Food Pathways of Salmonella and Its Ability to Cause Gastroenteritis in North Africa
Abstract
:1. Introduction
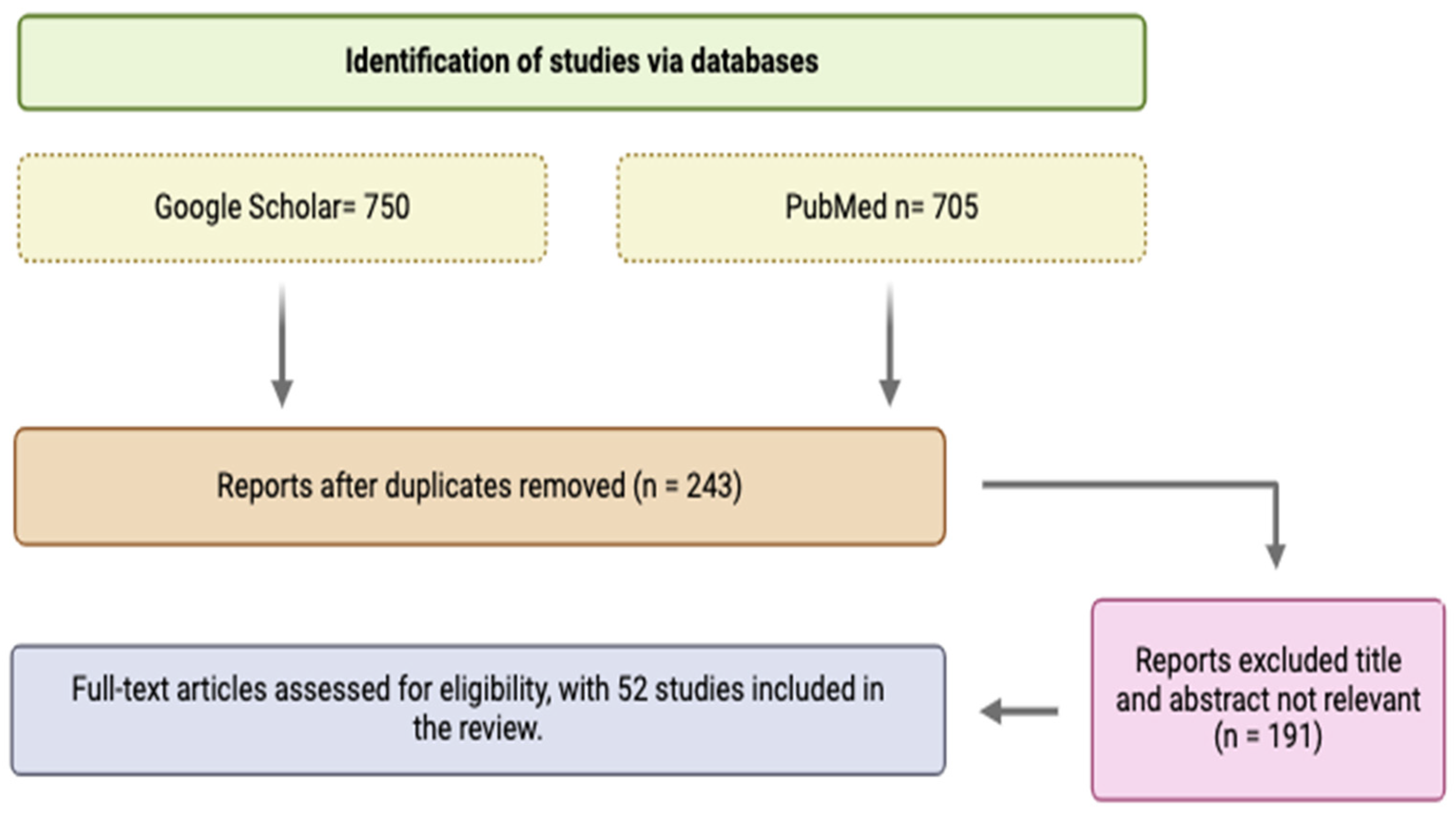
2. Study Design and Methodology
3. Current Status of Human Salmonellosis in North Africa
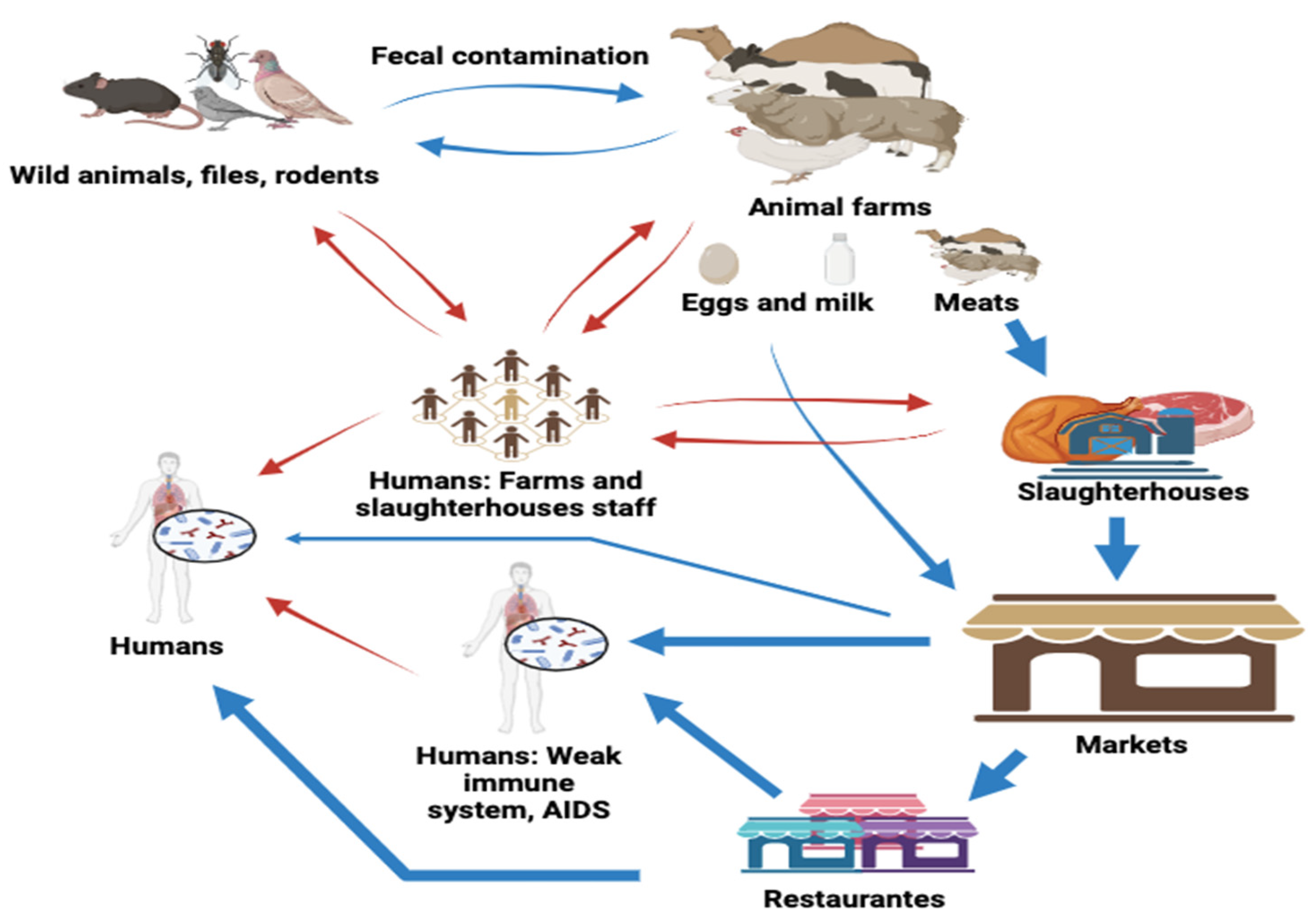
4. Source and Modes of Transmission to Humans in North African Countries
| Country | Tested Food Samples (Total Number) | % (Out of Total Number) of NTS Positive Samples | Salmonella Serotypes (%) * | Confirmation Methods (If PCR Targets Genes) and References |
|---|---|---|---|---|
| Algeria | Chicken meats (n = 60) | 50 | ND | API 20E identification kit [26] |
| Chicken meats (n = 70) | 57.14 | ND | MALDI-TOF MS [25] | |
| Frozen beef liver (n = 50) | 4 | ND | Biochemical test [49] | |
| Chicken Liver (n = 25) | 4 | Kentucky (100) | MALDI-TOF MS [23] | |
| Fruit and vegetable (n = 181) | 0 (not detected) | ND | ND [33] | |
| Eggs (n = 45) | 4.4 | Bradford (100), Entritidis (0) | API-10S identification kit [50] | |
| Eggs (n = 180) | 7.22 | ND | Biochemical test [41] | |
| Beef (n = 190) | 22.52 | ND | PCR technique (dNTPs) [51] | |
| Mutton (n = 251) | 12.74 | ND | ||
| Dairy products (n = 310) | ND | ND | Biochemical test [52] | |
| Raw red meat and meat products (n = 144) | 23.6 | Agona (1.6), Albany (3.1), Altona (12.5), Anatum (14.0), Corvallis (7.8), Enteritidis (7.8), Hadar (1.6), Heidelberg (4.7), Indiana (4.7), Infantis (1.6), Kedougou (1.6), Lexington (1.6), Liverpool (1.6), Mbandaka (4.7), Montevideo (6.3). | API 20E identification kit [14] | |
| Egypt | Chicken burger (n = 30) | 10 | Typhimurium (0), Infantis (100), Kentucky (0) | PCR technique (hilA, invA, stn) [43] |
| Chicken filet (n = 30), | 23.33 | Typhimurium (28.57), Infantis (42.86), Kentucky (28.57) | ||
| Chicken luncheon (n = 30), Chicken nuggets (n = 30), and Chicken panne (n = 30) | 0 | ND | ||
| Market raw milk (n = 30) | 6.7 | Enteritidis (7.1), Typhimurium (7.1), Virchow (7.1), Larochelle (0), Apeyeme (0) | Biochemical test [4] | |
| Bulk tank milk (n = 30) | 6.7 | Enteritidis (7.1), Typhimurium (7.1), Virchow (0), Larochelle (7.1), Apeyeme (0) | ||
| Pasteurized milk (n = 30) | 0 | ND | ||
| Kariesh cheese (n = 30) | 16.7 | Enteritidis (14.9), Typhimurium (7.1), Virchow (14.9), Larochelle (7.1), Apeyeme (7.1) | ||
| White soft cheese (n = 30) | 3.3 | Enteritidis (0), Typhimurium (7.1), Virchow (0), Larochelle (0), Apeyeme (0) | ||
| Raw/fresh food (n = 300) | 7.67 | ND | ||
| Ready to eat/drink food (n = 56) | 8.93 | ND | MALDI-TOF MS [53] | |
| Buffalo meat (n = 100) | 25 | Enteritidis (20.8), Typhimurium (17), Montevideo (11.3), Rissen (9.4), Infantis (9.4), Virchow (7.5), Essen (5.7), Anatum (3.8), Dublin (3.8), Tsevie (3.8), Chester (1.9), Derby (1.9), Papuana (1.9), Saintpaul (1.9). | PCR technique (invA) [27] | |
| Fresh poultry (n = 60) | 10 | Enteritidis (3.3), Typhimurium (3.3), Kentucky (3.3), Paratyphi A (6.7) | PCR technique (ompC) [54] | |
| Frozen poultry (n = 30) | 3.3 | |||
| Fresh beef (n = 60) | 11.7 | Enteritidis (3.3), Typhimurium (6.7), Kentucky (0), Paratyphi A (3.3) | ||
| Frozen beef (n = 30) | 3.3 | |||
| Fresh vegetables and ready-to-eat salads (n = 121) | 0 (not detected) | ND | ND [55] | |
| Raw chicken meat (n = 100) | 5 | Typhimurium (60.0), Enteritidis (40.0). | MALDI-TOF MS [56] | |
| Poultry products (n = 75) | 6.6 | Enteritidis (40), Typhimurium (40), Kentucky (20) | PCR technique (invA) [57] | |
| Fresh chicken meat (n = 200) | 3.5 | Enteritidis (14.3), Typhimurium (71.4), Kentucky (14.3). | PCR technique (invA) [58] | |
| Ready-to-eat chicken meat (n = 100) | ND | |||
| Raw egg yolk (n = 30) | ND | Enteritidis (3.1), Typhimurium (0), Kentucky (41.5), Other types (55.4) | API 20E identification kit [42] | |
| Eggshell (n = 30) | ND | |||
| Mixed chicken meat samples (n = 62) | 60 | |||
| Chicken skin (n = 22) | 64 | |||
| Chicken carcasses (n = 50) | 16 | Enteritidis (37.4), Typhimurium (30.1), Kentucky (10.8), Muenster (8.4), Virchow (4.8), Anatum (4.8), Haifa (1.2), Other types (2.4) | API 20E identification kit [59] | |
| Frozen beef (n = 160) | 2.5 | Enteritidis (32.1), Typhimurium (41.5), Infantis (20.8) | PCR technique (ompC) [60] | |
| Fresh beef (n = 80) | 18.7 | |||
| Beef carcasses (n = 240) | 8.3 | |||
| Chicken breast (n = 160) | 1.25 | |||
| Chicken legs (n = 160) | 7.5 | |||
| Raw milk (Buffalo) (n = 240) | 3.3 | |||
| Raw milk (Cow) (n = 240) | 1.6 | |||
| Cheese (Kareish) (n = 120) | 2.5 | |||
| Cheese (Domiati) (n = 120) | 0.83 | |||
| Yogurt (n = 80) | ND | |||
| Frozen chicken breast filets (n = 25) | 52 | Enteritidis (100) | PCR technique (Rfbj, Flic, Fljb, Sdf) [61] | |
| Frozen chicken legs (n = 25) | 36 | Enteritidis (100) | ||
| Minced frozen meats (n = 25) | 20 | Kentucky (100) | ||
| Morocco | Egg content (n = 290) | 2 | ND | Biochemical test [62] |
| Eggshell (n = 290) | 0 | ND | ||
| Broiler Chicken Meat (n = 540) | 7.40 | ND | API 20E identification kit [45] | |
| Chicken breast cut (n = 128) | 12.5 | Hadar (23.80), Typhimurium(19.05), Chester (19.05) Schwarzengrund (14.28), Kentucky (9.52), Bredeney (9.52), Saintpaul (4.76) | PCR technique (invA) [5] | |
| Viscera (liver, gizzard, heart) (n = 197) | 7.92 | |||
| Cut meats (skewers samples) spiced (n = 101) | 5.94 | |||
| Cut meats (skewers samples) not spiced (n = 80) | 11.25 | |||
| Minced meat (n = 55) | 12.73 | |||
| Chicken sausages (n = 54) | 11.11 | |||
| Traditional chicken mortadella (n = 5) | 40 | |||
| Shellfish (n = 150) | 12.7 | Chester (73.7), Hadar (15.8), Typhimurium (5.7), Kentucky (5.7) | PCR technique (invA) [63] | |
| Traditional cheese (n = 51) | 5.9 | Typhimurium (6.2), Enteritidis (4.2), Kentucky (22.9), Montevideo (6.2), Agona (16.7), Reading (12.5), Corvallis (8.3), Saintpaul (8.3), Israel (2.0), Hadar (2.0), Branderup (2.0) | API 20E identification kit [44] | |
| Minced meat (n = 138) | 12.3 | |||
| Sausage (n = 20) | 5 | |||
| Chicken (n = 86) | 20 | |||
| Turkey (n = 17) | 52.9 | |||
