Microalga Nannochloropsis gaditana as a Sustainable Source of Bioactive Peptides: A Proteomic and In Silico Approach
Abstract
1. Introduction
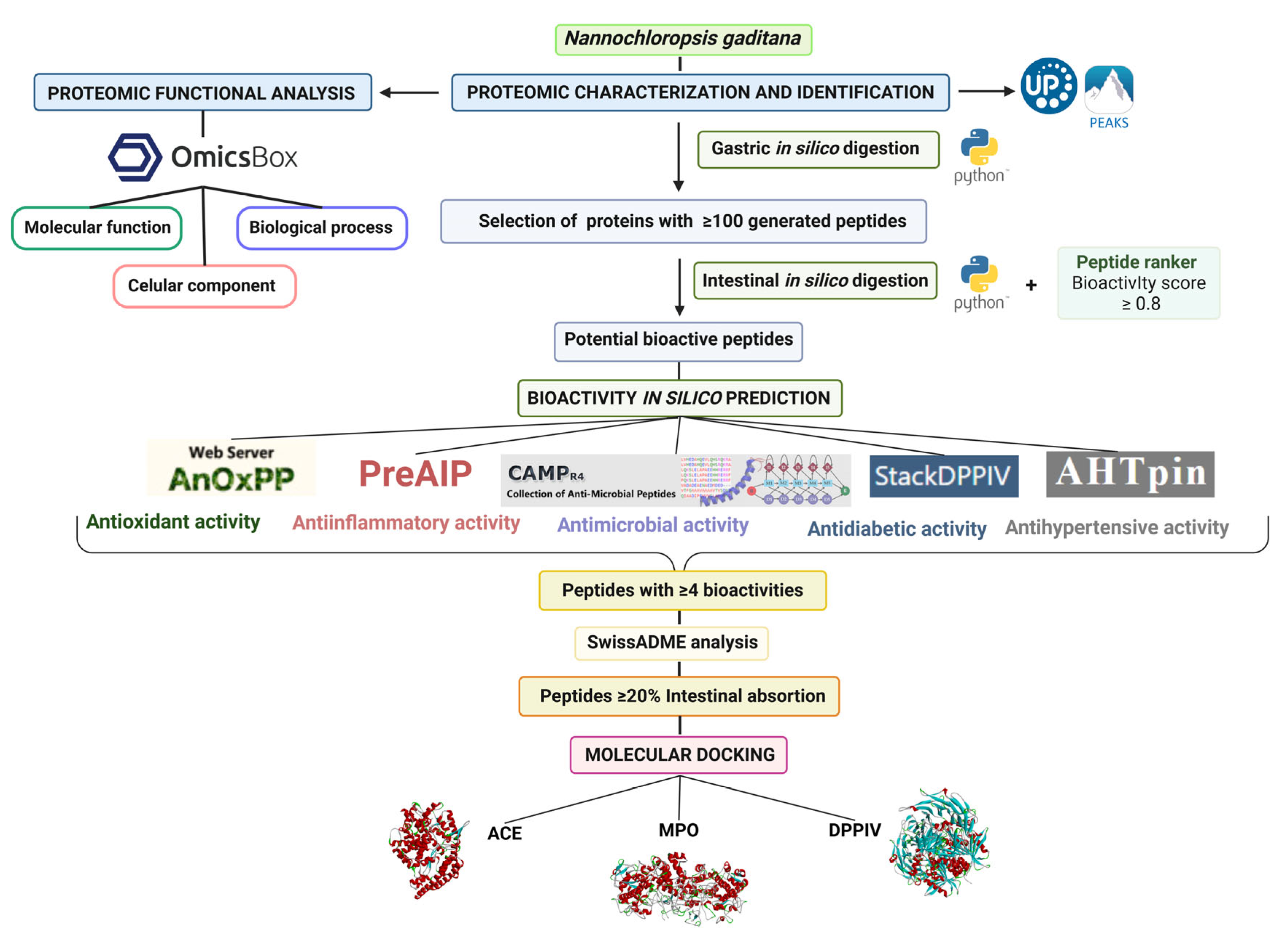
2. Materials and Methods
2.1. Samples and Reagents
2.2. Proteomic Identification and Characterization
2.2.1. Protein Content and AA Profile
2.2.2. In-Gel Digestion (Stacking Gel)
2.2.3. Reverse-Phase Liquid Chromatography (RP-LC-MS/MS) Analysis (Dynamic Exclusion Mode)
2.2.4. Data Processing
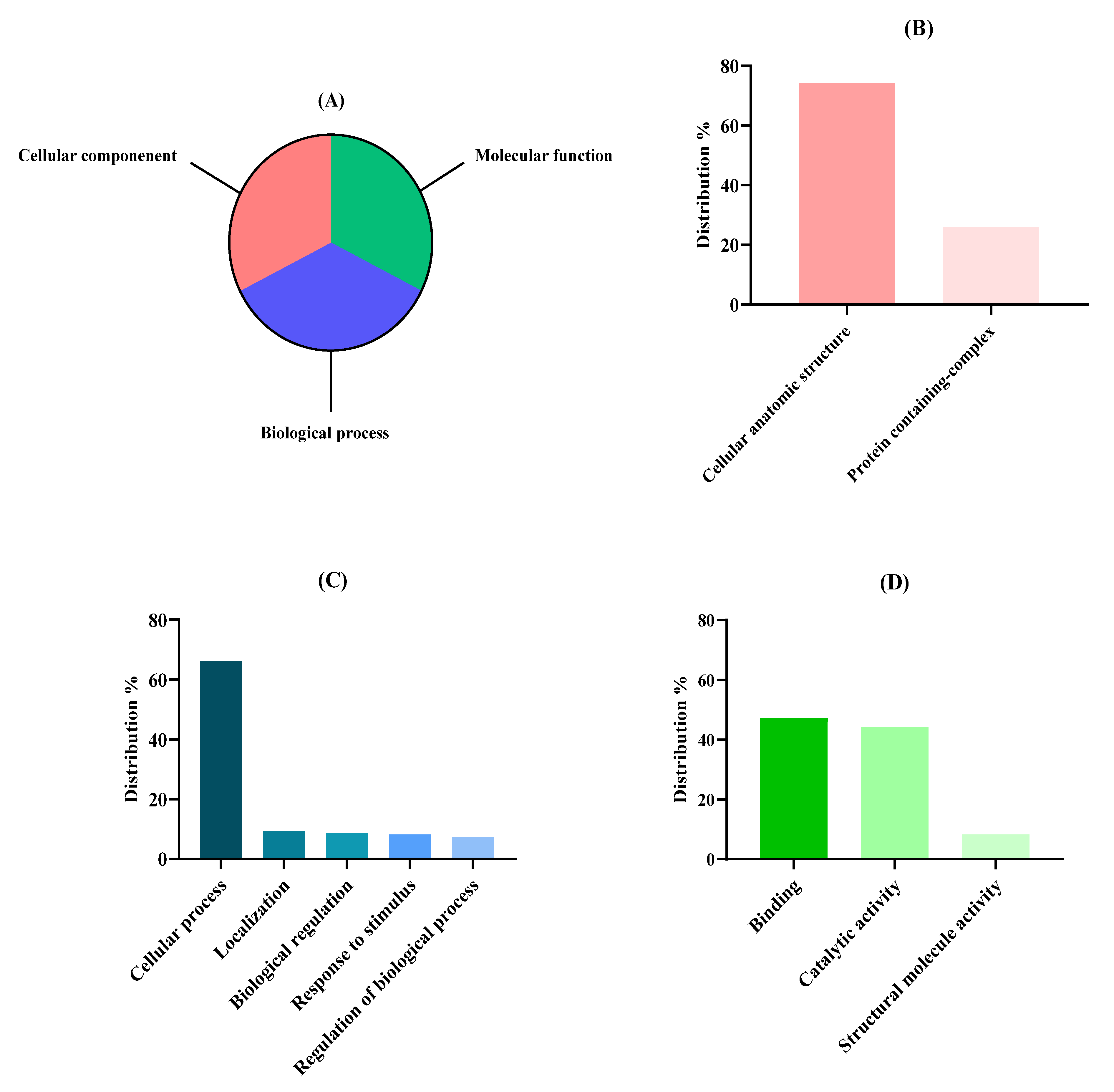
2.3. Proteomic Functional Analysis
2.4. In Silico Gastrointestinal Digestion of Microalga Proteins
2.5. Bioactivity Prediction by In Silico Analysis
2.6. Physicochemical and Pharmacokinetic Analysis
2.7. Peptide Molecular Docking
3. Results and Discussion
3.1. Proteome of the Microalga Nannochloropsis gaditana
3.2. Functional Analysis of Nannochloropsis gaditana Proteome
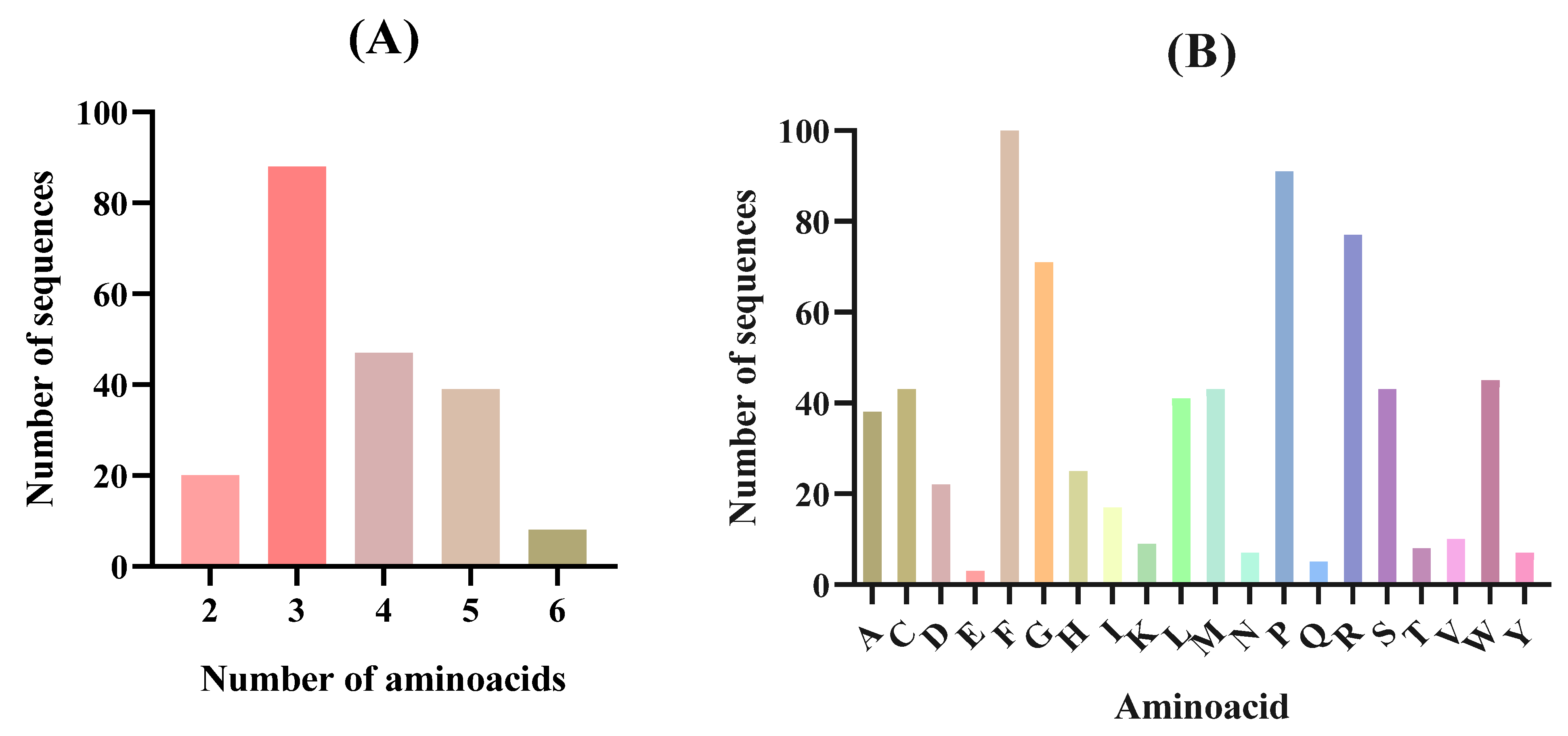
3.3. In Silico Gastrointestinal Digestion
| Peptide | Molecular Weight (Da) | Lipophilicity (MLogP) a | Bioavailability Score b | Water Solubility (log mol/L) c | % Intestinal Absorption d | AMES Toxicity e |
|---|---|---|---|---|---|---|
| FK | 293.36 | 0.6 | 0.55 | −2.818 | 35.3 | No |
| FR | 321.37 | 0.15 | 0.55 | −2.643 | 21.61 | No |
| GF | 222.24 | 0.34 | 0.55 | −1.85 | 41.89 | No |
| RF | 321.37 | 0.14 | 0.55 | −2.617 | 21.43 | No |
| ARF 1 | 60.06 | −1.6 | 0.55 | 0.824 | 71.496 | No |
| DPMP | 458.53 | −1.11 | 0.11 | −2.27 | 0 | No |
| FHPR | 555.63 | −1.46 | 0.17 | −2.875 | 6.011 | No |
| FSPR | 505.57 | −1.56 | 0.17 | −2.835 | 0 | No |
| HPKF | 527.62 | −1.11 | 0.17 | −2.815 | 17.04 | No |
| MPPR | 499.63 | −1.01 | 0.17 | −2784 | 6.78 | No |
| VPGF | 418.49 | −0.1 | 0.55 | −2.524 | 28.49 | No |
| APMRP | 570.71 | −1.53 | 0.17 | −2.922 | 0 | No |
| FIPGL 1,2 | 545.67 | 0.15 | 0.17 | −3.174 | 21.02 | No |
| GPGCG | 389.43 | −2.73 | 0.55 | −2.516 | 2.96 | No |
| KAPPF | 558.67 | −0.6 | 0.17 | −2.833 | 16.56 | No |
| KSPGW1 | 573.64 | −2.17 | 0.17 | −2.862 | 0.134 | No |
| PCMIR | 618.81 | −1.32 | 0.17 | −2.889 | 0 | No |
| PFGNR | 589.64 | −2.49 | 0.17 | −2.889 | 0 | No |
