Profiles of Killer Systems and Volatile Organic Compounds of Rowanberry and Rosehip-Inhabiting Yeasts Substantiate Implications for Biocontrol
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Cultures and Media
2.2. Sampling, Enrichment, and Identification of Cultivable Yeast
2.3. Analysis of Killing/Susceptibility Phenotype
2.4. Double-Stranded RNA Isolation from Yeast
2.5. Analysis of Biocontrol Properties of Killer Yeasts in Food System
2.6. Sampling and Analysis of Volatile Organic Compounds Produced by Yeasts
2.7. Statistical Analysis
3. Results
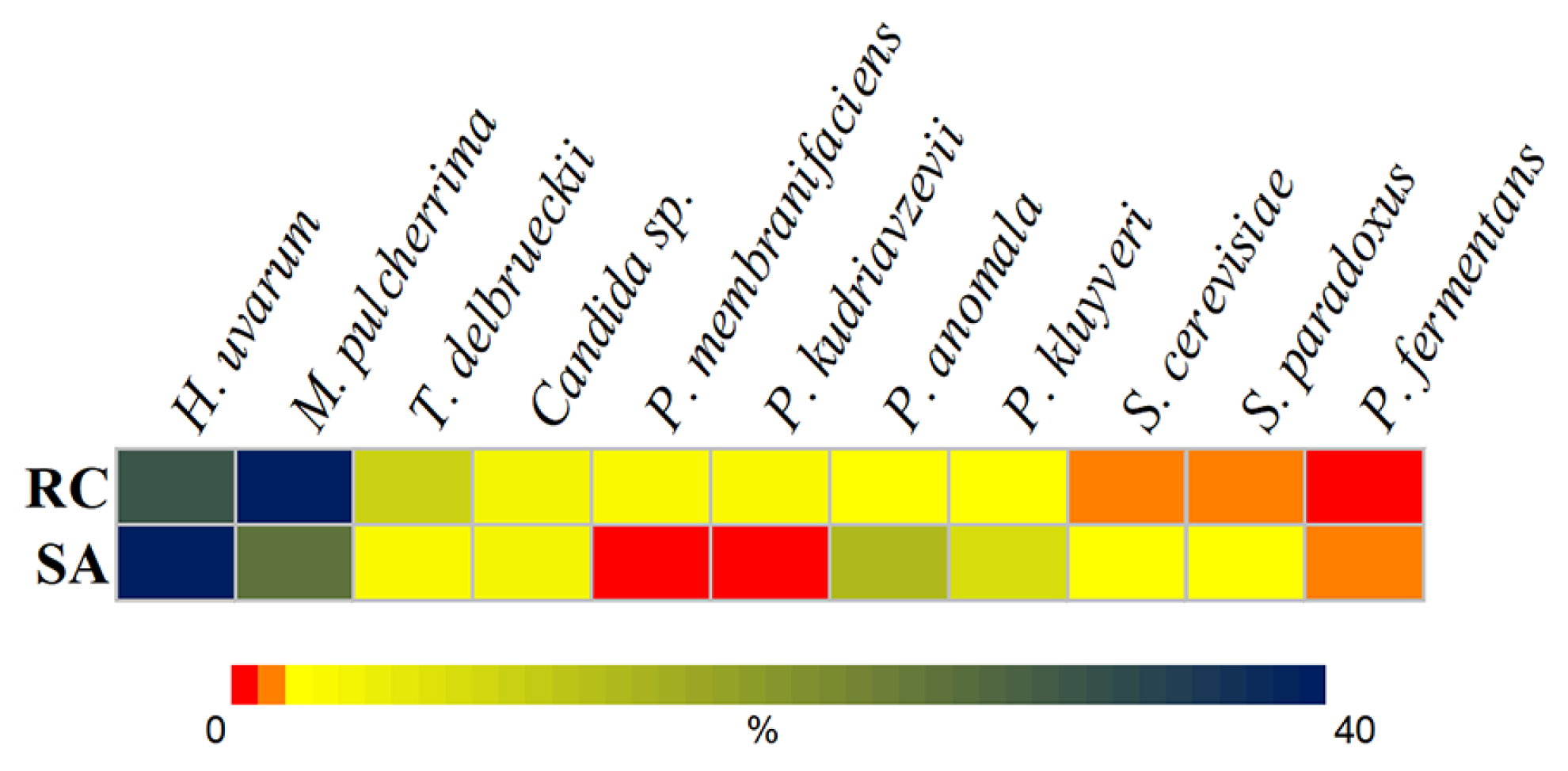
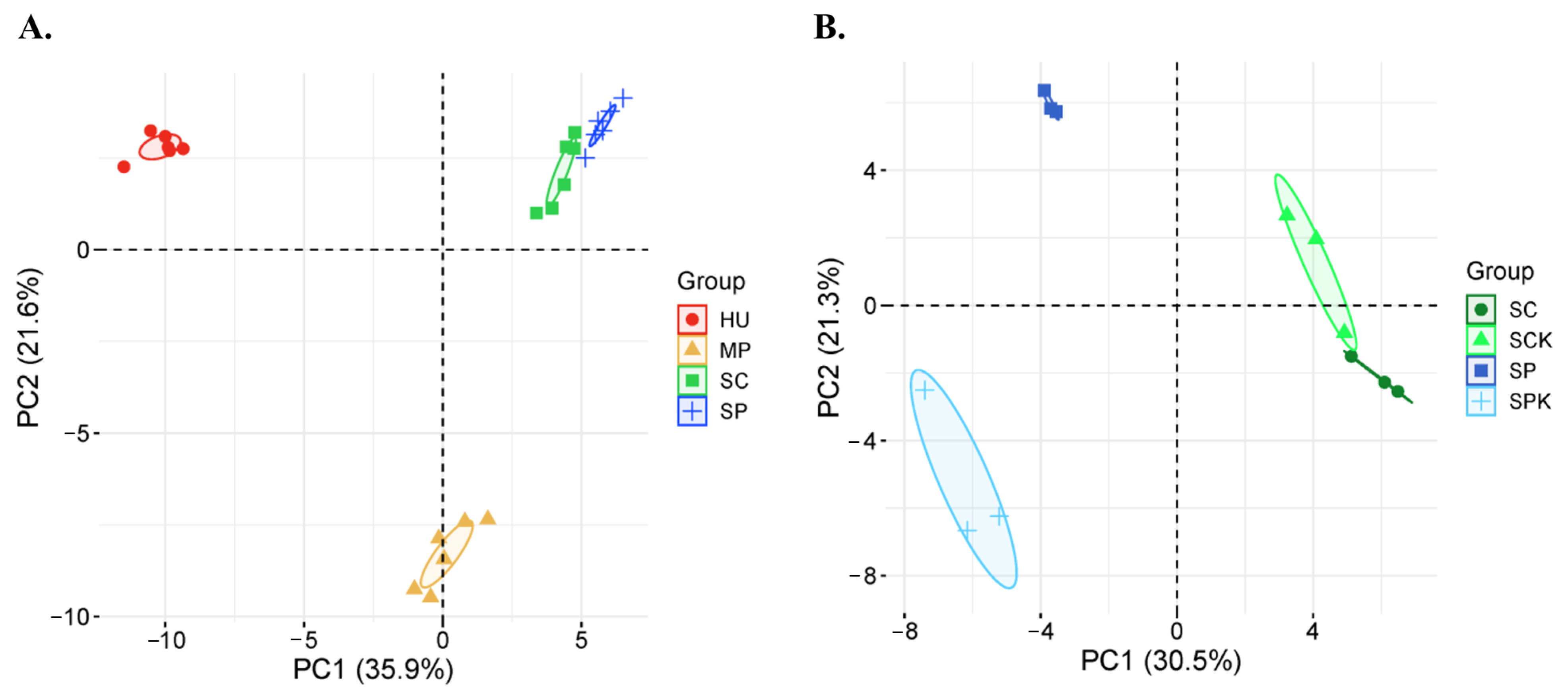
3.1. Cultured Yeast in Spontaneous Fermentations of Rowanberries and Rosehips
3.2. Antagonistic Activity of Yeasts
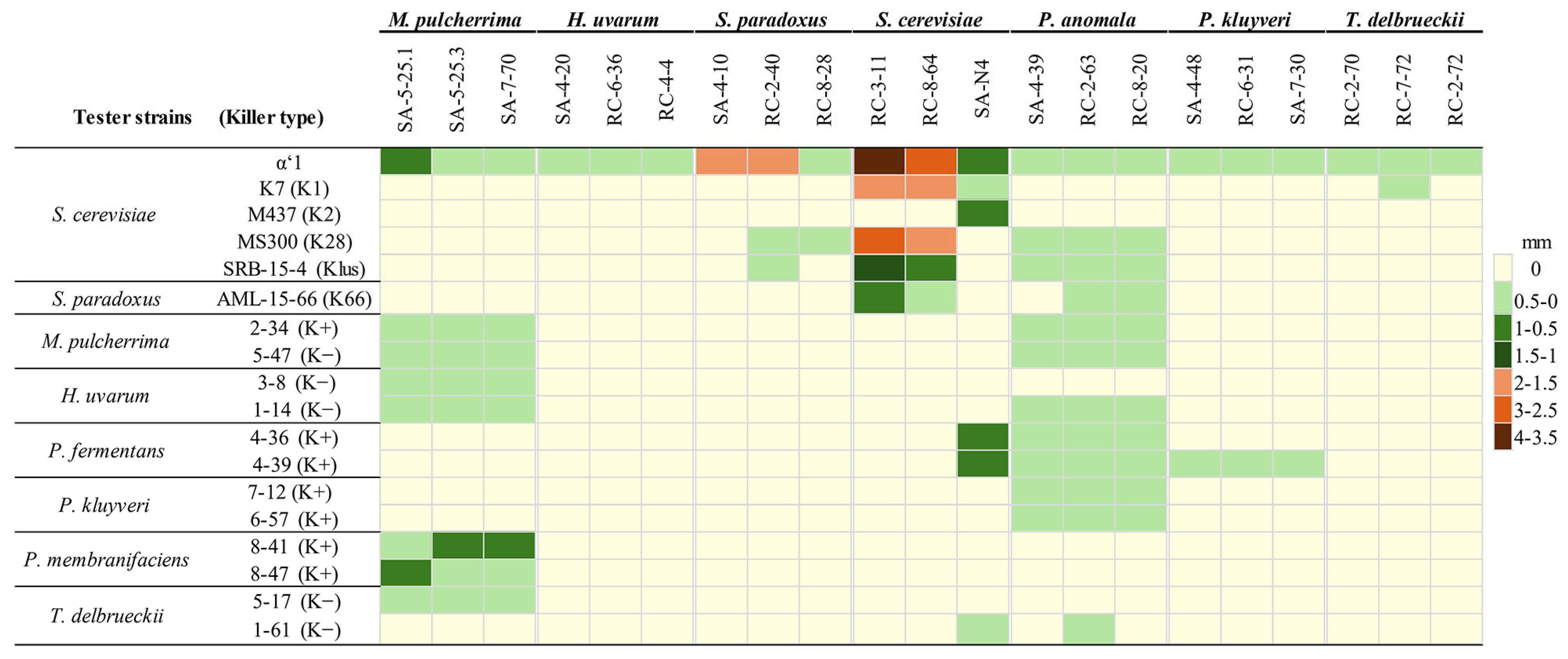
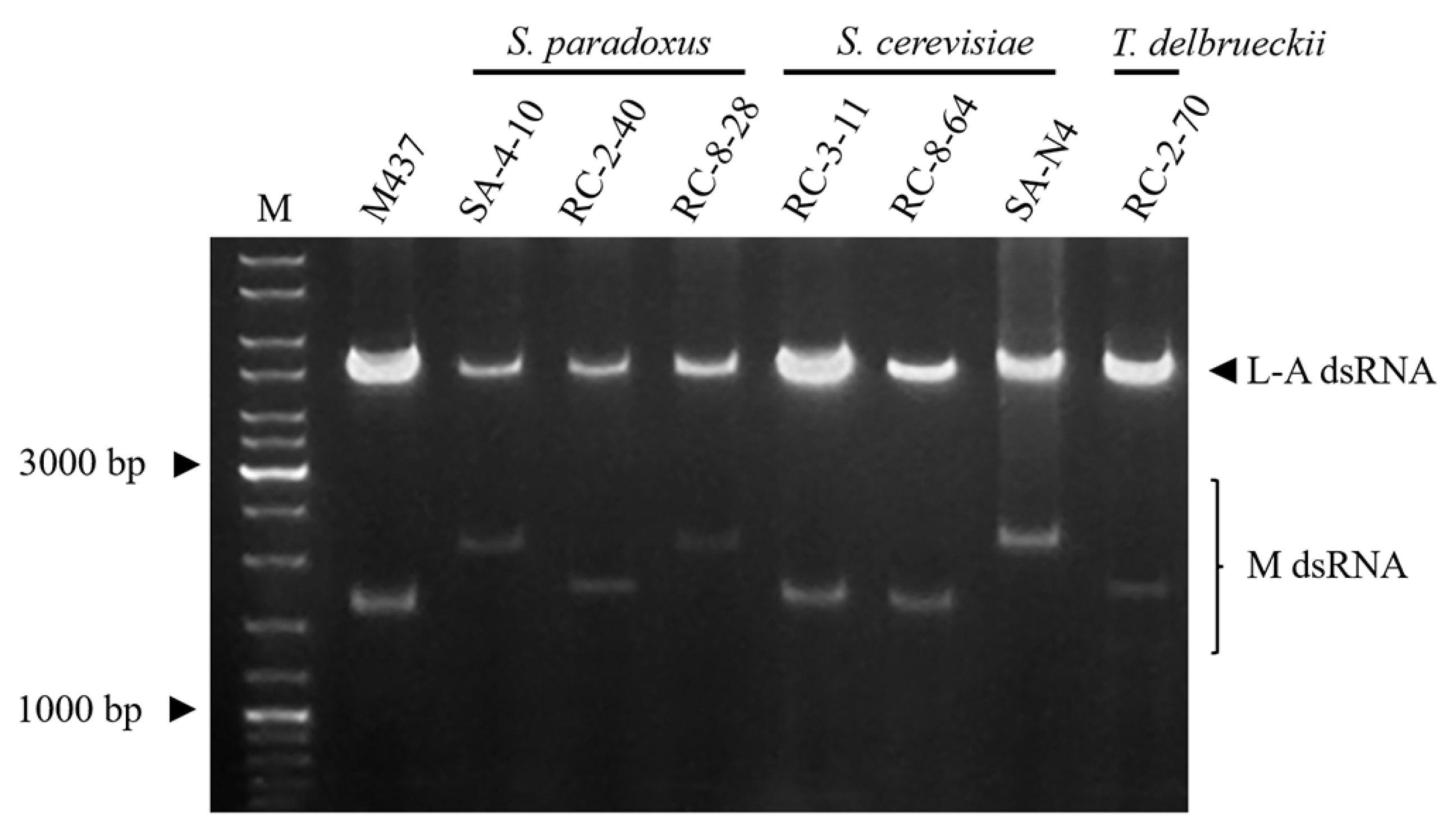
3.3. DsRNA-Encoded Killer Phenotype-Possessing Yeasts
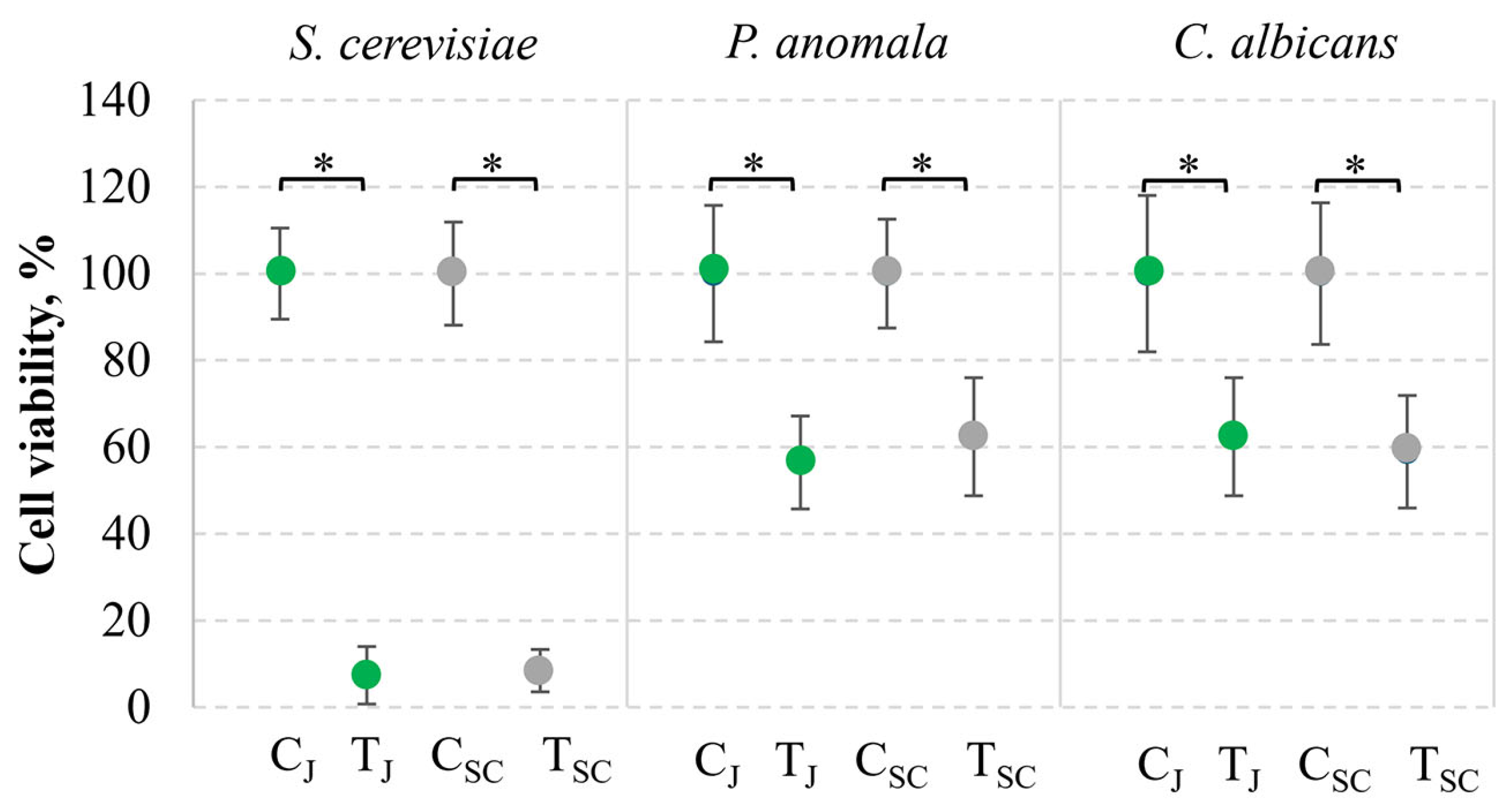
3.4. Application of Killer Yeasts in the Food System
3.5. Volatiles Produced by Different Yeast Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agirman, B.; Carsanba, E.; Settanni, L.; Erten, H. Exploring Yeast-Based Microbial Interactions: The next Frontier in Postharvest Biocontrol. Yeast 2023, 40, 457–475. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, J.; Krzymińska, J.; Tyburski, J. Yeasts as a Potential Biological Agent in Plant Disease Protection and Yield Improvement—A Short Review. Agriculture 2022, 12, 1404. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, M.; Qin, X.; Dong, Q.; Li, Z. Antimicrobial Function of Yeast against Pathogenic and Spoilage Microorganisms via Either Antagonism or Encapsulation: A Review. Food Microbiol. 2023, 112, 104242. [Google Scholar] [CrossRef] [PubMed]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol Yeasts: Mechanisms and Applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef]
- Oufensou, S.; Ul Hassan, Z.; Balmas, V.; Jaoua, S.; Migheli, Q. Perfume Guns: Potential of Yeast Volatile Organic Compounds in the Biological Control of Mycotoxin-Producing Fungi. Toxins 2023, 15, 45. [Google Scholar] [CrossRef]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol Ability and Action Mechanism of Food-Isolated Yeast Strains against Botrytis cinerea Causing Post-Harvest Bunch Rot of Table Grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef]
- Sui, Y.; Wisniewski, M.; Droby, S.; Liu, J. Responses of Yeast Biocontrol Agents to Environmental Stress. Appl. Environ. Microbiol. 2015, 81, 2968–2975. [Google Scholar] [CrossRef]
- Vepštaitė-Monstavičė, I.; Lukša, J.; Strazdaitė-Žielienė, Ž.; Serva, S.; Servienė, E. Distinct Microbial Communities Associated with Health-relevant Wild Berries. Environ. Microbiol. Rep. 2024, 16, e70048. [Google Scholar] [CrossRef]
- Maksimova, I.A.; Yurkov, A.M.; Chernov, I.Y. Spatial Structure of Epiphytic Yeast Communities on Fruits of Sorbus aucuparia L. Biol. Bull. 2009, 36, 613–618. [Google Scholar] [CrossRef]
- Rovná, K.; Ivanišová, E.; Žiarovská, J.; Ferus, P.; Terentjeva, M.; Kowalczewski, P.Ł.; Kačániová, M. Characterization of Rosa canina Fruits Collected in Urban Areas of Slovakia. Genome Size, IPBS Profiles and Antioxidant and Antimicrobial Activities. Molecules 2020, 25, 1888. [Google Scholar] [CrossRef]