| Milk and other derivatives (n = 152) | ND. | |||
| Turkey sausages (n = 60) | 23.3 | Typhimurium (5.9), Agona (2.9), Saintpaul (2.9), Mbandaka (11.8), Montevideo (8.8), Livingstone (2.9), Corvallis (23.5), Kentucky (17.6), Bovismorbificans (5.9), Anatum (2.9), Give (11.8), Muenster (2.9) | PCR technique (invA, spvC) [64] | |
| Beef sausages (n = 60) | 15 | |||
| Artisanal sausages (n = 36) | 30.6 | |||
| Cereal products (n = 60) | ND | - | API 20E identification kit [65] | |
| Chicken meat, eggs, and visceral organs (n = 432) | 0.7 | Hadar (9.1), Corvallis (18.2), Mbandaka (18.2), Ouakam (18.2), Tm var. cop (9.1), Virchow (18.2), Altona (9.1). | API 20E identification kit [66] | |
| Mussels (n = 279) | 10 | Kentucky (57.1), Blockley (42.9), Senftenberg (0) | PCR technique (invA) [67] | |
| Cooked meat (n = 2952) | 0.7 | Anatum (3.8), Bareilley (1.0), Berta (1.9), Blokley (10.4), Brenderup (6.6), Bredeney (12.3), Enteritidis (2.8), Hadar (3.8), Infantis (23.8), Kiambu (5.7) | API 20E identification kit [46] | |
| Sausages (n = 2052) | 0.1 | |||
| Chicken meat (n = 1200) | 0.4 | |||
| Pastry (n = 2232) | 0.2 | |||
| Chopped meat (n = 196) | 2.4 | |||
| Sea food (n = 562) | 1.8 | |||
| Spices (n = 80) | 1.3 | |||
| Beef meat (n = 2122) | 3.5 | Labadi (1.9), MBandaka (7.6), Montevideo (3.8), Typhimurium (8.5), Salamae type II (1.0), Non-typeable (2.8) | ||
| Raw ground beef (n = 150) | 2 | Entritidis (33.3), Typhimurium (33.3). | API 20E identification kit [68] | |
| Fresh sausage (n = 100) | 4 | Anatum (33.3), Bareilly: (14.3) | ||
| Chicken breast, legs, liver, gizzard (n = 576) | 10 | Typhimurium (40.4), Newport (26.3), Montevideo (17.5), Heidelberg (15.8) | Biochemical test [69] | |
| Vegetable samples (n = 50) | 2 | Arizona (100) | API 20E identification kit [70] | |
| Libya | Raw cow milk (n = 56) | 16 | ND | PCR technique (16S rDNA) [13] |
| Fermented raw milk (n = 18) | 11 | ND | ||
| Milk powder (n = 18) | 0 | ND | ||
| Maasora cheese (n = 13) | 22 | ND | ||
| Ricotta cheese (n = 13) | 46 | ND | ||
| Ice cream (n = 13) | 0 | ND | ||
| Minced meat (n = 50) | 6 | Typhimurium (33.3), Enteritidis (Hamad and Saleh, 2019) (3) | PCR technique (invA) [47] | |
| Beef burger (n = 50) | 4 | Typhimurium (50), Enteritidis (50), Inganda (0) | ||
| Uncooked chicken burger (n = 56) | 12.5 | ND | API 20E identification kit [28] | |
| Cooked spiced chicken burger (n = 64) | 1.6 | ND | ||
| Tunisia | Chicken products (n = 1288) | 5.7 | Enteritidis (22), Kentucky (19), Anatum (18), Infantis (16), Mbandaka (8), Zanzibar (8), Hadar (5), Agona (4) | API 20E identification kit [15] |
| Clams (n = 20) | 35 | Irenea (28.6), London (28.6), Enteritidis (14.3), Poona (14.3), Brandcaster (14.3) | PCR technique (siiA) [48] | |
| Chicken (n = 97) | 28.9 | Enteritidis (64.3), Kentucky (35.7) | ||
| Cow’s milk (n = 80) | 12.5 | Kentucky (80), Anatum (20) | ||
| Cooked dishes (n = 150) | 21.3 | Zanzibar (0.08), Kentucky (28.0), Manchester (12.0), Schwarzengrund (0.08), Bredeney (4.0), Altona (12.0), Anatum (20.0), Amsterdam (4.0), Orion (4.0) | API 10S identification kit [29] | |
| Raw milk (n = 93) | 33.3 | |||
| Dairy products (n = 22) | 22.7 | |||
| Vegetables salad (n = 70) | 12.8 | |||
| Seafood (n = 46) | 23.9 | |||
| Raw poultry meat including (n = 45) | 60 | |||
| Cakes (n = 41) | 26.8 | |||
| Salami and sausage (n = 20) | 25 | |||
| Raw red meat (n = 13) | 35.5 | |||
| Chicken carcasses (n = 50) | 16 | Enteritidis (100) | API 20E identification kit [71] | |
| Cuts of beef (n = 144) | 29.8 | Typhimurium (35.0), Kentucky (17.5), Suberu (15.0), Newlands (8.8), Zanzibar (13.8), Orion (7.5), Enteritidis (3.8), Neumuenster (1.3) | PCR technique (invA) [72] | |
| Raw chicken (n = 60) | 48.3 | |||
| Portions of minced meat (n = 56) | 10.7 | |||
| Cuts of lamb (n = 33) | 6 | |||
| Merguez (sausages) (n = 10) | ND | |||
| Fish (n = 12) | ND |
5. Virulence Factors
6. Antibiotic Resistance
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- ECDC. EFSA The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2019–2020. EFSA J. 2022, 20, e07209. [Google Scholar] [CrossRef]
- WHO. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Bur-Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015; ISBN 9789241565165. [Google Scholar]
- Elafify, M.; Darwish, W.S.; El-Toukhy, M.; Badawy, B.M.; Mohamed, R.E.; Shata, R.R. Prevalence of Multidrug Resistant Salmonella spp. in Dairy Products with the Evaluation of the Inhibitory Effects of Ascorbic Acid, Pomegranate Peel Extract, and D-Tryptophan against Salmonella Growth in Cheese. Int. J. Food Microbiol. 2022, 364, 109534. [Google Scholar] [CrossRef] [PubMed]
- Zahli, R.; Scheu, A.K.; Abrini, J.; Copa-Patiño, J.L.; Nadia, A.; Nadia, S.S.; Soliveri, J. Salmonella spp: Prevalence, Antimicrobial Resistance and Molecular Typing of Strains Isolated from Poultry in Tetouan-Morocco. LWT 2022, 153, 112359. [Google Scholar] [CrossRef]
- Teklemariam, A.D.; Al-Hindi, R.R.; Albiheyri, R.S.; Alharbi, M.G.; Alghamdi, M.A.; Filimban, A.A.R.; Al Mutiri, A.S.; Al-Alyani, A.M.; Alseghayer, M.S.; Almaneea, A.M.; et al. Human Salmonellosis: A Continuous Global Threat in the Farm-to-Fork Food Safety Continuum. Foods 2023, 12, 1756. [Google Scholar] [CrossRef]
- Habib, I.; Mohamed, M.Y.I.; Khan, M. Current State of Salmonella, Campylobacter and Listeria in the Food Chain across the Arab Countries: A Descriptive Review. Foods 2021, 10, 2369. [Google Scholar] [CrossRef] [PubMed]
- El Hanafi, M.; Nourredine, B.; Saadia, N.; Hakim, K. Occurrence of Antibiotic Resistance in Salmonella Serotypes Isolated from Environment, Humans, Animals, and Animal Products in Morocco: A Systematic Review. World’s Vet. J. 2023, 13, 32–44. [Google Scholar] [CrossRef]
- Al-Rifai, R.H.; Chaabna, K.; Denagamage, T.; Alali, W.Q. Prevalence of Enteric Non-typhoidal Salmonella in Humans in the Middle East and North Africa: A Systematic Review and Meta-analysis. Zoonoses Public Health 2019, 66, 701–728. [Google Scholar] [CrossRef] [PubMed]