| PRPMR | 655.81 | −1.98 | 0.17 | −2.889 | 0 | No |
| RRCLF 1 | 693.86 | −1.22 | 0.17 | −2.894 | 0 | No |
| WWGGV | 603.67 | −0.74 | 0.17 | −3.015 | 19.76 | No |
| YLPPR2 | 644.76 | −0.84 | 0.17 | −2.904 | 19.333 | No |
| AVMPIF | 676.87 | −0.07 | 0.17 | −3.012 | 11.265 | No |
| EFPMIR | 791.96 | −1.31 | 0.17 | −2.894 | 0 | No |
| FARPGL 1 | 659.78 | −1.51 | 0.17 | −2.922 | 0 | No |
| FGPQGG 1 | 561.59 | −2.77 | 0.17 | −2.977 | 0.112 | No |
| FLPPAL 1 | 656.81 | 0.12 | 0.17 | −3.29 | 17.79 | No |
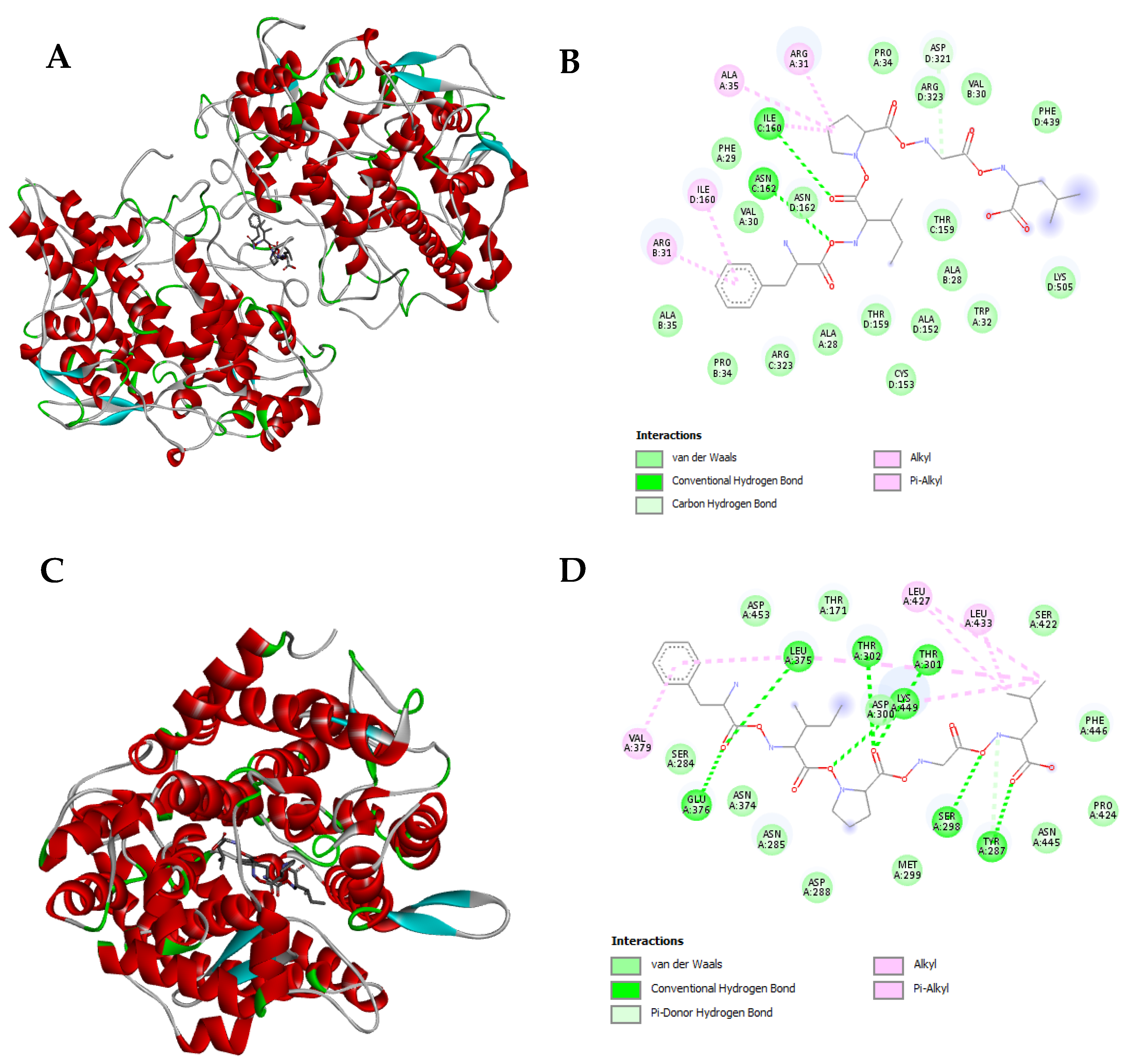
3.4. Molecular Docking of Nannochloropsis gaditana Potential Bioactive Peptides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bennett, J.M.; Reeves, G.; Billman, G.E.; Sturmberg, J.P. Inflammation–Nature’s Way to Efficiently Respond to All Types of Challenges: Implications for Understanding and Managing “the Epidemic” of Chronic Diseases. Front. Med. 2018, 5, 316. [Google Scholar] [CrossRef] [PubMed]
- Budreviciute, A.; Damiati, S.; Sabir, D.K.; Onder, K.; Schuller-Goetzburg, P.; Plakys, G.; Katileviciute, A.; Khoja, S.; Kodzius, R. Management and Prevention Strategies for Non-Communicable Diseases (NCDs) and Their Risk Factors. Front. Public Health 2020, 8, 574111. [Google Scholar] [CrossRef] [PubMed]
- Floret, C.; Monnet, A.F.; Micard, V.; Walrand, S.; Michon, C. Replacement of Animal Proteins in Food: How to Take Advantage of Nutritional and Gelling Properties of Alternative Protein Sources. Crit. Rev. Food Sci. Nutr. 2023, 63, 920–946. [Google Scholar] [CrossRef] [PubMed]
- Calvez, J.; Azzout-Marniche, D.; Tomé, D. Protein Quality, Nutrition and Health. Front. Nutr. 2024, 11, 1406618. [Google Scholar] [CrossRef]
- Zhuang, D.; He, N.; Khoo, K.S.; Ng, E.P.; Chew, K.W.; Ling, T.C. Application Progress of Bioactive Compounds in Microalgae on Pharmaceutical and Cosmetics. Chemosphere 2022, 291, 132932. [Google Scholar] [CrossRef]
- Mosibo, O.K.; Ferrentino, G.; Udenigwe, C.C. Microalgae Proteins as Sustainable Ingredients in Novel Foods: Recent Developments and Challenges. Foods 2024, 13, 733. [Google Scholar] [CrossRef]
- Chronakis, I.S.; Madsen, M. Algal Proteins. In Handbook of Food Proteins; Woodhead Publishing: Cambridge, UK, 2011; pp. 353–394. [Google Scholar] [CrossRef]
