Using Three Different Typing Techniques. Microorganisms 2020, 8, 1057. [Google Scholar] [CrossRef]
- Reindl, A.; Dzieciol, M.; Hein, I.; Wagner, M.; Zangerl, P. Enumeration of clostridia in goat milk using an optimized membrane filtration technique. J. Dairy Sci. 2014, 97, 6036–6045. [Google Scholar] [CrossRef]
- Garde, S.; Arias, R.; Gaya, P.; Nuñez, M. Occurrence of Clostridium spp. in ovine milk and Manchego cheese with late blowing defect: Identification and characterization of isolates. Int. Dairy J. 2011, 21, 272–278. [Google Scholar] [CrossRef]
- Klijn, N.; Nieuwenhof, F.F.J.; Hoolwerf, J.D.; van der Waals, C.B.; Weerkamp, A.H. Identification of Clostridium tyrobutyricum as the causative agent of late blowing in cheese by species-specific PCR amplification. Appl. Environ. Microbiol. 1995, 61, 2919–2924. [Google Scholar] [CrossRef]
- Bassi, D.; Puglisi, E.; Cocconcelli, P.S. Understanding the bacterial communities of hard cheese with blowing defect. Food Microbiol. 2015, 52, 106–118. [Google Scholar] [CrossRef]
- Haas, K.N.; Blanchard, J.L. Reclassification of the Clostridium clostridioforme and Clostridium sphenoides clades as Enterocloster gen. nov. and Lacrimispora gen. nov., including reclassification of 15 taxa. Int. J. Syst. Evol. Microbiol. 2020, 70, 23–34. [Google Scholar] [CrossRef]
- Sasi Jyothsna, T.S.; Tushar, L.; Sasikala, C.; Ramana, C.V. Paraclostridium benzoelyticum gen. nov., sp. nov., isolated from marine sediment and reclassification of Clostridium bifermentans as Paraclostridium bifermentans comb. nov. Proposal of a new genus Paeniclostridium gen. nov. to accommodate Clostridium sordellii and Clostridium ghonii. Int. J. Syst. Evol. Microbiol. 2016, 66, 1268–1274. [Google Scholar] [CrossRef]
- Hurley, M.A.; Roscoe, M.E. Automated statistical analysis of microbial enumeration by dilution series. J. Appl. Bacteriol. 1983, 55, 159–164. [Google Scholar] [CrossRef]
- CNERNA. Centre National d’Études et de Recommandations sur la Nutrition et l’Alimentation, Recommandations pour l’estimation de la Contamination du Lait en Spores de Clostridia par la Méthode de Culture en Milieu Liquide. Rev. Laitière Française 1986, 451, 39–45. [Google Scholar]
- Travers, R.S.; Martin, P.A.; Reichelderfer, C.F. Selective Process for Efficient Isolation of Soil Bacillus spp. Appl. Environ. Microbiol. 1987, 53, 1263–1266. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis. Curr. Dir. Psychol. Sci. 1992, 1, 98–101. [Google Scholar] [CrossRef]
- Kim, H.W.; Kim, N.K.; Thompson, J.; de Jesus, M.; Rehberger, J.; Rehberger, T.; Smith, A.H.; Mackie, R.I. Effects of dosing non-toxigenic Clostridia on the bacterial populations and immunological responses in the intestinal tract of lactating dairy cows. Front. Microbiol. 2023, 14, 1107964. [Google Scholar] [CrossRef]
- Borreani, G.; Ferrero, F.; Nucera, D.; Casale, M.; Piano, S.; Tabacco, E. Dairy farm management practices and the risk of contamination of tank milk from Clostridium spp. and Paenibacillus spp. spores in silage, total mixed ration, dairy cow feces, and raw milk. J. Dairy Sci. 2019, 102, 8273–8289. [Google Scholar] [CrossRef]
- Gopal, N.; Hill, C.; Ross, P.R.; Beresford, T.P.; Fenelon, M.A.; Cotter, P.D. The Prevalence and Control of Bacillus and Related Spore-Forming Bacteria in the Dairy Industry. Front. Microbiol. 2015, 6, 1418. [Google Scholar] [CrossRef]
- Carroll, L.M.; Cheng, R.A.; Wiedmann, M.; Kovac, J. Keeping up with the Bacillus cereus group: Taxonomy through the genomics era and beyond. Crit. Rev. Food Sci. Nutr. 2022, 62, 7677–7702. [Google Scholar] [CrossRef] [PubMed]
- Mezian, L.; Chincha, A.I.; Vecchione, A.; Ghelardi, E.; Bonatto, J.M.C.; Marsaioli, A.J.; Campelo, P.H.; Benamar, I.; Moussaoui, A.A.; Sant’Ana, A.S.; et al. Aerobic spore-forming bacteria in powdered infant formula: Enumeration, identification by MALDI-TOF mass spectrometry (MS), presence of toxin genes and rpoB gene typing. Int. J. Food Microbiol. 2022, 368, 109613. [Google Scholar] [CrossRef]
- de Jonghe, V.; Coorevits, A.; de Block, J.; van Coillie, E.; Grijspeerdt, K.; Herman, L.; de Vos, P.; Heyndrickx, M. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int. J. Food Microbiol. 2010, 136, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, F.; Hoolwerf, J.; Rademaker, J.L.W. Concurrence of spores of Clostridium tyrobutyricum, Clostridium beijerinckii and Paenibacillus polymyxa in silage, dairy cow faeces and raw milk. Int. Dairy J. 2016, 63, 70–77. [Google Scholar] [CrossRef]