- Clare, B. Inflammasome Activation by Salmonella. Curr. Opin. Microbiol. 2021, 64, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Fattinger, S.A.; Sellin, M.E.; Hardt, W.-D. Epithelial Inflammasomes in the Defense against Salmonella Gut Infection. Curr. Opin. Microbiol. 2021, 59, 86–94. [Google Scholar] [CrossRef]
- Takaya, A.; Yamamoto, T.; Tokoyoda, K. Humoral Immunity vs. Salmonella. Front. Immunol. 2020, 10, 3155. [Google Scholar] [CrossRef] [PubMed]
- Garbaj, A.M.; Ben Gawella, T.B.; Sherif, J.A.; Naas, H.T.; Eshamah, H.L.; Azwai, S.M.; Gammoudi, F.T.; Abolghait, S.K.; Moawad, A.A.; Barbieri, I.; et al. Occurrence and antibiogram of multidrug-resistant Salmonella enterica Isolated from Dairy Products in Libya. Vet. World 2022, 15, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Mezali, L.; Hamdi, T.M. Prevalence and Antimicrobial Resistance of Salmonella Isolated from Meat and Meat Products in Algiers (Algeria). Foodborne Pathog. Dis. 2012, 9, 522–529. [Google Scholar] [CrossRef]
- Oueslati, W.; Rjeibi, M.R.; Benyedem, H.; Mamlouk, A.; Souissi, F.; Selmi, R.; Ettriqui, A. Salmonella Broiler Meat’s Contamination in Tunisia: Prevalence, Serotypes, Antimicrobial Resistance and Molecular Characterization of Isolated Strains. Curr. Microbiol. 2022, 79, 208. [Google Scholar] [CrossRef]
- Al-Gallas, N.; Belghouthi, K.; Barratt, N.A.; Ghedira, K.; Hotzel, H.; Tomaso, H.; El-Adawy, H.; Neubauer, H.; Laouini, D.; Zarrouk, S.; et al. Identification and Characterization of Multidrug-resistant ESBL-producing Salmonella enterica Serovars Kentucky and Typhimurium Isolated in Tunisia CTX-M-61/TEM-34, a Novel Cefotaxime-hydrolysing Β-lactamase of Salmonella. J. Appl. Microbiol. 2022, 132, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Pinedo, C.L.; Mughini-Gras, L.; Franz, E.; Hald, T.; Pires, S.M. Sources and Trends of Human Salmonellosis in Europe, 2015–2019: An Analysis of Outbreak Data. Int. J. Food. Microbiol. 2022, 379, 109850. [Google Scholar] [CrossRef]
- Kebede, M.T.; Getu, A.A. Assessment of Bacteriological Quality and Safety of Raw Meat at Slaughterhouse and Butchers’ Shop (Retail Outlets) in Assosa Town, Beneshangul Gumuz Regional State, Western Ethiopia. BMC Microbiol. 2023, 23, 403. [Google Scholar] [CrossRef]
- Oueslati, W.; Ridha Rjeibi, M.; Benyedem, H.; Jebali, M.; Souissi, F.; Selmi, R.; El Asli, M.S.; Barguellil, F.; Ettriqui, A. Serotype Occurrence, Virulence Profiles, Antimicrobial Resistance and Molecular Characterization of Salmonella Isolated from Hospitalized Patients with Gastroenteritis in Great Tunisia between 2010 and 2020. Antibiotics 2023, 12, 526. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.A.; Heyderman, R.S. A Perspective on Invasive Salmonella Disease in Africa. Clin. Infect. Dis. 2015, 61, S235–S240. [Google Scholar] [CrossRef]
- Paré, G.; Trudel, M.C.; Jaana, M.; Kitsiou, S. Synthesizing Information Systems Knowledge: A Typology of Literature Reviews. Inf. Manag. 2015, 52, 183–199. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef] [PubMed]
- Djeffal, S.; Bakour, S.; Mamache, B.; Elgroud, R.; Agabou, A.; Chabou, S.; Hireche, S.; Bouaziz, O.; Rahal, K.; Rolain, J.M. Prevalence and Clonal Relationship of ESBL-Producing Salmonella Strains from Humans and Poultry in Northeastern Algeria. BMC Vet. Res. 2017, 13, 132. [Google Scholar] [CrossRef]
- Oueslati, W.; Rjeibi, M.R.; Benyedem, H.; Mamlouk, A.; Souissi, F.; Selmi, R.; Ettriqui, A. Prevalence, Risk Factors, Antimicrobial Resistance and Molecular Characterization of Salmonella in Northeast Tunisia Broiler Flocks. Vet. Sci. 2021, 9, 12. [Google Scholar] [CrossRef]
- Samia, D.; Bakir, M.; Rachid, E.; Chaffia, B.; Omar, B.; Rolain, J.M.; Diene, S.M. Prevalence and Genotypic Characterization of Salmonella spp. From Chicken Meats Marketed in the Province of Skikda, Algeria. J. Infect. Dev. Ctries. 2021, 15, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Guergueb, N.; Alloui, N.; Chachoua, I.; Ayachi, A.; Bennoun, O.; Aoun, L. Impact of Hygienic Slaughter Practices on Salmonella Contamination of Broiler Carcasses in Biskra, Algeria. Vet. Stanica 2020, 51, 463–470. [Google Scholar] [CrossRef]
- Abd-Elghany, S.M.; Fathy, T.M.; Zakaria, A.I.; Imre, K.; Morar, A.; Herman, V.; Pascalău, R.; Smuleac, L.; Morar, D.; Imre, M.; et al. Prevalence of Multidrug-Resistant Salmonella enterica Serovars in Buffalo Meat in Egypt. Foods 2022, 11, 2924. [Google Scholar] [CrossRef] [PubMed]
- El Shrek, Y.M.; Ali, M.R.M. Microbiological Study of Spiced Chicken Burgers in Tripoli City, Libya. East. Mediterr. Health J. 2012, 18, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Siala, M.; Barbana, A.; Smaoui, S.; Hachicha, S.; Marouane, C.; Kammoun, S.; Gdoura, R.; Messadi-Akrout, F. Screening and Detecting Salmonella in Different Food Matrices in Southern Tunisia Using a Combined Enrichment/Real-Time PCR Method: Correlation with Conventional Culture Method. Front. Microbiol. 2017, 8, 2416. [Google Scholar] [CrossRef]
- Mohamed, M.-Y.I.; Habib, I. Pathogenic E. coli in the Food Chain across the Arab Countries: A Descriptive Review. Foods 2023, 12, 3726. [Google Scholar] [CrossRef] [PubMed]
- Hawwas, H.A.E.H.; Aboueisha, A.K.M.; Fadel, H.M.; El-Mahallawy, H.S. Salmonella Serovars in Sheep and Goats and Their Probable Zoonotic Potential to Humans in Suez Canal Area, Egypt. Acta Vet. Scand. 2022, 64, 17. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Habib, I.; Shahabi, M.P.; Dybing, N.A.; Wang, P.; Bruce, M. A Review of the Public Health Challenges of Salmonella and Turtles. Vet. Sci. 2020, 7, 56. [Google Scholar] [CrossRef]
- EFSA. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Bui, H.T.; Truong, T.A.; Lam, D.N.; Ikeuchi, S.; Ly, L.K.T.; Hara-Kudo, Y.; Taniguchi, T.; Hayashidani, H. Retail Fresh Vegetables as a Potential Source of Salmonella Infection in the Mekong Delta, Vietnam. Int. J. Food Microbiol. 2021, 341, 109049. [Google Scholar] [CrossRef]