- Rzymski, P.; Budzulak, J.; Niedzielski, P.; Klimaszyk, P.; Proch, J.; Kozak, L.; Poniedziałek, B. Essential and Toxic Elements in Commercial Microalgal Food Supplements. J. Appl. Phycol. 2019, 31, 3567–3579. [Google Scholar] [CrossRef]
- Spolaore, P.; Joannis-Cassan, C.; Duran, E.; Isambert, A. Commercial Applications of Microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef]
- Guil-Guerrero, J.L.; Navarro-Juárez, R.; López-Martínez, J.C.; Campra-Madrid, P.; Rebolloso-Fuentes, M.M. Functional Properties of the Biomass of Three Microalgal Species. J. Food Eng. 2004, 65, 511–517. [Google Scholar] [CrossRef]
- Medina, C.; Rubilar, M.; Shene, C.; Torres, S.; Verdugo, M. Protein Fractions with Techno-Functional and Antioxidant Properties from Nannochloropsis Gaditana Microalgal Biomass. J. Biobased Mater. Bioenergy 2015, 9, 417–425. [Google Scholar] [CrossRef]
- Zaky, A.A.; Paterson, S.; Gómez-Cortés, P.; Hernández-Ledesma, B. Bioactive Peptides Released from Microalgae during Gastrointestinal Digestion. In Protein Digestion-Derived Peptides; Elsevier: Amsterdam, The Netherlands, 2024; pp. 335–352. [Google Scholar] [CrossRef]
- Du, C.; Gong, H.; Zhao, H.; Wang, P. Recent Progress in the Preparation of Bioactive Peptides Using Simulated Gastrointestinal Digestion Processes. Food Chem. 2024, 453, 139587. [Google Scholar] [CrossRef] [PubMed]
- Sheih, I.C.; Wu, T.K.; Fang, T.J. Antioxidant Properties of a New Antioxidative Peptide from Algae Protein Waste Hydrolysate in Different Oxidation Systems. Bioresour. Technol. 2009, 100, 3419–3425. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X. Isolation and Identification of Anti-Proliferative Peptides from Spirulina Platensis Using Three-Step Hydrolysis. J. Sci. Food Agric. 2017, 97, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, X. Separation, Antitumor Activities, and Encapsulation of Polypeptide from Chlorella Pyrenoidosa. Biotechnol. Prog. 2013, 29, 681–687. [Google Scholar] [CrossRef]