- Billerbeck, S.; Walker, R.S.K.; Pretorius, I.S. Killer Yeasts: Expanding Frontiers in the Age of Synthetic Biology. Trends Biotechnol. 2024, 42, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Büyüksırıt Bedir, T.; Kuleaşan, H. A Natural Approach, the Use of Killer Toxin Produced by Metschnikowia pulcherrima in Fresh Ground Beef Patties for Shelf Life Extention. Int. J. Food Microbiol. 2021, 345, 109154. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.L.; Leu, J.Y.; Chang, T.H. A Population Study of Killer Viruses Reveals Different Evolutionary Histories of Two Closely Related Saccharomyces Sensu Stricto Yeasts. Mol. Biol. 2015, 24, 4312–4322. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R.; López-Piñeiro, A.; Naranjo, B.; Roig, F.; Llorens, C. New Insights into the Genome Organization of Yeast Killer Viruses Based on “Atypical” Killer Strains Characterized by High-Throughput Sequencing. Toxins 2017, 9, 292. [Google Scholar] [CrossRef]
- Rodríguez-Cousiño, N.; Gómez, P.; Esteban, R. Variation and Distribution of L-A Helper Totiviruses in Saccharomyces Sensu Stricto Yeasts Producing Different Killer Toxins. Toxins 2017, 9, 313. [Google Scholar] [CrossRef]
- Vepštaitė-Monstavičė, I.; Lukša, J.; Konovalovas, A.; Ežerskytė, D.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Serva, S.; Servienė, E. Saccharomyces paradoxus K66 Killer System Evidences Expanded Assortment of Helper and Satellite Viruses. Viruses 2018, 10, 564. [Google Scholar] [CrossRef]
- Villalba, M.L.; Susana Sáez, J.; del Monaco, S.; Lopes, C.A.; Sangorrín, M.P. TdKT, a New Killer Toxin Produced by Torulaspora delbrueckii Effective against Wine Spoilage Yeasts. Int. J. Food Microbiol. 2016, 217, 94–100. [Google Scholar] [CrossRef]
- Rodriguez-Cousino, N.; Gomez, P.; Esteban, R. Expression of the K74 Killer Toxin from Saccharomyces paradoxus Is Modulated by the Toxin-Encoding M74 Double- Stranded RNA 59 Untranslated Terminal Region. Appl. Environ. Microbiol. 2022, 88, e0203021. [Google Scholar] [CrossRef]
- Lukša, J.; Podoliankaitė, M.; Vepštaitė, I.; Strazdaitė-Žielienė, Ž.; Urbonavičius, J.; Servienė, E. Yeast β-1,6-Glucan Is a Primary Target for the Saccharomyces cerevisiae K2 Toxin. Eukaryot. Cell 2015, 14, 406–414. [Google Scholar] [CrossRef]
- Martinac, B.; Zhu, H.; Kubalski, A.; Zhou, X.L.; Culbertson, M.; Bussey, H.; Kung, C. Yeast K1 Killer Toxin Forms Ion Channels in Sensitive Yeast Spheroplasts and in Artificial Liposomes. Proc. Natl. Acad. Sci. USA 1990, 87, 6228–6232. [Google Scholar] [CrossRef]
- Schmitt, M.J.; Breinig, F. The Viral Killer System in Yeast: From Molecular Biology to Application. FEMS Microbiol. Rev. 2002, 26, 257–276. [Google Scholar] [CrossRef] [PubMed]
- Prins, R.C.; Billerbeck, S. The Signal Sequence of Yeast Killer Toxin K2 Confers Producer Self-Protection and Allows Conversion into a Modular Toxin-Antitoxin System. Cell Rep. 2024, 43, 114449. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.; Schmitt, M.J. Yeast Killer Toxin K28: Biology and Unique Strategy of Host Cell Intoxication and Killing. Toxins 2017, 9, 333. [Google Scholar] [CrossRef]
- Reiter, J.; Herker, E.; Madeo, F.; Schmitt, M.J. Viral Killer Toxins Induce Caspase-Mediated Apoptosis in Yeast. J. Cell Biol. 2005, 168, 353–358. [Google Scholar] [CrossRef]
- Díaz, M.A.; Pereyra, M.M.; Picón-Montenegro, E.; Meinhardt, F.; Dib, J.R. Killer Yeasts for the Biological Control of Postharvest Fungal Crop Diseases. Microorganisms 2020, 8, 1680. [Google Scholar] [CrossRef]
- Di Canito, A.; Mateo-Vargas, M.A.; Mazzieri, M.; Cantoral, J.; Foschino, R.; Cordero-Bueso, G.; Vigentini, I. The Role of Yeasts as Biocontrol Agents for Pathogenic Fungi on Postharvest Grapes: A Review. Foods 2021, 10, 1650. [Google Scholar] [CrossRef]