- Habib, I.; Khan, M.; Mohamed, M.-Y.I.; Ghazawi, A.; Abdalla, A.; Lakshmi, G.; Elbediwi, M.; Al Marzooqi, H.M.; Afifi, H.S.; Shehata, M.G.; et al. Assessing the Prevalence and Potential Risks of Salmonella Infection Associated with Fresh Salad Vegetable Consumption in the United Arab Emirates. Foods 2023, 12, 3060. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.J.; Abdul-Aziz, S.; Bitrus, A.A.; Mohammed, D.G.; Abu, J.; Bejo, S.K.; Mohamed, M.A.; Mohamed, M.-Y.I. Occurrence of Multidrug Resistant (MDR) Campylobacter Species Isolated from Retail Chicken Meats in Selangor, Malaysia and Their Associated Risk Factors. Malays. J. Microbiol. 2018, 14, 261–272. [Google Scholar]
- ECDC. EFSA EMA (European Centre for Disease Prevention and Control, European Food Safety Authority and European Medicines Agency), 2015.” First Joint Report on the Integrated Analysis of the Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals (JIACRA). EFSA J. 2015, 13, 4006. [Google Scholar]
- Oludairo, O.; Kwaga, J.; Kabir, J.; Abdu, P.; Gitanjali, A.; Perrets, A.; Cibin, V.; Lettini, A.; Olaniyi Aiyedun, J.; Daodu, O.; et al. Transmission of Salmonella in Humans and Animals and Its Epidemiological Factors. Zagazig Vet. J. 2023, 51, 76–91. [Google Scholar] [CrossRef]
- Habib, I.; Mohteshamuddin, K.; Mohamed, M.-Y.I.; Lakshmi, G.B.; Abdalla, A.; Bakhit Ali Alkaabi, A. Domestic Pets in the United Arab Emirates as Reservoirs for Antibiotic-Resistant Bacteria: A Comprehensive Analysis of Extended-Spectrum Beta-Lactamase Producing Escherichia coli Prevalence and Risk Factors. Animals 2023, 13, 1587. [Google Scholar] [CrossRef]
- Djomgoue, N.G.; Fonbah, L.J.; Mbulli, A.I.; Ousenu, K.; Bonglavnyuy, T.C. Risk Factors and Associated Outcomes of Virulence Genes Eae, EntB, and PipD Carriage in Escherichia coli, Klebsiella pneumoniae, and Salmonella spp. From HIV-1 and HIV-Negative Gastroenteritis Patients in the Dschang Regional Hospital Annex. Cureus 2023, 15, e42329. [Google Scholar] [CrossRef]
- Merati, R.; Boudra, A. Detection and Antimicrobial Susceptibility of Salmonella spp. Isolated From Commercial Eggs in Tiaret Province, Algeria. World’s Vet. J. 2023, 13, 200–204. [Google Scholar] [CrossRef]
- Abdel-Maksoud, M.; Abdel-Khalek, R.; El-Gendy, A.; Gamal, R.F.; Abdelhady, H.M.; House, B.L. Genetic Characterisation of Multidrug-Resistant Salmonella enterica Serotypes Isolated from Poultry in Cairo, Egypt. Afr. J. Lab. Med. 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Morshdy, A.E.M.A.; Mahmoud, A.F.A.; Khalifa, S.M.; El-Dien, W.M.S.; Darwish, W.S.; El Bayomi, R. Prevalence of Staphylococcus aureus and Salmonella Species in Chicken Meat Products Retailed in Egypt. Slov. Vet. Res. 2023, 60, 425–432. [Google Scholar] [CrossRef]
- Amajoud, N.; Bouchrif, B.; El Maadoudi, M.; Skalli Senhaji, N.; Karraouan, B.; El Harsal, A.; El Abrini, J. Prevalence, Serotype Distribution, and Antimicrobial Resistance of Salmonella Isolated from Food Products in Morocco. J. Infect. Dev. Ctries. 2017, 11, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Nacer, S.; El Ftouhy, F.Z.; Derqaoui, S.; Khayli, M.; Nassik, S.; Lkhider, M. Prevalence and Antibiotic Resistance of Salmonella spp. and Staphylococcus aureus Isolated from Broiler Chicken Meat in Modern and Traditional Slaughterhouses of Morocco. World’s Vet. J. 2022, 4, 430–439. [Google Scholar] [CrossRef]
- Bouchrif, B.; Paglietti, B.; Murgia, M.; Piana, A.; Cohen, N.; Ennaji, M.M.; Rubino, S.; Timinouni, M. Prevalence and Antibiotic-Resistance of Salmonella Isolated from Food in Morocco. J. Infect. Dev. Ctries. 2009, 3, 35–40. [Google Scholar]
- Hamad, R.M.A.; Saleh, A.A.H. Incidence of Some Food Poisoning Bacteria in Raw Meat Products with Molecular Detection of Salmonella in Al Beida City, Libya. Alex. J. Vet. Sci. 2019, 61, 11–17. [Google Scholar] [CrossRef]
- Ben Hassena, A.; Siala, M.; Guermazi, S.; Zormati, S.; Gdoura, R.; Sellami, H. Occurrence and Phenotypic and Molecular Characterization of Antimicrobial Resistance of Salmonella Isolates from Food in Tunisia. J. Food Prot. 2019, 82, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Kirrella, G.A.K.; Deeb, A.M.M.; Abdallah, R.M.I. Safety of Frozen Liver for Human Consumption. J. Food Drug Anal. 2017, 25, 520–524. [Google Scholar] [CrossRef]
- Ayachi, A.; Bennoune, O.; Heleili, N.; Alloui, N. Minor Salmonella: Potential Pathogens in Eggs in Algeria. J. Infect. Dev. Ctries. 2015, 9, 1156–1160. [Google Scholar] [CrossRef]
- Nouichi, S.; Ouatouat, R.; Can, H.Y.; Mezali, L.; Belkader, C.; Ouar-Korichi, M.; Bertrand, S.; Cantekin, Z.; Hamdi, T.M. Prevalence and Antimicrobial Resistance of Salmonella Isolated from Bovine and Ovine Samples in Slaughterhouses of Algiers, Algeria. J. Hell. Vet. Med. Soc. 2018, 69, 863–872. [Google Scholar] [CrossRef]
- Adjlane-Kaouche, S.; Benhacine, R.; Ghozlane, F.; Mati, A. Nutritional and Hygienic Quality of Raw Milk in the Mid-Northern Region of Algeria: Correlations and Risk Factors. Sci. World J. 2014, 2014, 131593. [Google Scholar] [CrossRef] [PubMed]
- Sabeq, I.; Awad, D.; Hamad, A.; Nabil, M.; Aboubakr, M.; Abaza, M.; Fouad, M.; Hussein, A.; Shama, S.; Ramadan, H.; et al. Prevalence and Molecular Characterization of Foodborne and Human-derived Salmonella Strains for Resistance to Critically Important Antibiotics. Transbound. Emerg. Dis. 2022, 69, e2153–e2163. [Google Scholar] [CrossRef]
- Moawad, A.A.; Hotzel, H.; Awad, O.; Tomaso, H.; Neubauer, H.; Hafez, H.M.; El-Adawy, H. Occurrence of Salmonella enterica and Escherichia coli in Raw Chicken and Beef Meat in Northern Egypt and Dissemination of Their Antibiotic Resistance Markers. Gut Pathog. 2017, 9, 57. [Google Scholar] [CrossRef]
- Abaza, A.F. Bacteriological Assessment of Some Vegetables and Ready-to-Eat Salads in Alexandria, Egypt. J. Egypt. Public Health Assoc. 2017, 92, 177–187. [Google Scholar] [CrossRef]
- Tarabees, R.; Elsayed, M.S.A.; Shawish, R.; Basiouni, S.; Shehata, A.A. Isolation and Characterization of Salmonella enteritidis and Salmonella typhimurium from Chicken Meat in Egypt. J. Infect. Dev. Ctries. 2017, 11, 314–319. [Google Scholar] [CrossRef]