- Hayes, M.; Mora, L.; Lucakova, S. Identification of Bioactive Peptides from Nannochloropsis Oculata Using a Combination of Enzymatic Treatment, in Silico Analysis and Chemical Synthesis. Biomolecules 2022, 12, 1806. [Google Scholar] [CrossRef]
- Vo, T.S.; Ryu, B.M.; Kim, S.K. Purification of Novel Anti-Inflammatory Peptides from Enzymatic Hydrolysate of the Edible Microalgal Spirulina Maxima. J. Funct. Foods 2013, 5, 1336–1346. [Google Scholar] [CrossRef]
- Saleh, N.I.M.; Ghani, W.A.K.; Azlina, W.; Mohd Razif, H.; Kamal, M.; Mazlina, S. Optimization of Enzymatic Hydrolysis for the Production of Antioxidative Peptide from Nannochloropsis gaditana using Response Surface Methodology. Pertanika J. Sci. Technol. 2019, 27, 41–55. [Google Scholar] [CrossRef]
- Li, Y.; Aiello, G.; Fassi, E.M.A.; Boschin, G.; Bartolomei, M.; Bollati, C.; Roda, G.; Arnoldi, A.; Grazioso, G.; Lammi, C. Investigation of Chlorella Pyrenoidosa Protein as a Source of Novel Angiotensin I-Converting Enzyme (Ace) and Dipeptidyl Peptidase-Iv (Dpp-Iv) Inhibitory Peptides. Nutrients 2021, 13, 1624. [Google Scholar] [CrossRef]
- Pei, Y.; Cai, S.; Ryu, B.; Zhou, C.; Hong, P.; Qian, Z.J. An ACE Inhibitory Peptide from Isochrysis Zhanjiangensis Exhibits Antihypertensive Effect via Anti-Inflammation and Anti-Apoptosis in HUVEC and Hypertensive Rats. J. Funct. Foods 2022, 92, 105061. [Google Scholar] [CrossRef]
- Cavonius, L.R.; Albers, E.; Undeland, I. In Vitro Bioaccessibility of Proteins and Lipids of PH-Shift Processed Nannochloropsis Oculata Microalga. Food Funct. 2016, 7, 2016–2024. [Google Scholar] [CrossRef]
- Peredo-Lovillo, A.; Hernández-Mendoza, A.; Vallejo-Cordoba, B.; Romero-Luna, H.E. Conventional and in Silico Approaches to Select Promising Food-Derived Bioactive Peptides: A Review. Food Chem. X 2021, 13, 100183. [Google Scholar] [CrossRef] [PubMed]
- Paterson, S.; Fernández-Tomé, S.; Galvez, A.; Hernández-Ledesma, B. Evaluation of the Multifunctionality of Soybean Proteins and Peptides in Immune Cell Models. Nutrients 2023, 15, 1220. [Google Scholar] [CrossRef] [PubMed]
- Official Methods of Analysis, 22nd Edition (2023)—Aoac International. Available online: https://www.aoac.org/official-methods-of-analysis/ (accessed on 23 February 2024).