- Klassen, R.; Schaffrath, R.; Buzzini, P.; Ganter, P. Antagonistic Interactions and Killer Yeasts. In Yeasts in Natural Ecosystems: Ecology; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Cham, Switzerland, 2017; pp. 1–46. [Google Scholar]
- Schaffrath, R.; Meinhardt, F.; Klassen, R. Yeast Killer Toxins: Fundamentals and Applications. In Physiology and Genetics: Selected Basic and Applied Aspects; Anke, T., Schüffler, A., Eds.; Springer: Cham, Switzerland, 2018; pp. 87–118. [Google Scholar]
- Contarino, R.; Brighina, S.; Fallico, B.; Cirvilleri, G.; Parafati, L.; Restuccia, C. Volatile Organic Compounds (VOCs) Produced by Biocontrol Yeasts. Food Microbiol. 2019, 82, 70–74. [Google Scholar] [CrossRef]
- Farbo, M.G.; Urgeghe, P.P.; Fiori, S.; Marcello, A.; Oggiano, S.; Balmas, V.; Hassan, Z.U.; Jaoua, S.; Migheli, Q. Effect of Yeast Volatile Organic Compounds on Ochratoxin A-Producing Aspergillus carbonarius and A. Ochraceus. Int. J. Food Microbiol. 2018, 284, 1–10. [Google Scholar] [CrossRef]
- Sater, H.M.; Bizzio, L.N.; Tieman, D.M.; Muñoz, P.D. A Review of the Fruit Volatiles Found in Blueberry and Other Vaccinium Species. J. Agric. Food Chem. 2020, 68, 5777–5786. [Google Scholar] [CrossRef]
- Karsli, A.; Şahin, Y.S. The Role of Fungal Volatile Organic Compounds (FVOCs) in Biological Control. Türkiye Biyolojik Mücadele Derg. 2021, 12, 79–92. [Google Scholar] [CrossRef]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Performance Evaluation of Volatile Organic Compounds by Antagonistic Yeasts Immobilized on Hydrogel Spheres against Gray, Green and Blue Postharvest Decays. Food Microbiol. 2017, 63, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Mozūraitis, R.; Aleknavičius, D.; Vepštaitė-Monstavičė, I.; Stanevičienė, R.; Emami, S.N.; Apšegaitė, V.; Radžiutė, S.; Blažytė-Čereškienė, L.; Servienė, E.; Būda, V. Hippophae rhamnoides Berry Related Pichia kudriavzevii Yeast Volatiles Modify Behaviour of Rhagoletis batava Flies. J. Adv. Res. 2020, 21, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Mozūraitis, R.; Apšegaitė, V.; Radžiutė, S.; Aleknavičius, D.; Būdienė, J.; Stanevičienė, R.; Blažytė-Čereškienė, L.; Servienė, E.; Būda, V. Volatiles Produced by Yeasts Related to Prunus avium and P. cerasus Fruits and Their Potentials to Modulate the Behaviour of the Pest Rhagoletis cerasi Fruit Flies. J. Fungi 2022, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Lukša, J.; Vepštaitė-Monstavičė, I.; Apšegaitė, V.; Blažytė-Čereškienė, L.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Ravoitytė, B.; Aleknavičius, D.; Būda, V.; Mozūraitis, R.; et al. Fungal Microbiota of Sea Buckthorn Berries at Two Ripening Stages and Volatile Profiling of Potential Biocontrol Yeasts. Microorganisms 2020, 8, 456. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Li, G.Q.; Zhang, J.; Yang, L.; Che, H.J.; Jiang, D.H.; Huang, H.C. Control of Postharvest Botrytis Fruit Rot of Strawberry by Volatile Organic Compounds of Candida intermedia. Phytopathology 2011, 101, 859–869. [Google Scholar] [CrossRef]
- Lemos Junior, W.J.F.; Binati, R.L.; Felis, G.E.; Slaghenaufi, D.; Ugliano, M.; Torriani, S. Volatile Organic Compounds from Starmerella bacillaris to Control Gray Mold on Apples and Modulate Cider Aroma Profile. Food Microbiol. 2020, 89, 103446. [Google Scholar] [CrossRef]