- Abdel-Aziz, N.M. Detection of Salmonella Species in Chicken Carcasses Using Genus Specific Primer Belong to InvA Gene in Sohag City, Egypt. Vet. World 2016, 9, 1125–1128. [Google Scholar] [CrossRef] [PubMed]
- ElRahman, H.; El Rahman, A.E.-R.A.A. Molecular Characterization of Salmonella enterica Isolated from Chicken Meat and Its Products by Multiplex PCR. Alex. J. Vet. Sci. 2015, 46, 155. [Google Scholar] [CrossRef]
- Abd-Elghany, S.M.; Sallam, K.I.; Abd-Elkhalek, A.; Tamura, T. Occurrence, Genetic Characterization and Antimicrobial Resistance of Salmonella Isolated from Chicken Meat and Giblets. Epidemiol. Infect. 2015, 143, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Shimamoto, T. Isolation and Molecular Characterization of Salmonella enterica, Escherichia coli O157:H7 and Shigella spp. from Meat and Dairy Products in Egypt. Int. J. Food Microbiol. 2014, 168–169, 57–62. [Google Scholar] [CrossRef]
- Hassanein, R.; Fathi Hassan Ali, S.; Mohamed Abd El-Malek, A.; Mohamed, A.; Ibrahim Elsayh, K. Detection and Identification of Salmonella Species in Minced Beef and Chicken Meats by Using Multiplex PCR in Assiut City. Vet. World 2011, 4, 5–11. [Google Scholar] [CrossRef]
- El Ftouhy, F.Z.; Hmyene, A.; Nacer, S.; Kadiri, A.; Charrat, N.; Fagrach, A.; Derqaoui, S.; Nassik, S. Antibiotic Resistance of Escherichia coli and Salmonella Species Isolated from Table Eggs in Morocco. World’s Vet. J. 2023, 13, 167–174. [Google Scholar] [CrossRef]
- Zahli, R.; Soliveri, J.; Abrini, J.; Copa-Patiño, J.L.; Nadia, A.; Scheu, A.-K.; Nadia, S.S. Prevalence, Typing and Antimicrobial Resistance of Salmonella Isolates from Commercial Shellfish in the North Coast of Morocco. World J. Microbiol. Biotechnol. 2021, 37, 170. [Google Scholar] [CrossRef] [PubMed]
- Ed-dra, A.; Filali, F.R.; Karraouan, B.; El Allaoui, A.; Aboulkacem, A.; Bouchrif, B. Prevalence, Molecular and Antimicrobial Resistance of Salmonella Isolated from Sausages in Meknes, Morocco. Microb. Pathog. 2017, 105, 340–345. [Google Scholar] [CrossRef]
- Ennadir, J.; Hassikou, R.; Ohmani, F.; Hammamouchi, J.; Bouazza, F.; Qasmaoui, A.; Mennane, Z.; Touhami, A.O.; Charof, R.; Khedid, K. Qualité Microbiologique Des Farines de Blé Consommées Au Maroc. Can. J. Microbiol. 2012, 58, 145–150. [Google Scholar] [CrossRef]
- Ammari, S.; Laglaoui, A.; En-Nanei, L.; Bertrand, S.; Wildemauwe, C.; Abid, M. Isolation, Drug Resistance and Molecular Characterization of Salmonella Isolates in Northern Morocco. J. Infect. Dev. Ctries. 2009, 3, 041–049. [Google Scholar] [CrossRef] [PubMed]
- Setti, I.; Rodriguez-Castro, A.; Pata, M.P.; Cadarso-Suarez, C.; Yacoubi, B.; Bensmael, L.; Moukrim, A.; Martinez-Urtaza, J. Characteristics and Dynamics of Salmonella Contamination along the Coast of Agadir, Morocco. Appl. Environ. Microbiol. 2009, 75, 7700–7709. [Google Scholar] [CrossRef]
- Cohen, N.; Filhol, I.; Karraouan, B.; Badri, S.; Carle, I.; Ennaji, H.; Bouchrif, B.; Hassar, M.; Hakim Karib, P. Microbial Quality Control of Raw Ground Beef and Fresh Sausage in Casablanca (Morocco). J. Environ. Health 2008, 71, 51–55. [Google Scholar]
- Abdellah, C.; Fouzia, R.F.; Abdelkader, C.; Rachida, S.B.; Mouloud, Z. Occurrence of Salmonella in Chicken Carcasses and Giblets in Meknès-Morocco. Pak. J. Nutr. 2008, 7, 231–233. [Google Scholar] [CrossRef]
- Ibenyassine, K.; Mhand, R.A.; Karamoko, Y.; Anajjar, B.; Chouibani, M.; Ennaji, M. Bacterial Pathogens Recovered from Vegetables Irrigated by Wastewater in Morocco. J. Environ. Health 2007, 69, 47–51. [Google Scholar]
- Gritli, A.; Daboussi, T.; Ben Moussa, M.; Abassi, M. Prevalence and Characterizaton of Salmonella in Chicken Consumed in Military Cantines. JS-INAT 2015, 12, 908–914. [Google Scholar]
- Abbassi-Ghozzi, I.; Jaouani, A.; Hammami, S.; Martinez-Urtaza, J.; Boudabous, A.; Gtari, M. Molecular Analysis and Antimicrobial Resistance of Salmonella Isolates recovered from Raw Meat Marketed in the Area of ‘“Grand Tunis”’, Tunisia. Pathol. Biol. 2012, 60, e49–e54. [Google Scholar] [CrossRef]
- Vilela, F.P.; dos Prazeres Rodrigues, D.; Costa, R.G.; Casas, M.R.T.; Falcão, J.P.; Campioni, F. High Similarity and High Frequency of Virulence Genes among Salmonella Dublin Strains Isolated over a 33-Year Period in Brazil. Braz. J. Microbiol. 2020, 51, 497–509. [Google Scholar] [CrossRef]
- Mohamed, M.-Y.I.; Abu, J.; Aziz, S.A.; Zakaria, Z.; Khan, A.R.; Habib, I. Occurrence of Antibiotic Resistant C. jejuni and E. coli in Wild Birds, Chickens, Humans, and the Environment in Malay Villages, Kedah, Malaysia. Vet. Med. 2022, 67, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Hull, D.M.; Erin, H.; Lyndy, H.; Siddhartha, T. Multidrug Resistance and Virulence Genes Carried by Mobile Genomic Elements in Salmonella enterica Isolated from Live Food Animals, Processed, and Retail Meat in North Carolina, 2018–2019. Int. J. Food Microbiol. 2022, 2, 109821. [Google Scholar] [CrossRef] [PubMed]
- Khalefa, H.S.; Ahmed, Z.S.; Abdel-Kader, F.; Ismail, E.M.; Elshafiee, E.A. Sequencing and Phylogenetic Analysis of the Stn Gene of Salmonella Species Isolated from Different Environmental Sources at Lake Qarun Protectorate: The Role of Migratory Birds and Public Health Importance. Vet. World 2021, 14, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.-Y.I.; Habib, I.; Khalifa, H.O. Salmonella in the Food Chain within the Gulf Cooperation Council Countries. AIMS Microbiol. 2024, 10, 468–488. [Google Scholar] [CrossRef] [PubMed]
- Kombade, S.; Kaur, N. Pathogenicity Island in Salmonella. In Salmonella spp.—A Global Challenge; IntechOpen: London, UK, 2021. [Google Scholar]
- Wang, M.; Qazi, I.H.; Wang, L.; Zhou, G.; Han, H. Salmonella Virulence and Immune Escape. Microorganisms 2020, 8, 407. [Google Scholar] [CrossRef]
