Bioprospecting Indigenous Oenococcus oeni Strains from Chinese Wine Regions: Multivariate Screening for Stress Tolerance and Aromatic Competence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains Isolation and Medium
2.2. Targeted Screening by PCR of Functional Genes
2.3. Stress Resistance Analysis of O. oeni Strains During MLF
2.4. MLF of O. oeni in Lab-Scale Vinification
2.5. Aroma Component Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Strain Screening Based on PCR of Functional Genes
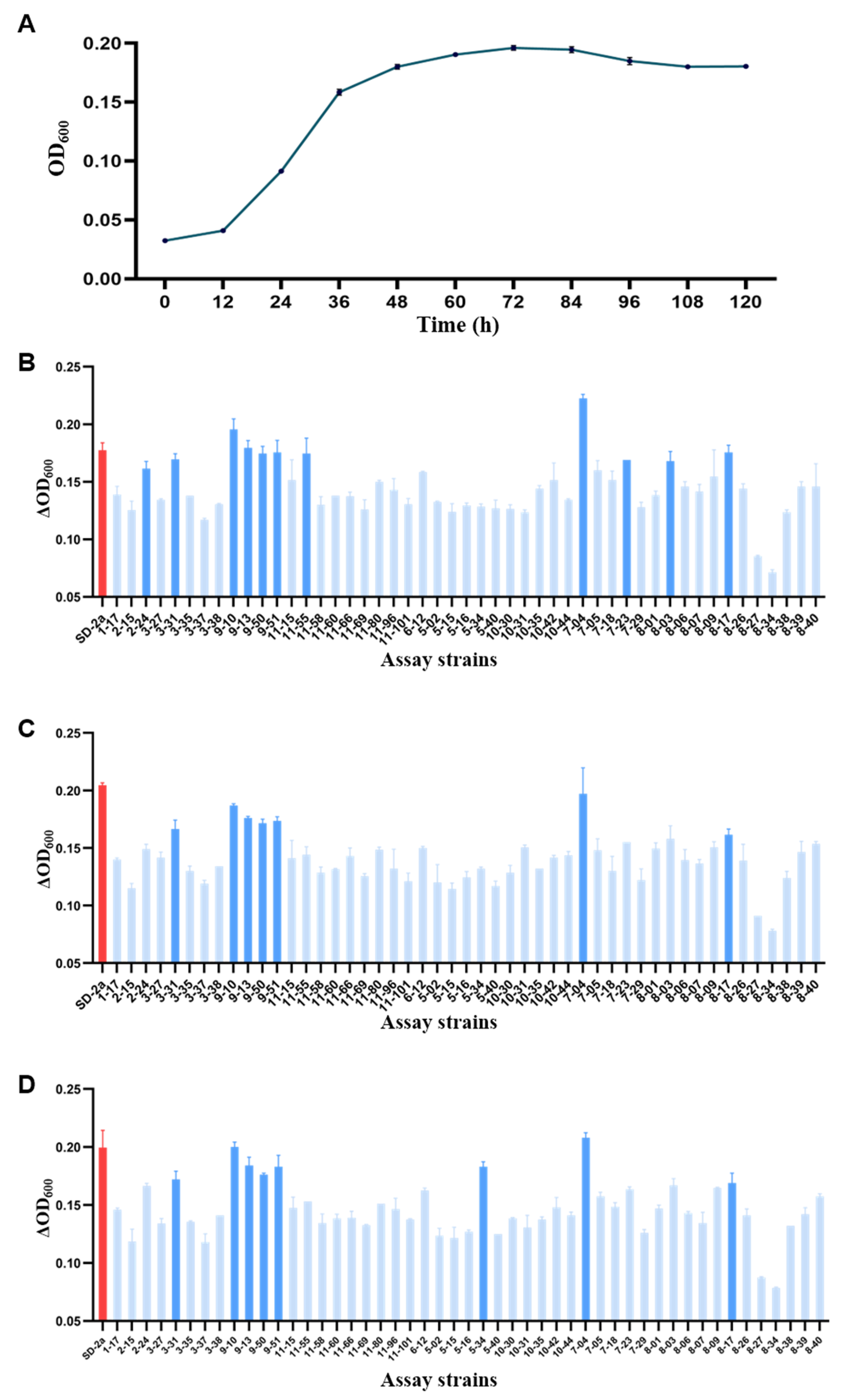
3.2. Screening of Superior Strains in Stress Conditions
3.3. Performance of O. oeni Strains During MLF
3.3.1. Consumption of L-Malic Acid in Wines
3.3.2. The Change of Physical and Chemical Indexes During MLF
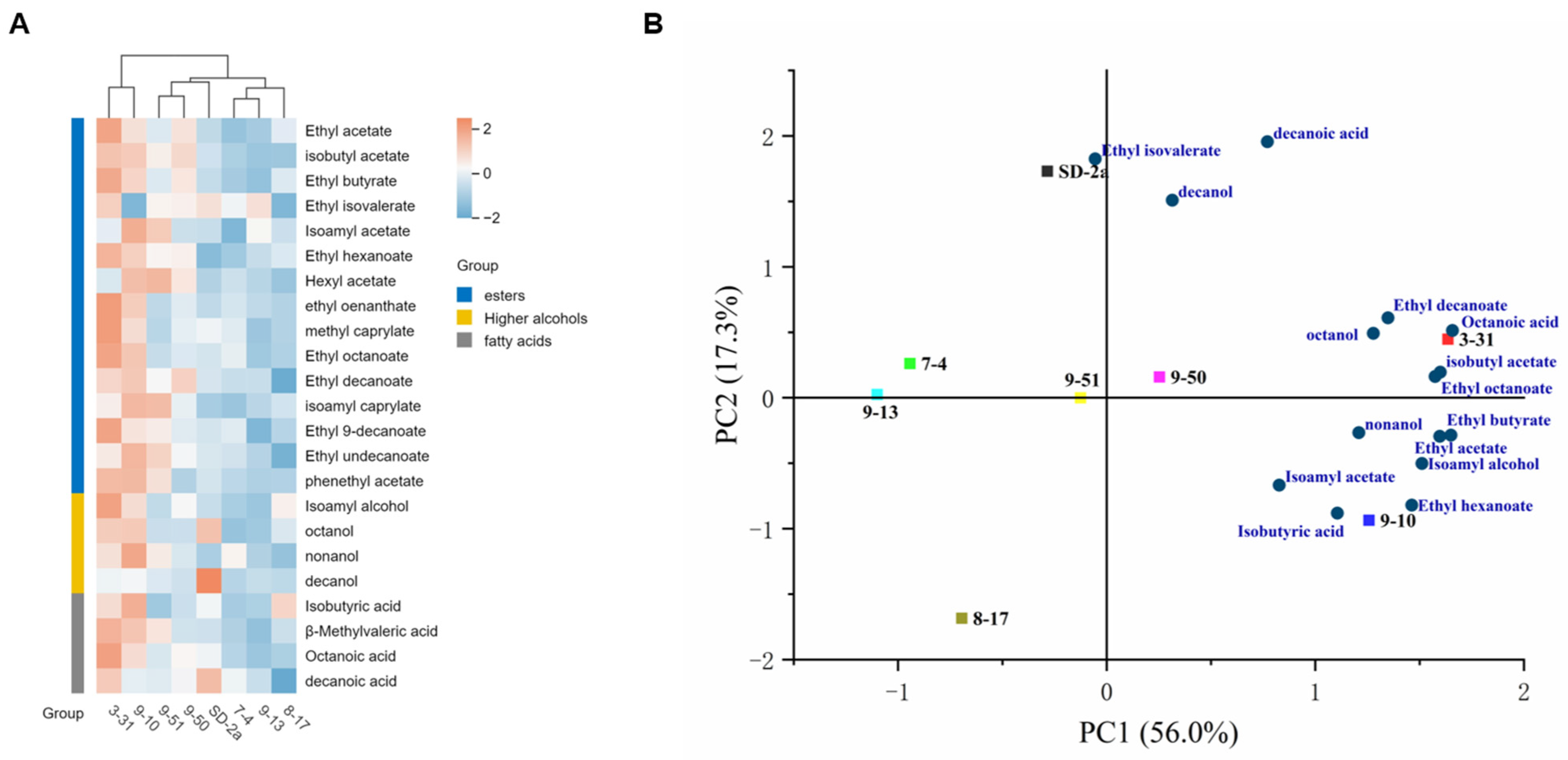
3.3.3. Aroma Components Analysis of Superior Indigenous Strains in Marselan Wines
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Versari, A.; Parpinello, G.P.; Cattaneo, M. Leuconostoc oenos and malolactic fermentation in wine: A review. J. Ind. Microbiol. Biotechnol. 1999, 23, 447–455. [Google Scholar]
- Nielsen, J.C.; Richelieu, M. Control of flavor development in wine during and after malolactic fermentation by Oenococcus oeni. Appl. Environ. Microbiol. 1999, 65, 740–745. [Google Scholar]
- Grandvalet, C.; Assad-García, J.S.; Chu-Ky, S.; Tollot, M.; Guzzo, J.; Gresti, J.; Tourdot-Maréchal, R. Changes in membrane lipid composition in ethanol- and acid-adapted Oenococcus oeni cells: Characterization of the cfa gene by heterologous complementation. Microbiology 2008, 154 Pt 9, 2611–2619. [Google Scholar] [CrossRef]
- Virdis, C.; Sumby, K.; Bartowsky, E.; Jiranek, V. Lactic Acid Bacteria in Wine: Technological Advances and Evaluation of Their Functional Role. Front. Microbiol. 2021, 11, 612118. [Google Scholar]
- Betteridge, A.; Grbin, P.; Jiranek, V. Improving Oenococcus oeni to overcome challenges of wine malolactic fermentation. Trends Biotechnol. 2015, 33, 547–553. [Google Scholar] [CrossRef]