- Huang, R.; Che, H.J.; Zhang, J.; Yang, L.; Jiang, D.H.; Li, G.Q. Evaluation of Sporidiobolus pararoseus Strain YCXT3 as Biocontrol Agent of Botrytis cinerea on Post-Harvest Strawberry Fruits. Biol. Control 2012, 62, 53–63. [Google Scholar] [CrossRef]
- Di Francesco, A.; Ugolini, L.; Lazzeri, L.; Mari, M. Production of Volatile Organic Compounds by Aureobasidium pullulans as a Potential Mechanism of Action against Postharvest Fruit Pathogens. Biol. Control 2015, 81, 8–14. [Google Scholar] [CrossRef]
- Aragno, J.; Fernandez-Valle, P.; Thiriet, A.; Grondin, C.; Legras, J.L.; Camarasa, C.; Bloem, A. Two-Stage Screening of Metschnikowia spp. Bioprotective Properties: From Grape Juice to Fermented Must by Saccharomyces cerevisiae. Microorganisms 2024, 12, 1659. [Google Scholar] [CrossRef]
- Moore, G.G.; Lebar, M.D.; Carter-Wientjes, C.H. Cumulative Effects of Non-Aflatoxigenic Aspergillus flavus Volatile Organic Compounds to Abate Toxin Production by Mycotoxigenic Aspergilli. Toxins 2022, 14, 340. [Google Scholar] [CrossRef]
- Sipiczki, M. Metschnikowia pulcherrima and Related Pulcherrimin-Producing Yeasts: Fuzzy Species Boundaries and Complex Antimicrobial Antagonism. Microorganisms 2020, 8, 1029. [Google Scholar] [CrossRef] [PubMed]
- Lukša, J.; Ravoitytė, B.; Konovalovas, A.; Aitmanaitė, L.; Butenko, A.; Yurchenko, V.; Serva, S.; Servienė, E. Different Metabolic Pathways Are Involved in Response of Saccharomyces cerevisiae to L-A and M Viruses. Toxins 2017, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Gulbiniene, G.; Kondratiene, L.; Jokantaite, T.; Serviene, E.; Melvydas, V.; Petkuniene, G. Killer Yeast Strains in Wine Yeast Population. Food Technol. Biotechnol. 2004, 42, 159–163. [Google Scholar]
- Vepštaitė-Monstavičė, I.; Lukša, J.; Stanevičienė, R.; Strazdaitė-Žielienė, Ž.; Yurchenko, V.; Serva, S.; Servienė, E. Distribution of Apple and Blackcurrant Microbiota in Lithuania and the Czech Republic. Microbiol. Res. 2018, 206, 1–8. [Google Scholar] [CrossRef]
- Stanevičienė, R.; Lukša, J.; Strazdaitė-Žielienė, Ž.; Ravoitytė, B.; Losinska-Sičiūnienė, R.; Mozūraitis, R.; Servienė, E. Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts. Microorganisms 2021, 9, 1423. [Google Scholar] [CrossRef]
- Servienė, E.; Lukša, J.; Orentaitė, I.; Lafontaine, D.L.J.; Urbonavičius, J. Screening the Budding Yeast Genome Reveals Unique Factors Affecting K2 Toxin Susceptibility. PLoS ONE 2012, 7, e50779. [Google Scholar] [CrossRef]
- Fried, H.M.; Fink, G.R. Electron Microscopic Heteroduplex Analysis of “Killer” Double-Stranded RNA Species from Yeast. Proc. Natl. Acad. Sci. USA 1978, 75, 4224–4228. [Google Scholar] [CrossRef]
- Grybchuk, D.; Akopyants, N.S.; Kostygov, A.Y.; Konovalovas, A.; Lye, L.-F.; Dobson, D.E.; Zangger, H.; Fasel, N.; Butenko, A.; Frolov, A.O.; et al. Viral Discovery and Diversity in Trypanosomatid Protozoa with a Focus on Relatives of the Human Parasite Leishmania. Proc. Natl. Acad. Sci. USA 2018, 115, E506–E515. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. The Python Language Reference; Python Software Foundation: Wilmington, DE, USA, 2015. [Google Scholar]
- Wilcox, R.R. Introduction to Robust Estimation and Hypothesis Testing, 3rd ed.; Academic Press: Cambridge, MA, USA, 2011; ISBN 9780123869838. [Google Scholar]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A Free Online Platform for Data Visualization and Graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; López-Piñeiro, A.; Ribas, J.C. A New Wine Torulaspora delbrueckii Killer Strain with Broad Antifungal Activity and Its Toxin-Encoding Double-Stranded RNA Virus. Front. Microbiol. 2015, 6, 893. [Google Scholar] [CrossRef]