- Borah, P.; Dutta, R.; Das, L.; Hazarika, G.; Choudhury, M.; Deka, N.K.; Malakar, D.; Hussain, M.I.; Barkalita, L.M. Prevalence, Antimicrobial Resistance and Virulence Genes of Salmonella Serovars Isolated from Humans and Animals. Vet. Res. Commun. 2022, 46, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Nikiema, M.E.M.; Kakou-ngazoa, S.; Ky/Ba, A.; Sylla, A.; Bako, E.; Addablah, A.Y.A.; Ouoba, J.B.; Sampo, E.; Gnada, K.; Zongo, O.; et al. Characterization of Virulence Factors of Salmonella Isolated from Human Stools and Street Food in Urban Areas of Burkina Faso. BMC Microbiol. 2021, 21, 338. [Google Scholar] [CrossRef] [PubMed]
- Habib, I.; Elbediwi, M.; Ghazawi, A.; Mohamed, M.Y.I.; Lakshmi, G.B.; Khan, M. First Report from Supermarket Chicken Meat and Genomic Characterization of Colistin Resistance Mediated by Mcr-1.1 in ESBL-Producing, Multidrug-Resistant Salmonella Minnesota. Int. J. Food Microbiol. 2022, 379, 109835. [Google Scholar] [CrossRef]
- Lozano-Villegas, K.J.; Herrera-Sánchez, M.P.; Beltrán-Martínez, M.A.; Cárdenas-Moscoso, S.; Rondón-Barragán, I.S. Molecular Detection of Virulence Factors in Salmonella Serovars Isolated from Poultry and Human Samples. Vet. Med. Int. 2023, 2023, 1875253. [Google Scholar] [CrossRef] [PubMed]
- Dougnon, T.V.; Legba, B.; Deguenon, E.; Hounmanou, G.; Agbankpe, J.; Amadou, A.; Fabiyi, K.; Assogba, P.; Hounsa, E.; Aniambossou, A.; et al. Pathogenicity, Epidemiology and Virulence Factors of Salmonella Species: A Review. Not. Sci. Biol. 2017, 9, 460–466. [Google Scholar] [CrossRef]
- Cerny, O.; Holden, D.W. Salmonella SPI-2 Type III Secretion System-Dependent Inhibition of Antigen Presentation and T Cell Function. Immunol. Lett. 2019, 215, 35–39. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; MacKenzie, K.D.; Cameron, A.D.S. Salmonella Pathogenicity Island 1 (SPI-1): The Evolution and Stabilization of a Core Genomic Type Three Secretion System. Microorganisms 2020, 8, 576. [Google Scholar] [CrossRef] [PubMed]
- Wemyss, M.A.; Pearson, J.S. Host Cell Death Responses to Non-Typhoidal Salmonella Infection. Front. Immunol. 2019, 10, 1758. [Google Scholar] [CrossRef] [PubMed]
- Hasan, T.O. Genomic Diversity Analysis of Salmonella spp. from Broiler and Layer Flocks and Their Feed and Water in Karbala. Ph.D. Thesis, University of Baghdad, Baghdad, Iraq, 2021. [Google Scholar]
- Wang, Y.; Liu, G.; Zhang, J.; Gu, D.; Hu, M.; Zhang, Y.; Pan, Z.; Geng, S.; Jiao, X. WbaP Is Required for Swarm Motility and Intramacrophage Multiplication of Salmonella enteritidis SpiC Mutant by Glucose Use Ability. Microbiol. Res. 2021, 245, 126686. [Google Scholar] [CrossRef] [PubMed]
- Albanwawy, J.N.A.; Abdul-Lateef, L.A. Molecular Detection of Some of the Salmonella Typhi Virulence Genes Isolated in the Province of Babylon/Iraq. Ann. Rom. Soc. Cell Biol. 2021, 25, 679–685. [Google Scholar]
- Hsu, C.-H.; Li, C.; Hoffmann, M.; McDermott, P.; Abbott, J.; Ayers, S.; Tyson, G.H.; Tate, H.; Yao, K.; Allard, M.; et al. Comparative Genomic Analysis of Virulence, Antimicrobial Resistance, and Plasmid Profiles of Salmonella Dublin Isolated from Sick Cattle, Retail Beef, and Humans in the United States. Microb. Drug Resist. 2019, 25, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Wohlleben, W. Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity. Med. Sci. 2022, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Liu, X.; Li, C.; Jiang, Z.; Li, D.; Zhu, L. Genomic Virulence Genes Profile Analysis of Salmonella enterica Isolates from Animal and Human in China from 2004 to 2019. Microb. Pathog. 2022, 173, 105808. [Google Scholar] [CrossRef] [PubMed]
- Karabasanavar, N.S.; Madhavaprasad, C.B.; Gopalakrishna, S.A.; Hiremath, J.; Patil, G.S.; Barbuddhe, S.B. Prevalence of Salmonella Serotypes S. Enteritidis and S. Typhimurium in Poultry and Poultry Products. J. Food Saf. 2020, 40, e12852. [Google Scholar] [CrossRef]
- Nouichi, S.; Mezali, L.; Hamdi, T.M. Distribution of Salmonella Virulence Factors Originated from Sheep and Cattle in Algerian Slaughterhouses. J. Hell. Vet. Med. Soc. 2022, 73, 5013–5020. [Google Scholar] [CrossRef]
- Helal, G.; Tarabees, R.; Younis, G. Journal of Current Veterinary Research Molecular Characterization of Virulence Genes Associated with Salmonella spp. Isolated From Poultry. J. Curr. Vet. Res. 2019, 2, 36–46. [Google Scholar]
- Diab, M.S.; Thabet, A.S.; Elsalam, M.A.; Ewida, R.M.; Sotohy, S.A. Detection of Virulence and β-Lactamase Resistance Genes of Non-Typhoidal Salmonella Isolates from Human and Animal Origin in Egypt “One Health Concern”. Gut Pathog. 2023, 15, 16. [Google Scholar] [CrossRef]
- Ed-Dra, A.; Filali, F.R.; Khayi, S.; Oulghazi, S.; Bouchrif, B.; El Allaoui, A.; Ouhmidou, B.; Moumni, M. Antimicrobial Resistance, Virulence Genes, and Genetic Diversity of Salmonella enterica Isolated from Sausages. Eur. J. Microbiol. Immunol. 2019, 9, 56–61. [Google Scholar] [CrossRef]
- Ben Hassena, A.; Belmabrouk, S.; Amor, M.G.-B.; Zormati, S.; Guermazi-Toumi, S.; Siala-Trigui, M.; Gdoura, R. Study of Virulence Genes, Antimicrobial Resistance, and Genetic Relatedness of Foodborne Salmonella Isolates from Tunisia. J. Food Prot. 2022, 85, 1779–1789. [Google Scholar] [CrossRef] [PubMed]
- Habib, I.; Elbediwi, M.; Mohamed, M.-Y.I.; Ghazawi, A.; Abdalla, A.; Khalifa, H.O.; Khan, M. Enumeration, Antimicrobial Resistance and Genomic Characterization of Extended-Spectrum β-Lactamases Producing Escherichia coli from Supermarket Chicken Meat in the United Arab Emirates. Int. J. Food Microbiol. 2023, 398, 110224. [Google Scholar] [CrossRef]
- Haseeb, A.; Faidah, H.S.; Alghamdi, S.; Alotaibi, A.F.; Elrggal, M.E.; Mahrous, A.J.; Abuhussain, S.S.A.; Obaid, N.A.; Algethamy, M.; AlQarni, A.; et al. Dose Optimization of β-Lactams Antibiotics in Pediatrics and Adults: A Systematic Review. Front. Pharmacol. 2022, 13, 964005. [Google Scholar] [CrossRef] [PubMed]