- Torres-Sánchez, E.; Morato, E.; Hernández-Ledesma, B.; Gutiérrez, L.F. Proteomic Analysis of the Major Alkali-Soluble Inca Peanut (Plukenetia volubilis) Proteins. Foods 2024, 13, 3275. [Google Scholar] [CrossRef] [PubMed]
- Sanchiz, Á.; Morato, E.; Rastrojo, A.; Camacho, E.; la Fuente, S.G.; Marina, A.; Aguado, B.; Requena, J.M. The Experimental Proteome of Leishmania Infantum Promastigote and Its Usefulness for Improving Gene Annotations. Genes 2020, 11, 1036. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Alonso, R.; Pisa, D.; Marina, A.I.; Morato, E.; Rábano, A.; Rodal, I.; Carrasco, L. Evidence for Fungal Infection in Cerebrospinal Fluid and Brain Tissue from Patients with Amyotrophic Lateral Sclerosis. Int. J. Biol. Sci. 2015, 11, 546. [Google Scholar] [CrossRef]
- Tran, N.H.; Qiao, R.; Xin, L.; Chen, X.; Liu, C.; Zhang, X.; Shan, B.; Ghodsi, A.; Li, M. Deep Learning Enables de Novo Peptide Sequencing from Data-Independent-Acquisition Mass Spectrometry. Nat. Methods 2019, 16, 63–66. [Google Scholar] [CrossRef]
- Tran, N.H.; Zhang, X.; Xin, L.; Shan, B.; Li, M. De Novo Peptide Sequencing by Deep Learning. Proc. Natl. Acad. Sci. USA 2017, 114, 8247–8252. [Google Scholar] [CrossRef]
- Tran, N.H.; Rahman, M.Z.; He, L.; Xin, L.; Shan, B.; Li, M. Complete De Novo Assembly of Monoclonal Antibody Sequences. Sci. Rep. 2016, 6, 31730. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Maillet, N. Rapid Peptides Generator: Fast and Efficient in Silico Protein Digestion. NAR Genom. Bioinform. 2020, 2, 1l. [Google Scholar] [CrossRef] [PubMed]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Jiao, L.; Wang, R.; Zhao, Y.; Hao, Y.; Liang, G. Prediction of Antioxidant Peptides Using a Quantitative Structure-Activity Relationship Predictor (AnOxPP) Based on Bidirectional Long Short-Term Memory Neural Network and Interpretable Amino Acid Descriptors. Comput. Biol. Med. 2023, 154, 106591. [Google Scholar] [CrossRef] [PubMed]
- Khatun, M.S.; Hasan, M.M.; Kurata, H. PreAIP: Computational Prediction of Anti-Inflammatory Peptides by Integrating Multiple Complementary Features. Front. Genet. 2019, 10, 129. [Google Scholar] [CrossRef]
- Gawde, U.; Chakraborty, S.; Waghu, F.H.; Barai, R.S.; Khanderkar, A.; Indraguru, R.; Shirsat, T.; Idicula-Thomas, S. CAMPR4: A Database of Natural and Synthetic Antimicrobial Peptides. Nucleic Acids Res. 2023, 51, D377–D383. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Singh Chauhan, J.; Nagpal, G.; Kumar, R.; Sharma, M.; Raghava, G.P.S. An in Silico Platform for Predicting, Screening and Designing of Antihypertensive Peptides. Sci. Rep. 2015, 5, 12512. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Nantasenamat, C.; Hasan, M.M.; Moni, M.A.; Lio’, P.; Manavalan, B.; Shoombuatong, W. StackDPPIV: A Novel Computational Approach for Accurate Prediction of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides. Methods 2022, 204, 189–198. [Google Scholar] [CrossRef]
- Marin, D.E.; Taranu, I. Using In Silico Approach for Metabolomic and Toxicity Prediction of Alternariol. Toxins 2023, 15, 421. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. Sect. D Struct. Biol. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Luna-Vital, D.; Weiss, M.; Gonzalez de Mejia, E. Anthocyanins from Purple Corn Ameliorated Tumor Necrosis Factor-α-Induced Inflammation and Insulin Resistance in 3T3-L1 Adipocytes via Activation of Insulin Signaling and Enhanced GLUT4 Translocation. Mol. Nutr. Food Res. 2017, 61, 1700362. [Google Scholar] [CrossRef] [PubMed]
- Villaseñor, V.M.; Navat Enriquez-Vara, J.; Urías-Silva, J.E.; del Carmen Lugo-Cervantes, E.; Luna-Vital, D.A.; Mojica, L. Mexican Grasshopper (Sphenarium purpurascens) as Source of High Protein Flour: Techno-Functional Characterization, and in Silico and in Vitro Biological Potential. Food Res. Int. 2022, 162, 112048. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, A.J.; Sáez, M.I.; Martínez, T.F.; Acién, F.G.; Alarcón, F.J. Differential Hydrolysis of Proteins of Four Microalgae by the Digestive Enzymes of Gilthead Sea Bream and Senegalese Sole. Algal Res. 2019, 37, 145–153. [Google Scholar] [CrossRef]
- Levasseur, W.; Perré, P.; Pozzobon, V. A Review of High Value-Added Molecules Production by Microalgae in Light of the Classification. Biotechnol. Adv. 2020, 41, 107545. [Google Scholar] [CrossRef] [PubMed]
- Paterson, S.; Gómez-Cortés, P.; de la Fuente, M.A.; Hernández-Ledesma, B. Bioactivity and Digestibility of Microalgae Tetraselmis Sp. and Nannochloropsis Sp. as Basis of Their Potential as Novel Functional Foods. Nutrients 2023, 15, 477. [Google Scholar] [CrossRef]
- Pereira, H.; Silva, J.; Santos, T.; Gangadhar, K.N.; Raposo, A.; Nunes, C.; Coimbra, M.A.; Gouveia, L.; Barreira, L.; Varela, J. Nutritional Potential and Toxicological Evaluation of Tetraselmis Sp. CtP4 Microalgal Biomass Produced in Industrial Photobioreactors. Molecules 2019, 24, 3192. [Google Scholar] [CrossRef]
- Millward, D.J.; Layman, D.K.; Tomé, D.; Schaafsma, G. Protein Quality Assessment: Impact of Expanding Understanding of Protein and Amino Acid Needs for Optimal Health. Am. J. Clin. Nutr. 2008, 87, 1576S–1581S. [Google Scholar] [CrossRef]