- Aurori, M.; Niculae, M.; Hanganu, D.; Pall, E.; Cenariu, M.; Vodnar, D.C.; Fiţ, N.; Andrei, S. The Antioxidant, Antibacterial and Cell-Protective Properties of Bioactive Compounds Extracted from Rowanberry (Sorbus aucuparia L.) Fruits In Vitro. Plants 2024, 13, 538. [Google Scholar] [CrossRef] [PubMed]
- Igual, M.; García-Herrera, P.; Cámara, R.M.; Martínez-Monzó, J.; García-Segovia, P.; Cámara, M. Bioactive Compounds in Rosehip (Rosa canina) Powder with Encapsulating Agents. Molecules 2022, 27, 4737. [Google Scholar] [CrossRef] [PubMed]
- Arvinte, O.M.; Senila, L.; Becze, A.; Amariei, S. Rowanberry—A Source of Bioactive Compounds and Their Biopharmaceutical Properties. Plants 2023, 12, 3225. [Google Scholar] [CrossRef] [PubMed]
- Winther, K.; Campbell-Tofte, J.; Vinther Hansen, A.S. Bioactive Ingredients of Rose Hips (Rosa canina L.) with Special Reference to Antioxidative and Anti-Inflammatory Properties: In Vitro Studies. Botanics 2016, 11, 11–23. [Google Scholar] [CrossRef]
- Muccilli, S.; Restuccia, C. Bioprotective Role of Yeasts. Microorganims 2015, 3, 588–611. [Google Scholar] [CrossRef]
- Bajaj, B.K.; Singh, S. Biology of Killer Yeast and Technological Implications. In Yeast Diversity in Human Welfare; Satyanarayana, T., Kunze, G., Eds.; Springer: Singapore, 2017; pp. 163–190. [Google Scholar]
- Vijayraghavan, S.; Kozmin, S.G.; Strope, P.K.; Skelly, D.A.; Magwene, P.M.; Dietrich, F.S.; McCusker, J.H. RNA Viruses, M Satellites, Chromosomal Killer Genes, and Killer/Nonkiller Phenotypes in the 100-Genomes S. cerevisiae Strains. G3 Genes. Genom. Genet. 2023, 13, jkad167. [Google Scholar] [CrossRef]
- Travers-Cook, T.J.; Jokela, J.; Buser, C.C. The Evolutionary Ecology of Fungal Killer Phenotypes. Proc. Roy. Soc. B-Biol. Sci. 2023, 290, 20231108. [Google Scholar] [CrossRef]
- Boynton, P.J. The Ecology of Killer Yeasts: Interference Competition in Natural Habitats. Yeast 2019, 36, 473–485. [Google Scholar] [CrossRef]
- Giovati, L.; Ciociola, T.; De Simone, T.; Conti, S.; Magliani, W. Wickerhamomyces Yeast Killer Toxins’ Medical Applications. Toxins 2021, 13, 655. [Google Scholar] [CrossRef]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A New Wine Saccharomyces cerevisiae Killer Toxin (Klus), Encoded by a Double-Stranded RNA Virus, with Broad Antifungal Activity Is Evolutionarily Related to a Chromosomal Host Gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef]
- Crabtree, A.M.; Taggart, N.T.; Lee, M.D.; Boyer, J.M.; Rowley, P.A. The Prevalence of Killer Yeasts and Double-Stranded RNAs in the Budding Yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2023, 23, foad046. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, R.; Zamora, E.; Alvarez, M.L.; Ramirez, M. Using Torulaspora delbrueckii Killer Yeasts in the Elaboration of Base Wine and Traditional Sparkling Wine. Int. J. Food Microbiol. 2019, 289, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Puyo, M.; Simonin, S.; Bach, B.; Klein, G.; Alexandre, H.; Tourdot-Maréchal, R. Bio-protection in Enology by Metschnikowia pulcherrima: From Field Results to Scientific Inquiry. Front. Microbiol. 2023, 14, 1252973. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Metschnikowia Strains Isolated from Botrytized Grapes Antagonize Fungal and Bacterial Growth by Iron Depletion. Appl. Environ. Microbiol. 2006, 72, 6716–6724. [Google Scholar] [CrossRef]