- González-Arenzana, L.; Santamaría, P.; López, R.; López-Alfaro, I. Indigenous lactic acid bacteria communities in alcoholic and malolactic fermentations of Tempranillo wines elaborated in ten wineries of La Rioja (Spain). Food Res. Int. 2013, 50, 438–445. [Google Scholar]
- Sumby, K.M.; Bartle, L.; Grbin, P.R.; Jiranek, V. Measures to improve wine malolactic fermentation. Appl. Microbiol. Biotechnol. 2019, 103, 2033–2051. [Google Scholar]
- Sumby, K.M.; Grbin, P.R.; Jiranek, V. Implications of new research and technologies for malolactic fermentation in wine. Appl. Microbiol. Biotechnol. 2014, 98, 8111–8132. [Google Scholar]
- Mas, A.; Padilla, B.; Esteve-Zarzoso, B.; Beltran, G.; Reguant, C.; Bordons, A. Taking Advantage of Natural Biodiversity for Wine Making: The WILDWINE Project. Agric. Agric. Sci. Procedia 2016, 8, 4–9. [Google Scholar]
- Berbegal, C.; Benavent-Gil, Y.; Navascués, E.; Calvo, A.; Albors, C.; Pardo, I.; Ferrer, S. Lowering histamine formation in a red Ribera del Duero wine (Spain) by using an indigenous O. oeni strain as a malolactic starter. Int. J. Food Microbiol. 2017, 244, 11–18. [Google Scholar]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; Curilén, Y.; Delfederico, L.; Caballero, A.; Semorile, L.; Pozo-Bayón, M.Á.; Tymczyszyn, E.E. Advantages of Using Blend Cultures of Native L. plantarum and O. oeni Strains to Induce Malolactic Fermentation of Patagonian Malbec Wine. Front. Microbiol. 2018, 9, 2109. [Google Scholar]
- Franquès, J.; Araque, I.; El Khoury, M.; Lucas, P.; Reguant, R.; Bordons, A. Selection and characterization of autochthonous strains of Oenococcus oeni for vinification in Priorat (Catalonia, Spain). OENO One 2018, 52, 45–56. [Google Scholar]
- Garofalo, C.; El Khoury, M.; Lucas, P.; Bely, M.; Russo, P.; Spano, G.; Capozzi, V. Autochthonous starter cultures and indigenous grape variety for regional wine production. J. Appl. Microbiol. 2015, 118, 1395–1408. [Google Scholar]
- Romero, J.; Ilabaca, C.; Ruiz, M.; Jara, C. Oenococcus oeni in Chilean Red Wines: Technological and Genomic Characterization. Front. Microbiol. 2018, 9, 90. [Google Scholar]