- Xie, W.; Li, X.; Xu, H.; Chen, F.; Cheng, K.W.; Liu, H.; Liu, B. Optimization of Heterotrophic Culture Conditions for the Microalgae Euglena gracilis to Produce Proteins. Mar. Drugs 2023, 21, 519. [Google Scholar] [CrossRef]
- Cai, Y.; Zhai, L.; Fang, X.; Wu, K.; Liu, Y.; Cui, X.; Wang, Y.; Yu, Z.; Ruan, R.; Liu, T.; et al. Effects of C/N Ratio on the Growth and Protein Accumulation of Heterotrophic Chlorella in Broken Rice Hydrolysate. Biotechnol. Biofuels Bioprod. 2022, 15, 102. [Google Scholar] [CrossRef]
- Pruteanu, L.L.; Bailey, D.S.; Grădinaru, A.C.; Jäntschi, L. The Biochemistry and Effectiveness of Antioxidants in Food, Fruits, and Marine Algae. Antioxidants 2023, 12, 860. [Google Scholar] [CrossRef]
- Han, Y.; Ma, B.; Zhang, K. Spider: Software for Protein Identification from Sequence Tags with de Novo Sequencing Error. J. Bioinform. Comput. Biol. 2011, 3, 697–716. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.U.; Yi, E.K.J.; Amin, N.D.M.; Ismail, M.N. An Empirical Analysis of Sacha Inchi (Plantae: Plukenetia volubilis L.) Seed Proteins and Their Applications in the Food and Biopharmaceutical Industries. Appl. Biochem. Biotechnol. 2024, 196, 4823–4836. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Acero, F.J.; Amil-Ruiz, F.; Durán-Peña, M.J.; Carrasco, R.; Fajardo, C.; Guarnizo, P.; Fuentes-Almagro, C.; Vallejo, R.A. Valorisation of the Microalgae Nannochloropsis gaditana Biomass by Proteomic Approach in the Context of Circular Economy. J. Proteom. 2019, 193, 239–242. [Google Scholar] [CrossRef]
- Ferreira, M.; Teixeira, C.; Abreu, H.; Silva, J.; Costas, B.; Kiron, V.; Valente, L.M.P. Nutritional Value, Antimicrobial and Antioxidant Activities of Micro- and Macroalgae, Single or Blended, Unravel Their Potential Use for Aquafeeds. J. Appl. Phycol. 2021, 33, 3507–3518. [Google Scholar] [CrossRef]
- Hulatt, C.J.; Smolina, I.; Dowle, A.; Kopp, M.; Vasanth, G.K.; Hoarau, G.G.; Wijffels, R.H.; Kiron, V. Proteomic and Transcriptomic Patterns during Lipid Remodeling in Nannochloropsis gaditana. Int. J. Mol. Sci. 2020, 21, 6946. [Google Scholar] [CrossRef]
- Sun, D.; Wu, S.; Li, X.; Ge, B.; Zhou, C.; Yan, X.; Ruan, R.; Cheng, P. The Structure, Functions and Potential Medicinal Effects of Chlorophylls Derived from Microalgae. Mar. Drugs 2024, 22, 65. [Google Scholar] [CrossRef]
- Meyer, J.G. In Silico Proteome Cleavage Reveals Iterative Digestion Strategy for High Sequence Coverage. Int. Sch. Res. Not. 2014, 2014, 960902. [Google Scholar] [CrossRef]
- Kruchinin, A.G.; Bolshakova, E.I.; Barkovskaya, I.A. Bioinformatic Modeling (In Silico) of Obtaining Bioactive Peptides from the Protein Matrix of Various Types of Milk Whey. Fermentation 2023, 9, 380. [Google Scholar] [CrossRef]
- Akbarian, M.; Khani, A.; Eghbalpour, S.; Uversky, V.N. Bioactive Peptides: Synthesis, Sources, Applications, and Proposed Mechanisms of Action. Int. J. Mol. Sci. 2022, 23, 1445. [Google Scholar] [CrossRef]
- Han, X.; Liu, Y.; Chen, Z. Zinc Finger Protein LipR Represses Docosahexaenoic Acid and Lipid Biosynthesis in Schizochytrium sp. Appl. Environ. Microbiol. 2022, 88, 6. [Google Scholar] [CrossRef]
- Wang, R.; Li, J.; Zhang, F.; Miao, X. Non-Tandem CCCH-Type Zinc-Finger Protein CpZF_CCCH1 Improves Fatty Acid Desaturation and Stress Tolerance in Chlamydomonas reinhardtii. J. Agric. Food Chem. 2023, 71, 17188–17201. [Google Scholar] [CrossRef] [PubMed]
- Ashtiani, F.R.; Jalili, H.; Rahaie, M.; Sedighi, M.; Amrane, A. Effect of Mixed Culture of Yeast and Microalgae on Acetyl-CoA Carboxylase and Glycerol-3-Phosphate Acyltransferase Expression. J. Biosci. Bioeng. 2021, 131, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Bardani, E.; Kallemi, P.; Tselika, M.; Katsarou, K.; Kalantidis, K. Spotlight on Plant Bromodomain Proteins. Biology 2023, 12, 1076. [Google Scholar] [CrossRef]
- Lin, Y.; Li, Y.; Wu, X.; Xu, W.; Zhang, Z.; Zhu, H.; Zhou, H. NgAP2a Targets KCS Gene to Promote Lipid Accumulation in Nannochloropsis gaditana. Int. J. Mol. Sci. 2024, 25, 10305. [Google Scholar] [CrossRef]
- Heda, R.; Toro, F.; Tombazzi, C.R. Physiology, Pepsin; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Salelles, L.; Floury, J.; Le Feunteun, S. Pepsin Activity as a Function of PH and Digestion Time on Caseins and Egg White Proteins under Static in Vitro Conditions. Food Funct. 2021, 12, 12468–12478. [Google Scholar] [CrossRef]
- Purohit, K.; Reddy, N.; Sunna, A. Exploring the Potential of Bioactive Peptides: From Natural Sources to Therapeutics. Int. J. Mol. Sci. 2024, 25, 1391. [Google Scholar] [CrossRef]
- Yao, S.; Agyei, D.; Udenigwe, C.C. Structural Basis of Bioactivity of Food Peptides in Promoting Metabolic Health. Adv. Food Nutr. Res. 2018, 84, 145–181. [Google Scholar] [CrossRef]
- Bhuyan, B.J.; Mugesh, G. Antioxidant Activity of Peptide-Based Angiotensin Converting Enzyme Inhibitors. Org. Biomol. Chem. 2012, 10, 2237–2247. [Google Scholar] [CrossRef]