- Gore-Lloyd, D.; Sumann, I.; Brachmann, A.O.; Schneeberger, K.; Ortiz-Merino, R.A.; Moreno-Beltrán, M.; Schläfli, M.; Kirner, P.; Santos Kron, A.; Rueda-Mejia, M.P.; et al. Snf2 Controls Pulcherriminic Acid Biosynthesis and Antifungal Activity of the Biocontrol Yeast Metschnikowia pulcherrima. Mol. Microbiol. 2019, 112, 317–332. [Google Scholar] [CrossRef]
- Hicks, R.H.; Moreno-Beltrán, M.; Gore-Lloyd, D.; Chuck, C.J.; Henk, D.A. The Oleaginous Yeast Metschnikowia pulcherrima Dislays Killer Activity against Avian-Derived Pathogenic Bacteria. Biology 2021, 10, 1227. [Google Scholar] [CrossRef]
- Schneider, J.; Rupp, O.; Trost, E.; Jaenicke, S.; Passoth, V.; Goesmann, A.; Tauch, A.; Brinkrolf, K. Genome Sequence of Wickerhamomyces Anomalus DSM 6766 Reveals Genetic Basis of Biotechnologically Important Antimicrobial Activities. FEMS Yeast Res. 2012, 12, 382–386. [Google Scholar] [CrossRef]
- Grzegorczyk, M.; Żarowska, B.; Restuccia, C.; Cirvilleri, G. Postharvest Biocontrol Ability of Killer Yeasts against Monilinia fructigena and Monilinia fructicola on Stone Fruit. Food Microbiol. 2017, 61, 93–101. [Google Scholar] [CrossRef]
- Villalba, M.L.; Mazzucco, M.B.; Lopes, C.A.; Ganga, M.A.; Sangorrín, M.P. Purification and Characterization of Saccharomyces eubayanus Killer Toxin: Biocontrol Effectiveness against Wine Spoilage Yeasts. Int. J. Food Microbiol. 2020, 331, 108714. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Alonso, A.; Marquina, D.; Santos, A. The Biology of Pichia membranifaciens Killer Toxins. Toxins 2017, 9, 112. [Google Scholar] [CrossRef]
- da Cruz Almeida, E.T.; de Medeiros Barbosa, I.; Tavares, J.F.; Barbosa-Filho, J.M.; Magnani, M.; de Souza, E.L. Inactivation of Spoilage Yeasts by Mentha spicata L. and M. × villosa Huds. Essential Oils in Cashew, Guava, Mango, and Pineapple Juices. Front. Microbiol. 2018, 9, 1111. [Google Scholar] [CrossRef]
- Lukša, J.; Serva, S.; Servienė, E. Saccharomyces cerevisiae K2 Toxin Requires Acidic Environment for Unidirectional Folding Into Active State. Mycoscience 2017, 57, 51–57. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, J.; Tian, R.; Liu, Y. Microbial Volatile Organic Compounds: Antifungal Mechanisms, Applications, and Challenges. Front. Microbiol. 2022, 13, 922450. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.M.; Guo, J.H.; Cheng, Y.J.; Luo, L.; Liu, P.; Wang, B.Q.; Deng, B.X.; Long, C.A. Control of Gray Mold of Grape by Hanseniaspora uvarum and Its Effects on Postharvest Quality Parameters. Ann. Microbiol. 2010, 60, 31–35. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Tworkoski, T.J.; Kurtzman, C.P. Biocontrol Potential of Metchnikowia pulcherrima Strains Against Blue Mold of Apple. Phytopatology 2001, 91, 1098–1108. [Google Scholar] [CrossRef]
- Rosend, J.; Kuldjärv, R.; Rosenvald, S.; Paalme, T. The Effects of Apple Variety, Ripening Stage, and Yeast Strain on the Volatile Composition of Apple Cider. Heliyon 2019, 5, e01953. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, J.; Yan, H.; Li, D.; Wang, X.; Liang, W.; Wang, G. Ethyl Acetate Produced by Hanseniaspora uvarum Is a Potential Biocontrol Agent against Tomato Fruit Rot Caused by Phytophthora nicotianae. Front. Microbiol. 2022, 13, 978920. [Google Scholar] [CrossRef]