- Judge, A.; Hu, L.; Sankaran, B.; Van Riper, J.; Venkataram Prasad, B.V.; Palzkill, T. Mapping the Determinants of Catalysis and Substrate Specificity of the Antibiotic Resistance Enzyme CTX-M β-Lactamase. Commun. Biol. 2023, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Krajnc, A.; Gobec, S. Conjugates of Monocyclic β-Lactams and Siderophore Mimetics: A Patent Evaluation (WO2023023393). Expert. Opin. Ther. Pat. 2023, 33, 471–476. [Google Scholar] [CrossRef]
- Habib, I.; Elbediwi, M.; Mohteshamuddin, K.; Mohamed, M.-Y.I.; Lakshmi, G.B.; Abdalla, A.; Anes, F.; Ghazawi, A.; Khan, M.; Khalifa, H. Genomic Profiling of Extended-Spectrum β-Lactamase-Producing Escherichia coli from Pets in the United Arab Emirates: Unveiling Colistin Resistance Mediated by Mcr-1.1 and Its Probable Transmission from Chicken Meat—A One Health Perspective. J. Infect. Public Health 2023, 16, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Rahal, K.; Reghal, A. A Nosocomial Epidemic of Salmonella Mbandaka Which Produces Various Broad Spectrum Beta-Lactamases: Preliminary Results. Med. Trop. 1994, 54, 227–230. [Google Scholar]
- Khalifa, H.O.; Shikoray, L.; Mohamed, M.-Y.I.; Habib, I.; Matsumoto, T. Veterinary Drug Residues in the Food Chain as an Emerging Public Health Threat: Sources, Analytical Methods, Health Impacts, and Preventive Measures. Foods 2024, 13, 1629. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, N.; Balestracci, K.; Tovar, A.; Corcoran, C.; Watts, D.-J.; Tobar, F.; Samson, M.; Amin, S. Multi-Prong Formative Evaluation of a Pediatric Clinical-Community Food Access and Nutrition Education Intervention. Am. J. Health Promot. 2024. [Google Scholar] [CrossRef]
- Mohamed, M.-Y.I. Campylobacteriosis in North Africa. AIMS Agric. Food 2024, 9, 801–821. [Google Scholar] [CrossRef]
- Eckstein, S.; Ehmann, R.; Gritli, A.; Ben Rhaiem, M.; Ben Yahia, H.; Diehl, M.; Wölfel, R.; Handrick, S.; Ben Moussa, M.; Stoecker, K. Viral and Bacterial Zoonotic Agents in Dromedary Camels from Southern Tunisia: A Seroprevalence Study. Microorganisms 2022, 10, 727. [Google Scholar] [CrossRef] [PubMed]
- Habib, I.; Mohamed, M.Y.I. Foodborne Infections in the Middle East. In Food Safety in the Middle East; Elsevier: Amsterdam, The Netherlands, 2022; pp. 71–107. ISBN 9780128224175. [Google Scholar]
- Mohamed, M.-Y.I.; Abu, J.; Zakaria, Z.; Khan, A.R.; Abdul Aziz, S.; Bitrus, A.A.; Habib, I. Multi-Drug Resistant Pathogenic Escherichia coli Isolated from Wild Birds, Chicken, and the Environment in Malaysia. Antibiotics 2022, 11, 1275. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, S.; Chhetri, R. A Literature Review on Impact of COVID-19 Pandemic on Teaching and Learning. High. Educ. Future 2021, 8, 133–141. [Google Scholar] [CrossRef]
- Adil, M.T.; Rahman, R.; Whitelaw, D.; Jain, V.; Al-Taan, O.; Rashid, F.; Munasinghe, A.; Jambulingam, P. SARS-CoV-2 and the Pandemic of COVID-19. Postgrad. Med. J. 2021, 97, 110–116. [Google Scholar] [CrossRef]
| Country | Geographical Distribution | Tested Food Samples or Human | Trend Over Time | Salmonella Serotypes (Total Number) | Virulence Genes (%) * | Techniques Used for Virulence Genes Detection and References |
|---|---|---|---|---|---|---|
| Algeria | El-Harrach and Hussein-Dey slaughterhouses in Algiers, | Sheep and cattle | Between 2013 and 2014 | Muenster (n = 33) | invA (63.6), pefA (0), sefA (0), pipB (0), sseC (63.6), ssaP (63.6), spvC (63.6), iroB (63.6) | PCR technique [95] |
| Kentucky (n = 13) | invA (76.9), pefA (0), sefA (0), pipB (0), sseC (38.7), ssaP (38.7), spvC (38.7), iroB (38.7) | |||||
| Infantis (n = 12) | invA (91.7), pefA (0), sefA (0), pipB (0), sseC (66.7), ssaP (66.7), spvC (66.7), iroB (66.7) | |||||
| Anatum (n = 11) | invA (81.8), pefA (0), sefA (0), pipB (0), sseC (72.7), ssaP (72.7), spvC (72.7), iroB (72.7) | |||||
| Richmond (n = 4) | invA (100), pefA (0), sefA (0), pipB (0), sseC (100), ssaP (100), spvC (100), iroB (100) | |||||
| Havana (n = 3) | invA (66.7), pefA (0), sefA (0), pipB (0), sseC (0), ssaP (0), spvC (0), iroB (0) | |||||
| Typhimurium (n = 3) | invA (100), pefA (0), sefA (100), pipB (100), sseC (100), ssaP (100), spvC (100), iroB (100) | |||||
| Montevideo (n = 3) | invA (100), pefA (0), sefA (0), pipB (0), sseC (66.7), ssaP (66.7), spvC (66.7), iroB (66.7) | |||||
| Virginia (n = 1) | invA (100), pefA (0), sefA (0), pipB (0), sseC (0), ssaP (0), spvC (0), iroB (0) | |||||
| Braenderup (n = 1) | invA (100), pefA (0), sefA (0), pipB (0), sseC (100), ssaP (100), spvC (100), iroB (100) | |||||
| Egypt | Different poultry farms located in El-Minufyia and El-Gharbia governorates | Liver, yolk sac and spleen | Between February 2017 to December 2017 | Sinchem (n = 1), Gallinarum (n = 1), S. enterica subsp. Salamae (n = 1), Kentucky (n = 1), Entertidis (n = 1), Typhimurium (n = 1), Heidelberg (n = 1), Hydra (n = 1), Virchow (n = 1), Farsta (n = 1) | pagC (100), msgA (100), spiA (100), invA (100), prgH (100), orgA (100), sipB (100), tolC (100), iroN (100), lpfC (100), pefA (100), sitC (100), sifA (100), sopB (100), spvB (70), cdtB (30) | PCR technique [96] |
| Different localities in the New Valley and Assiut Governorates | Raw milk, kareish cheese, Damietta cheese, yogurt, ice cream, animal fecal swabs, human fecal swabs and hand swabs | Between April 2020 and May 2021 | Enteritidis (n = 4), Typhimurium (n = 4), Infantis (n = 4), Tsevie (n = 4), Larochelle (n = 4), Virchow (n = 3), Haifa (n = 3), Molade (n = 3), Heidelberg (n = 2), Essen (n = 2), Shubra (n = 1), Alfort (n = 1), Apeyeme (n = 1) | invA (100), hilA (94.4), stn (72.2), spvC (30.6) | PCR technique [97] | |
| Suez Canal Area | Humans, sheep, and goats | 2021 | Typhimurium (n = 13) | invA (61.5), sopB (92.3), stn (92.3), spvC (92.3) | PCR technique [31] | |
| Enteritidis (n = 12) | invA (25), sopB (100), stn (100), spvC (100) | |||||
| Heidelberg (n = 4) | invA (0), sopB (100), stn (100), spvC (100) | |||||
| Dublin (n = 5) | invA (40), sopB (100), stn (100), spvC (100) | |||||
| Montevideo (n = 5) | invA (40), sopB (100), stn (100), spvC (100) | |||||
| Saintpaul (n = 6) | invA (50), sopB (100), stn (100), spvC (100) | |||||