- Ruiz, P.; Izquierdo, P.M.; Seseña, S.; Palop, M.L. Selection of autochthonous Oenococcus oeni strains according to their oenological properties and vinification results. Int. J. Food Microbiol. 2010, 137, 230–235. [Google Scholar] [PubMed]
- Battistelli, N.; Perpetuini, G.; Perla, C.; Arfelli, G.; Zulli, C.; Rossetti, A.P.; Tofalo, R. Characterization of natural Oenococcus oeni strains for Montepulciano d’Abruzzo organic wine production. Eur. Food Res. Technol. 2020, 246, 1031–1039. [Google Scholar]
- Swiegers, J.; Bartowsky, E.; Henschke, P.A.; Pretorius, I. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar]
- Gagné, S.; Lucas, P.M.; Perello, M.C.; Claisse, O.; Lonvaud-Funel, A.; de Revel, G. Variety and variability of glycosidase activities in an Oenococcus oeni strain collection tested with synthetic and natural substrates. J. Appl. Microbiol. 2011, 110, 218–228. [Google Scholar]
- Mateo, J.J.; Di, S.R. Description of the β-glucosidase activity of wine yeasts. Food Microbiol. 1997, 14, 583–591. [Google Scholar]
- Palmeri, R.; Spagna, G. β-Glucosidase in cellular and acellular form for winemaking application. Enzym. Microb. Technol. 2007, 40, 382–389. [Google Scholar]
- Iorizzo, M.; Testa, B.; Lombardi, S.J.; García-Ruiz, A.; Muñoz-González, C.; Bartolomé, B.; Moreno-Arribas, M.V. Selection and technological potential of Lactobacillus plantarum bacteria suitable for wine malolactic fermentation and grape aroma release. LWT Food Sci. Technol. 2016, 73, 557–566. [Google Scholar]
- Michlmayr, H.; Eder, R.; Kulbe, K.; del Hierro, A. β-Glycosidase activities of Oenococcus oeni: Current state of research and future challenges. Mitt. Klosterneubg. 2012, 62, 87–96. [Google Scholar]
- Olguín, N.; Alegret, J.O.; Bordons, A.; Reguant, R. β-Glucosidase Activity and bgl Gene Expression of Oenococcus oeni Strains in Model Media and Cabernet Sauvignon Wine. Am. J. Enol. Vitic. 2011, 62, 99. [Google Scholar]
- Pérez-Martín, F.; Seseña, S.; Izquierdo, P.M.; Martín, R.; Palop, M.L. Screening for glycosidase activities of lactic acid bacteria as a biotechnological tool in oenology. World J. Microbiol. Biotechnol. 2012, 28, 1423–1432. [Google Scholar]