- Ruiz-Moyano, S.; Hernández, A.; Galvan, A.I.; Córdoba, M.G.; Casquete, R.; Serradilla, M.J.; Martín, A. Selection and Application of Antifungal VOCs-Producing Yeasts as Biocontrol Agents of Grey Mould in Fruits. Food Microbiol. 2020, 92, 103556. [Google Scholar] [CrossRef]
- Hua, S.S.T.; Beck, J.J.; Sarreal, S.B.L.; Gee, W. The Major Volatile Compound 2-Phenylethanol from the Biocontrol Yeast, Pichia Anomala, Inhibits Growth and Expression of Aflatoxin Biosynthetic Genes of Aspergillus flavus. Mycotoxin Res. 2014, 30, 71–78. [Google Scholar] [CrossRef]
- Di Francesco, A.; Zajc, J.; Gunde-Cimerman, N.; Aprea, E.; Gasperi, F.; Placì, N.; Caruso, F.; Baraldi, E. Bioactivity of Volatile Organic Compounds by Aureobasidium Species against Gray Mold of Tomato and Table Grape. World J. Microbiol. Biotechnol. 2020, 36, 171. [Google Scholar] [CrossRef]
- Khunnamwong, P.; Lertwattanasakul, N.; Jindamorakot, S.; Suwannarach, N.; Matsui, K.; Limtong, S. Evaluation of Antagonistic Activity and Mechanisms of Endophytic Yeasts against Pathogenic Fungi Causing Economic Crop Diseases. Folia Microbiol. 2020, 65, 573–590. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Hernandez, E.; Gonzalez-Garcia, V.; Palacio-Bielsa, A.; Casanova-Gascon, J.; Navas-Gracia, L.M.; Martin-Gil, J.; Martin-Ramos, P. Phytochemical Constituents and Antimicrobial Activity of Euphorbia serrata L. Extracts for Borago officinalis L. Crop Protection. Horticulture 2023, 9, 652. [Google Scholar] [CrossRef]
- Le Dare, B.; Lagente, V.; Gicquel, T. Ethanol and Its Metabolites: Update on Toxicity, Benefits, and Focus on Immunomodulatory Effects. Drug Metab. Rev. 2019, 51, 545–561. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, J.H.; Jin, H.J.; Lee, S.Y. Antimicrobial Activity of Ethanol Extracts of Laminaria japonica against Oral Microorganisms. Anaerobe 2013, 21, 34–38. [Google Scholar] [CrossRef]
- Turu, D.; Bozyel, M.E.; Candan, K.; Yakan, M.A.; Eray Bozyel, M.; Benek, A.; Canli, K. In Vitro Antimicrobial and Antioxidant Activities of Pyracantha coccinea Fruits Ethanol Extract. Int. J. Acad. Multidiscip. Res. 2020, 4, 89–93. [Google Scholar]
- Watts, S.; Ramstedt, M.; Salentinig, S. Ethanol Inactivation of Enveloped Viruses: A Structural and Surface Chemistry Insight on Phi6. J. Phys. Chem. Lett. 2021, 12, 9557–9563. [Google Scholar] [CrossRef]
- Peters, B.M.; Ward, R.M.; Rane, H.S.; Lee, S.A.; Noverr, M.C. Efficacy of Ethanol against Candida albicans and Staphylococcus aureus Polymicrobial Biofilms. Antimicrob. Agents Chemother. 2013, 57, 74–82. [Google Scholar] [CrossRef]
- Fialho, M.B.; de Moraes, M.H.D.; Tremocoldi, A.R.; Pascholati, S.F. Potential of Antimicrobial Volatile Organic Compounds to Control Sclerotinia sclerotiorum in Bean Seeds. Pesqui. Agropecu. Bras. 2011, 46, 137–142. [Google Scholar] [CrossRef]
- Josselin, L.; De Clerck, C.; De Boevre, M.; Moretti, A.; Jijakli, M.H.; Soyeurt, H.; Fauconnier, M.-L. Volatile Organic Compounds Emitted by Aspergillus flavus Strains Producing or Not Aflatoxin B1. Toxins 2021, 13, 705. [Google Scholar] [CrossRef]
- Caceres, I.; Al Khoury, A.; El Khoury, R.; Lorber, S.; Oswald, I.P.; El Khoury, A.; Atoui, A.; Puel, O.; Bailly, J.D. Aflatoxin Biosynthesis and Genetic Regulation: A Review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef]

| No | Compound | CAS No | RI | GR | Control | HU SA | HU RC | MP SA | MP RC | SP SA | SP RC | SC SA | SC RC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ethyl acetate | 141-78-6 | nd | ES | 15.1 ± 12.3 | 131.5 ± 51.7 | 110.3 ± 14.2 | 280.5 ± 12.7 | 220.3 ± 55.8 | 20.9 ± 15.9 ns | 30.4 ± 17.2 ns | 51.2 ± 24.9 ns | 33.6 ± 23.6 ns |