- Gupta, T.B.; Brightwell, G. Identification and Characterisation of Spore-Forming Bacteria in Bovine Raw Milk Collected from Four Dairy Farms in New Zealand. Dairy 2023, 4, 650–671. [Google Scholar] [CrossRef]
- Podrzaj, L.; Burtscher, J.; Küller, F.; Domig, K.J. Strain-Dependent Cheese Spoilage Potential of Clostridium tyrobutyricum. Microorganisms 2020, 8, 1836. [Google Scholar] [CrossRef] [PubMed]
- Ruusunen, M.; Surakka, A.; Korkeala, H.; Lindstrom, M. Clostridium tyrobutyricum strains show wide variation in growth at different NaCl, pH, and temperature conditions. J. Food Prot. 2012, 75, 1791–1795. [Google Scholar] [CrossRef]
- Quiberoni, A.; Guglielmotti, D.; Reinheimer, J. New and classical spoilage bacteria causing widespread blowing in Argentinean soft and semihard cheeses. Int. J. Dairy Technol. 2008, 61, 358–363. [Google Scholar] [CrossRef]
- Amrein, R.; Winkler, H.; Jakob, E. So Sichert Sich der Käser Gegen Schäden ab.; ALP Forum: Bern, Switzerland, 2008. [Google Scholar]
- Jakob, E. Analytik rund um die Buttersäuregärung. ALP Forum 2011, 85, 1–20. [Google Scholar]
- Lycken, L.; Borch, E. Characterization of Clostridium spp. isolated from spoiled processed cheese products. J. Food Prot. 2006, 69, 1887–1891. [Google Scholar] [CrossRef]
- Arias, C.; Oliete, B.; Seseña, S.; Jimenez, L.; Pérez-Guzmán, M.D.; Arias, R. Importance of on-farm management practices on lactate-fermenting Clostridium spp. spore contamination of Manchega ewe milk: Determination of risk factors and characterization of Clostridium population. Small Rumin. Res. 2013, 111, 120–128. [Google Scholar] [CrossRef]
- Branska, B.; Pechacova, Z.; Kolek, J.; Vasylkivska, M.; Patakova, P. Flow cytometry analysis of Clostridium beijerinckii NRRL B-598 populations exhibiting different phenotypes induced by changes in cultivation conditions. Biotechnol. Biofuels 2018, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Brändle, J.; Fraberger, V.; Berta, J.; Puglisi, E.; Jami, M.; Kneifel, W.; Domig, K.J. Butyric acid producing clostridia in cheese—Towards the completion of knowledge by means of an amalgamate of methodologies. Int. Dairy J. 2018, 86, 86–95. [Google Scholar] [CrossRef]
- Le Bourhis, A.G.; Saunier, K.; Doré, J.; Carlier, J.P.; Chamba, J.F.; Popoff, M.R.; Tholozan, J.L. Development and validation of PCR primers to assess the diversity of Clostridium spp. in cheese by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 2005, 71, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Ansorge-Schumacher, M.B.; Fritsch, M.; Hartmeier, W. Influence of hydrogenase overexpression on hydrogen production of Clostridium acetobutylicum DSM 792. Enzym. Microb. Technol. 2010, 46, 384–390. [Google Scholar] [CrossRef]
- Burtscher, J.; Hobl, L.; Kneifel, W.; Domig, K.J. Short communication: Clostridial spore counts in vat milk of Alpine dairies. J. Dairy Sci. 2020, 103, 2111–2116. [Google Scholar] [CrossRef] [PubMed]
- Carminati, D.; Bonvini, B.; Francolino, S.; Ghiglietti, R.; Locci, F.; Tidona, F.; Mariut, M.; Abeni, F.; Zago, M.; Giraffa, G. Low-Level Clostridial Spores’ Milk to Limit the Onset of Late Blowing Defect in Lysozyme-Free, Grana-Type Cheese. Foods 2023, 12, 1880. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A.P.; Hull, R.R. Late blowing of Swiss cheese: Incidence of Clostridium tyrobutyricum in manufacturing milk. Aust. J. Dairy Technol. 1989, 44, 82–87. [Google Scholar]
- Vissers, M.M.; Driehuis, F.; Te Giffel, M.C.; de Jong, P.; Lankveld, J.M. Minimizing the level of butyric acid bacteria spores in farm tank milk. J. Dairy Sci. 2007, 90, 3278–3285. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burtscher, J.; Rudavsky, T.; Zitz, U.; Domig, K.J. Specificity of the AMP-6000 Method for Enumerating Clostridium Endospores in Milk. Foods 2024, 13, 1192. https://doi.org/10.3390/foods13081192
Burtscher J, Rudavsky T, Zitz U, Domig KJ. Specificity of the AMP-6000 Method for Enumerating Clostridium Endospores in Milk. Foods. 2024; 13(8):1192. https://doi.org/10.3390/foods13081192
Chicago/Turabian StyleBurtscher, Johanna, Tamara Rudavsky, Ulrike Zitz, and Konrad J. Domig. 2024. "Specificity of the AMP-6000 Method for Enumerating Clostridium Endospores in Milk" Foods 13, no. 8: 1192. https://doi.org/10.3390/foods13081192
APA StyleBurtscher, J., Rudavsky, T., Zitz, U., & Domig, K. J. (2024). Specificity of the AMP-6000 Method for Enumerating Clostridium Endospores in Milk. Foods, 13(8), 1192. https://doi.org/10.3390/foods13081192