| Anatum (n = 3) | invA (33.3), sopB (100), stn (100), spvC (100) | |||||
| Tsevie (n = 1) | invA (100), sopB (100), stn (100), spvC (100) | |||||
| Chester (n = 1) | invA (0), sopB (100), stn (100), spvC (100) | |||||
| Essen (n = 2) | invA (0), sopB (100), stn (100), spvC (100) | |||||
| Apeyeme (n = 1) | invA (100), sopB (100), stn (100), spvC (100) | |||||
| Infantis (n = 1) | invA (100), sopB (100), stn (100), spvC (100) | |||||
| Morocco | Meknes city | Sausages | 2018 | Typhimurium (n = 2), Kentucky (n = 6), Saintpaul (n = 1), Corvallis (n = 8), Montevideo (n = 3), Mbandaka (n = 4), Bovismorbificans (n = 2), Give (n = 4), Anatum (n = 1), Livingstone (n = 1), Muenster (n = 1), Agon (n = 1) | orgA (100), sipB (100), sitC (100), spiA (100) iroN (100) sifA (100) | PCR technique [98] |
| Tunisia | Bab Saadoun, Tunis. | Chicken consumed in military cantines | February to June 2013 | Enteritidis (n = 4) | invE/A (100), ttrC (100), mgtC (100), spvC (45.8) sopB (12.5), siiD (0) | PCR technique [71] |
| Sfax city | Foodborne Salmonella isolates | ND | Enteritidis (n = 27) | siiA (100), sopB (96.3), sopE (100), sopE2 (100), cat2 (100), safC (100), sefB (100), spvC (92.6) spvB (92.6) | PCR technique [99] | |
| London (n = 8) | siiA (100), sopB (100), sopE (0), sopE2 (100), cat2 (100), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Kentucky (n = 6) | siiA (100), sopB (100), sopE (0), sopE2 (0), cat2 (16.7), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Havana (n = 3) | siiA (100), sopB (100), sopE (0), sopE2 (0), cat2 (0), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Zanzibar (n = 2) | siiA (100), sopB (100), sopE (0), sopE2 (100), cat2 (100), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Anatum (n = 2) | siiA (100), sopB (100), sopE (0), sopE2 (100), cat2 (100), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Infantis (n = 1) | siiA (100), sopB (100), sopE (0), sopE2 (100), cat2 (100), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Manchester (n = 1) | siiA (100), sopB (0), sopE (0), sopE2 (100), cat2 (100), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Livingstone (n = 1) | siiA (100), sopB (100), sopE (0), sopE2 (100), cat2 (0), safC (100), sefB (0), spvC (0) spvB (0) | |||||
| Gabon (n = 1) | siiA (100), sopB (100), sopE (0), sopE2 (100), cat2 (100), safC (0), sefB (0), spvC (0) spvB (0) | |||||
| Northeast Tunisia | Broiler flocks | September 2019 and August 2020 | Kentucky (n = 13) | invA (100), spvC (0), hli (53.8) gipA (61.5), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | PCR technique [24] | |
| Mbandaka (n = 12) | invA (100), spvC (0), hli (33.3) gipA (83.3), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Anatum (n = 11) | invA (100), spvC (0), hli (36.4) gipA (72.7), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Zanzibar (n = 10) | invA (100), spvC (0), hli (80) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Enteritidis (n = 8) | invA (100), spvC (0), hli (75) gipA (87.5), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Infantis (n = 2) | invA (100), spvC (0), hli (0) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Indiana (n = 2) | invA (100), spvC (0), hli (50) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Corvallis (n = 1) | invA (100), spvC (0), hli (0) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Agona (n = 1) | invA (100), spvC (0), hli (0) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Hadar (n = 1) | invA (100), spvC (0), hli (100) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Montevideo (n = 1) | invA (100), spvC (0), hli (0) gipA (0), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Cerro (n = 1) | invA (100), spvC (0), hli (100) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Virginia (n = 1) | invA (100), spvC (0), hli (0) gipA (100), mgtC (100), trhH (0), sirA (100), pagK (100), sipA (0), sipD (0), sopD (0), SEN (0) | |||||
| Great Tunisia | Hospitalized patients with gastroenteritis | Between 2010 and 2020 | Enteritidis (n = 17) | invA (100), mgtC (100), sirA (100), gipA (76.5), pagK (100), hli (11.8), trhH (0), spvC (0), sipA (0), sipD (0), sopD (0), SEN 1417 (0) | PCR technique [19] | |
| Typhimurium (n = 16) | invA (100), mgtC (100), sirA (100), gipA (62.5), pagK (100), hli (93.8), trhH (0), spvC (0), sipA (0), sipD (0), sopD (0), SEN 1417 (0) | |||||
| Kentucky (n = 15) | invA (100), mgtC (100), sirA (100), gipA (60), pagK (93.3), hli (80), trhH (0), spvC (0), sipA (0), sipD (0), sopD (0), SEN 1417 (0) | |||||
| Anatum (n = 5) | invA (100), mgtC (100), sirA (100), gipA (40), pagK (80), hli (0), trhH (0), spvC (0), sipA (0), sipD (0), sopD (0), SEN 1417 (0) | |||||
| Infantis (n = 3) | invA (100), mgtC (100), sirA (100), gipA (0), pagK (100), hli (0), trhH (0), spvC (0), sipA (0), sipD (0), sopD (0), SEN 1417 (0) | |||||
| Muenster (n = 3) | invA (100), mgtC (100), sirA (100), gipA (33.3), pagK (66.7), hli (0), trhH (0), spvC (0), sipA (0), sipD (0), sopD (0), SEN 1417 (0) | |||||
| Mbandaka (n = 2) | invA (100), mgtC (100), sirA (100), gipA (100), pagK (100), hli (0), trhH (0), spvC (0), sipA (0), sipD (0), sopD (0), SEN 1417 (0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, M.-Y.I.; Khalifa, H.O.; Habib, I. Food Pathways of Salmonella and Its Ability to Cause Gastroenteritis in North Africa. Foods 2025, 14, 253. https://doi.org/10.3390/foods14020253
Mohamed M-YI, Khalifa HO, Habib I. Food Pathways of Salmonella and Its Ability to Cause Gastroenteritis in North Africa. Foods. 2025; 14(2):253. https://doi.org/10.3390/foods14020253
Chicago/Turabian StyleMohamed, Mohamed-Yousif Ibrahim, Hazim O. Khalifa, and Ihab Habib. 2025. "Food Pathways of Salmonella and Its Ability to Cause Gastroenteritis in North Africa" Foods 14, no. 2: 253. https://doi.org/10.3390/foods14020253
APA StyleMohamed, M.-Y. I., Khalifa, H. O., & Habib, I. (2025). Food Pathways of Salmonella and Its Ability to Cause Gastroenteritis in North Africa. Foods, 14(2), 253. https://doi.org/10.3390/foods14020253