- Li, Y.W.; Li, B.; He, J.; Qian, P. Quantitative Structure–Activity Relationship Study of Antioxidative Peptide by Using Different Sets of Amino Acids Descriptors. J. Mol. Struct. 2011, 998, 53–61. [Google Scholar] [CrossRef]
- Fernando, R.; Sun, X.; Rupasinghe, H.P.V. Production of Bioactive Peptides from Microalgae and Their Biological Properties Related to Cardiovascular Disease. Macromol 2024, 4, 582–596. [Google Scholar] [CrossRef]
- Pedroni, L.; Perugino, F.; Galaverna, G.; Dall’Asta, C.; Dellafiora, L. An In Silico Framework to Mine Bioactive Peptides from Annotated Proteomes: A Case Study on Pancreatic Alpha Amylase Inhibitory Peptides from Algae and Cyanobacteria. Nutrients 2022, 14, 4680. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- He, W.; Su, G.; Sun-Waterhouse, D.; Waterhouse, G.I.N.; Zhao, M.; Liu, Y. In Vivo Anti-Hyperuricemic and Xanthine Oxidase Inhibitory Properties of Tuna Protein Hydrolysates and Its Isolated Fractions. Food Chem. 2019, 272, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Guan-Jhong, H.; Lu, T.; Chiu, C.; Chen, H.; Wu, C.; Lin, Y.; Hsieh, W.; Liao, J.; Sheu, M.; Lin, Y. Sweet potato storage root defensin and its tryptic hydrolysates exhibited angiotensin converting enzyme inhibitory activity in vitro. Bot. Stud. 2011, 52, 257–264. [Google Scholar]
- Peng, H.; Carretero, O.A.; Vuljaj, N.; Liao, T.D.; Motivala, A.; Peterson, E.L.; Rhaleb, N.E. Angiotensin-Converting Enzyme Inhibitors: A New Mechanism of Action. Circulation 2005, 112, 2436–2445. [Google Scholar] [CrossRef] [PubMed]
- Lan, V.T.T.; Ito, K.; Ohno, M.; Motoyama, T.; Ito, S.; Kawarasaki, Y. Analyzing a Dipeptide Library to Identify Human Dipeptidyl Peptidase IV Inhibitor. Food Chem. 2015, 175, 66–73. [Google Scholar] [CrossRef]
- Cheung, H.S.; Wang, F.L.; Ondetti, M.A.; Sabo, E.F.; Cushman, D.W. Binding of Peptide Substrates and Inhibitors of Angiotensin-Converting Enzyme. Importance of the COOH-Terminal Dipeptide Sequence. J. Biol. Chem. 1980, 255, 401–407. [Google Scholar] [CrossRef]
- Dhanda, S.; Singh, J.; Singh, H. Hydrolysis of Various Bioactive Peptides by Goat Brain Dipeptidylpeptidase-III Homologue. Cell Biochem. Funct. 2008, 26, 339–345. [Google Scholar] [CrossRef]
- Saito, Y.; Nakamura, K.W.; Kawato, A.; Imayasu, S. Structure and Activity of Angiotensin I Converting Enzyme Inhibitory Peptides from Sake and Sake Lees. Biosci. Biotechnol. Biochem. 1994, 58, 1767–1771. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, H.; Bao, X.; Wu, J. Angiotensin-Converting Enzyme 2 Activation Is Not a Common Feature of Angiotensin-Converting Enzyme Inhibitory Peptides. J. Agric. Food Chem. 2023, 71, 8867–8876. [Google Scholar] [CrossRef]
- Senadheera, T.R.L.; Hossain, A.; Dave, D.; Shahidi, F. In Silico Analysis of Bioactive Peptides Produced from Underutilized Sea Cucumber By-Products—A Bioinformatics Approach. Mar. Drugs 2022, 20, 610. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Moriguchi, I.; Hirano, H.; Nakagome, I. Comparison of Reliability of Log P Values for Drugs Calculated by Several Methods. Chem. Pharm. Bull. 1994, 42, 976–978. [Google Scholar] [CrossRef]
- Moriguchi, I.; Hirono, S.; Liu, Q.; Nakagome, I.; Matsushita, Y. Simple Method of Calculating Octanol/Water Partition Coefficient. Chem. Pharm. Bull. 1992, 40, 127–130. [Google Scholar] [CrossRef]
- Martin, Y.C. A Bioavailability Score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef]
- Ji, D.; Xu, M.; Udenigwe, C.C.; Agyei, D. Physicochemical Characterisation, Molecular Docking, and Drug-Likeness Evaluation of Hypotensive Peptides Encrypted in Flaxseed Proteome. Curr. Res. Food Sci. 2020, 3, 41–50. [Google Scholar] [CrossRef]
- Nong, N.T.P.; Hsu, J.L. Bioactive Peptides: An Understanding from Current Screening Methodology. Processes 2022, 10, 1114. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Valadez-Cosmes, P.; Raftopoulou, S.; Mihalic, Z.N.; Marsche, G.; Kargl, J. Myeloperoxidase: Growing Importance in Cancer Pathogenesis and Potential Drug Target. Pharmacol. Ther. 2022, 236, 108052. [Google Scholar] [CrossRef]
- Vakhrusheva, T.V.; Sokolov, A.V.; Moroz, G.D.; Kostevich, V.A.; Gorbunov, N.P.; Smirnov, I.P.; Grafskaia, E.N.; Latsis, I.A.; Panasenko, O.M.; Lazarev, V.N. Effects of Synthetic Short Cationic Antimicrobial Peptides on the Catalytic Activity of Myeloperoxidase, Reducing Its Oxidative Capacity. Antioxidants 2022, 11, 2419. [Google Scholar] [CrossRef]
- Majumder, K.; Chakrabarti, S.; Morton, J.S.; Panahi, S.; Kaufman, S.; Davidge, S.T.; Wu, J. Egg-Derived ACE-Inhibitory Peptides IQW and LKP Reduce Blood Pressure in Spontaneously Hypertensive Rats. J. Funct. Foods 2015, 13, 50–60. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural Requirements of Angiotensin I-Converting Enzyme Inhibitory Peptides: Quantitative Structure-Activity Relationship Modeling of Peptides Containing 4-10 Amino Acid Residues. QSAR Comb. Sci. 2006, 25, 873–880. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.U.; Sturrock, E.D.; Acharya, K.R. Crystal Structure of the Human Angiotensin-Converting Enzyme-Lisinopril Complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vaquero, M.; Mora, L.; Hayes, M. In Vitro and in Silico Approaches to Generating and Identifying Angiotensin-Converting Enzyme I Inhibitory Peptides from Green Macroalga Ulva Lactuca. Mar. Drugs 2019, 17, 204. [Google Scholar] [CrossRef]