| 2 | Ethanol | 64-17-5 | nd | OH | 6.7 ± 3.7 | 27.6 ± 3.7 | 24.9 ± 2.5 | 34.9 ± 0.7 | 42.0 ± 2.9 | 31.2 ± 6.5 | 36.2 ± 5.8 | 33.9 ± 2.8 | 38.4 ± 1.5 |
| 3 | Ethyl propionate | 1105-37-3 | nd | ES | 4.6 ± 1.7 | 25.3 ± 7.9 | 25.0 ± 3.6 | 32.8 ± 5.4 | 24.9 ± 9.9 | 8.9 ± 1.2 | 11.2 ± 2.5 | 8.4 ± 2.9 ns | 11.8 ± 5.1 |
| 4 | Propyl acetate | 109-60-4 | nd | ES | 0.7 ± 0.6 | 6.1 ± 3.7 | 4.3 ± 0.9 | 15.3 ± 1.5 | 11.6 ± 3.5 | 1.5 ± 0.9 ns | 2.2 ± 0.3 | 4.2 ± 1.3 | 2.7 ± 1.6 ns |
| 5 | 2-Methyl-1-propanol | 78-83-1 | 1094 | OH | 0 | 3.1 ± 1.0 | 6.3 ± 3.2 | 5.8 ± 0.6 | 14.5 ± 3.7 | 12.3 ± 3.8 | 13.6 ± 3.8 | 10.2 ± 1.7 | 10.8 ± 1.5 |
| 6 | 3-Methylbutyl acetate | 123-92-2 | 1112 | ES | 1.5 ± 1.3 | 86.0 ± 20.2 | 88.2 ± 15.4 | 23.5 ± 2.3 | 25.3 ± 6.2 | 63.4 ± 2.8 | 67.6 ± 10.1 | 56.1 ± 8.4 | 59.5 ± 9.2 |
| 7 | 3-Methylbutyl propionate | 105-68-0 | 1180 | ES | 0 | 26.3 ± 8.6 | 23.5 ± 5.2 | 2.0 ± 0.3 | 1.1 ± 0.6 | 5.8 ± 1.3 | 12.8 ± 7.9 | 8.6 ± 2.6 | 8.7 ± 2.8 |
| 8 | 3-Methyl-1-butanol | 123-51-3 | 1211 | OH | 1.2 ± 1.2 | 59.3 ± 7.8 | 62.8 ± 3.4 | 53.9 ± 3.6 | 78.5 ± 11.2 | 118.7 ± 6.6 | 130.1 ± 8.1 | 157.8 ± 7.0 | 161.7 ± 19.5 |
| 9 | Acetic acid | 64-19-7 | 1441 | FA | 0.6 ± 1.1 | 23.9 ± 6.1 | 23.8 ± 6.0 | 2.5 ± 1.8 | 0.3 ± 0.5 ns | 0.9 ± 0.1 ns | 0.7 ± 0.2 ns | 0.7 ± 0.1 ns | 0.6 ± 0.1 ns |
| 10 | 3-Methylbutanoic acid | 503-74-2 | 1662 | FA | 2.8 ± 2.3 | 39.2 ± 3.5 | 41.7 ± 3.0 | 22.7 ± 3.6 | 11.1 ± 9.3 ns | 0.9 ± 0.3 | 0.9 ± 0.4 ns | 4.8 ± 2.1 ns | 4.6 ± 1.3 ns |
| 11 | 2-Phenylethyl acetate | 103-45-7 | 1804 | AR, ES | 0 | 22.1 ± 8.1 | 19.9 ± 3.6 | 2.9 ± 1.1 | 4.6 ± 3.8 | 2.7 ± 1.4 | 1.6 ± 0.4 | 5.8 ± 1.1 | 5.3 ± 1.7 |
| 12 | 2-Phenylethanol | 60-12-8 | 1895 | AR, OH | 1.9 ± 1.1 | 44.1 ± 4.9 | 45.7 ± 2.6 | 29.5 ± 7.8 | 34.0 ± 6.3 | 53.4 ± 10.9 | 43.9 ± 4.4 | 63.4 ± 4.2 | 66.2 ± 12.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vepštaitė-Monstavičė, I.; Lukša-Žebelovič, J.; Apšegaitė, V.; Mozūraitis, R.; Lisicinas, R.; Stanevičienė, R.; Blažytė-Čereškienė, L.; Serva, S.; Servienė, E. Profiles of Killer Systems and Volatile Organic Compounds of Rowanberry and Rosehip-Inhabiting Yeasts Substantiate Implications for Biocontrol. Foods 2025, 14, 288. https://doi.org/10.3390/foods14020288
Vepštaitė-Monstavičė I, Lukša-Žebelovič J, Apšegaitė V, Mozūraitis R, Lisicinas R, Stanevičienė R, Blažytė-Čereškienė L, Serva S, Servienė E. Profiles of Killer Systems and Volatile Organic Compounds of Rowanberry and Rosehip-Inhabiting Yeasts Substantiate Implications for Biocontrol. Foods. 2025; 14(2):288. https://doi.org/10.3390/foods14020288
Chicago/Turabian StyleVepštaitė-Monstavičė, Iglė, Juliana Lukša-Žebelovič, Violeta Apšegaitė, Raimondas Mozūraitis, Robertas Lisicinas, Ramunė Stanevičienė, Laima Blažytė-Čereškienė, Saulius Serva, and Elena Servienė. 2025. "Profiles of Killer Systems and Volatile Organic Compounds of Rowanberry and Rosehip-Inhabiting Yeasts Substantiate Implications for Biocontrol" Foods 14, no. 2: 288. https://doi.org/10.3390/foods14020288
APA StyleVepštaitė-Monstavičė, I., Lukša-Žebelovič, J., Apšegaitė, V., Mozūraitis, R., Lisicinas, R., Stanevičienė, R., Blažytė-Čereškienė, L., Serva, S., & Servienė, E. (2025). Profiles of Killer Systems and Volatile Organic Compounds of Rowanberry and Rosehip-Inhabiting Yeasts Substantiate Implications for Biocontrol. Foods, 14(2), 288. https://doi.org/10.3390/foods14020288