- Hayes, M.; Bastiaens, L.; Gouveia, L.; Gkelis, S.; Skomedal, H.; Skjanes, K.; Murray, P.; García-Vaquero, M.; Hosoglu, M.I.; Dodd, J.; et al. Microalgal Bioactive Compounds Including Protein, Peptides, and Pigments: Applications, Opportunities, and Challenges During Biorefinery Processes. Nov. Proteins Food Pharm. Agric. 2018, 12, 239–255. [Google Scholar] [CrossRef]
- Lucakova, S.; Branyikova, I.; Hayes, M. Microalgal Proteins and Bioactives for Food, Feed, and Other Applications. Appl. Sci. 2022, 12, 4402. [Google Scholar] [CrossRef]
| Amino Acid | Content | FAO Recommendation (g/100 g Protein) | |
|---|---|---|---|
| g/100 g Protein | g/100 g Biomass | ||
| Essential | |||
| Lysine (K) | 4.53 ± 0.09 | 2.01 ± 0.04 | 5.20 |
| Tryptophan (W) | n.d. | n.d. | 0.70 |
| Phenylalanine (F) | 3.62 ± 0.17 | 1.61 ± 0.08 | 4.60 a |
| Tyrosine (Y) | 2.42 ± 0.07 | 1.07 ± 0.03 | |
| Methionine (M) | 1.57 ± 0.01 | 0.70 ± 0.01 | 2.60 b |
| Cysteine (C) | 0.87 ± 0.12 | 0.38 ± 0.05 | |
| Threonine (T) | 3.40± 0.15 | 1.51 ± 0.06 | 2.70 |
| Leucine (L) | 5.97 ± 0.01 | 2.65 ± 0.00 | 6.30 |
| Isoleucine (I) | 2.53 ± 0.03 | 1.12 ± 0.01 | 3.10 |
| Valine (V) | 3.40 ± 0.02 | 1.51 ± 001 | 4.20 |
| Non-essential | |||
| Aspartic acid + Asparragine (D + N) | 6.44 ± 0.00 | 2.85 ± 0.00 | |
| Glutamic acid + Glutamine (E + Q) | 8.81 ± 0.09 | 3.91 ± 0.04 | |
| Serine (S) | 3.29 ± 0.15 | 1.46 ± 0.07 | |
| Histidine (H) | 1.38 ± 0.01 | 0.61 ± 0.00 | |
| Arginine (R) | 4.17 ± 0.00 | 1.85 ± 0.00 | |
| Alanine (A) | 5.02 ± 0.01 | 2.22 ± 0.01 | |
| Proline (P) | 6.44 ± 0.01 | 2.85 ± 0.00 | |
| Glycine (G) | 3.55 ± 0.02 | 1.57 ± 0.01 | |
| EAA | 28.31 | 14.13 | |
| NEAA | 39.01 | 17.28 | |
| TAA | 67.41 | 31.41 | |
| EAA × 100/TAA (%) | 42.00 | ||
| EAA × 100/NEAA (%) | 72.57 | ||
| HAA × 100/TAA (%) | 47.23 | ||
| AAA × 100/TAA (%) | 8.96 | ||
| Accession a | −10logP b | Description c | Average Mass (KDa) | Peptides Generated After In Silico Gastric Digestion |
|---|---|---|---|---|
| I2CQP5 | 426.87 | Acetyl-CoA carboxylase | 235,196 | 207 |
| W7U8G3 | 380.26 | ATP-citrate synthase | 120,317 | 102 |
| W7TN63 | 348.14 | Choline dehydrogenase | 138,839 | 113 |
| K8YSL1 | 287.43 | Aminopeptidase N | 137,581 | 102 |
| W7TQD1 | 264.44 | Pyruvate dehydrogenase E1 component subunit alpha | 120,314 | 103 |
| W7TTR4 | 192.91 | P-type atpase | 129,122 | 103 |
| W7UC18 | 192.19 | Phosphoribosylformylglycinamidine synthase | 145,158 | 113 |
| W7TNH0 | 182.55 | Pyruvate carboxilase | 134,055 | 102 |
| K8Z9A0 | 174.11 | Uncharacterized protein | 123,187 | 107 |
| W7TRK7 | 118.17 | Coatomer subunit alpha | 141,192 | 119 |
| W7UBG5 | 106.82 | Ubiquitin-activating enzyme e1 | 136,993 | 111 |
| W7U8K2 | 105.69 | Clathrin heavy chain | 196,113 | 163 |
| W7TM9 | 93.66 | Pentatricopeptide repeat containing protein | 171,849 | 132 |
| W7UBN0 | 87.72 | WD40-repeat-containing protein | 138,346 | 110 |
| W7U2D5 | 80.11 | Peptidase M16 | 143,228 | 109 |
| W7U0Z1 | 79.24 | Hydantoin utilization protein | 146,652 | 128 |
| W7U5E4 | 65.46 | Carbamoyl-phosphate synthase | 168,186 | 140 |
| W7U2B9 | 62.39 | Zinc finger. ZZ-type | 552,820 | 376 |
| K8YRI3 | 61.47 | DUF2428 domain-containing protein (Fragment) | 124,306 | 102 |
| W7TVB1 | 56.86 | Bromodomain containing 1 | 243,436 | 169 |
| W7TYI7 | 46.56 | Cytochrome p450 | 118,277 | 117 |
| W7U1T9 | 46.41 | Nuclear receptor corepressor 1-like protein | 164,110 | 122 |
| W7U7L8 | 45.19 | Protease-associated domain PA | 142,381 | 101 |
| W7TPR4 | 44.49 | Tubulin-specific chaperone d | 147,094 | 109 |
| TOTAL | 3160 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paterson, S.; Alonso-Pintre, L.; Morato-López, E.; González de la Fuente, S.; Gómez-Cortés, P.; Hernández-Ledesma, B. Microalga Nannochloropsis gaditana as a Sustainable Source of Bioactive Peptides: A Proteomic and In Silico Approach. Foods 2025, 14, 252. https://doi.org/10.3390/foods14020252
Paterson S, Alonso-Pintre L, Morato-López E, González de la Fuente S, Gómez-Cortés P, Hernández-Ledesma B. Microalga Nannochloropsis gaditana as a Sustainable Source of Bioactive Peptides: A Proteomic and In Silico Approach. Foods. 2025; 14(2):252. https://doi.org/10.3390/foods14020252
Chicago/Turabian StylePaterson, Samuel, Laura Alonso-Pintre, Esperanza Morato-López, Sandra González de la Fuente, Pilar Gómez-Cortés, and Blanca Hernández-Ledesma. 2025. "Microalga Nannochloropsis gaditana as a Sustainable Source of Bioactive Peptides: A Proteomic and In Silico Approach" Foods 14, no. 2: 252. https://doi.org/10.3390/foods14020252
APA StylePaterson, S., Alonso-Pintre, L., Morato-López, E., González de la Fuente, S., Gómez-Cortés, P., & Hernández-Ledesma, B. (2025). Microalga Nannochloropsis gaditana as a Sustainable Source of Bioactive Peptides: A Proteomic and In Silico Approach. Foods, 14(2), 252. https://doi.org/10.3390/foods14020252