- Aznar, M.; Arroyo, T. Analysis of wine volatile profile by purge-and-trap-gas chromatography-mass spectrometry. Application to the analysis of red and white wines from different Spanish regions. J. Chromatogr. A 2007, 1165, 151–157. [Google Scholar]
- Ferreira, V.; López, R.; Cacho, J.F. Quantitative determination of the odorants of young red wines from different grape varieties. J. Sci. Food Agric. 2000, 80, 1659–1667. [Google Scholar]
- Margalef-Català, M.; Felis, G.E.; Reguant, C.; Stefanelli, E.; Torriani, S.; Bordons, A. Identification of variable genomic regions related to stress response in Oenococcus oeni. Food Res. Int. 2017, 102, 625–638. [Google Scholar]
- Da Silveira, M.G.; Golovina, E.A.; Hoekstra, F.A.; Rombouts, F.M.; Abee, T. Membrane fluidity adjustments in ethanol-stressed Oenococcus oeni cells. Appl. Environ. Microbiol. 2003, 69, 5826–5832. [Google Scholar] [PubMed]
- Guzzo, J.; Cavin, J.F.; Divies, C. Induction of stress proteins in Leuconostoc oenos to perform direct inoculation of wine. Biotechnol. Lett. 1994, 16, 1189–1194. [Google Scholar]
- Vigentini, I.; Picozzi, C.; Tirelli, A.; Giugni, A.; Foschino, R. Survey on indigenous Oenococcus oeni strains isolated from red wines of Valtellina, a cold climate wine-growing Italian area. Int. J. Food Microbiol. 2009, 136, 123–128. [Google Scholar]
- Canas, B.J.; Joe, F.L., Jr.; Diachenko, G.W.; Burns, G. Determination of ethyl carbamate in alcoholic beverages and soy sauce by gas chromatography with mass selective detection: Collaborative study. J. AOAC Int. 1994, 77, 1530–1536. [Google Scholar] [PubMed]
- Lonvaud-Funel, A. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Leeuwenhoek 1999, 76, 317–331. [Google Scholar] [PubMed]
- Maintz, L.; Novak, N. Histamine and histamine intolerance. Am. J. Clin. Nutr. 2007, 85, 1185–1196. [Google Scholar] [PubMed]
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B.; et al. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64 (Suppl. 3), S95–S100. [Google Scholar]
- Liu, S.Q.; Pilone, G.J. A REVIEW: Arginine metabolism in wine lactic acid bacteria and its practical significance. J. Appl. Microbiol. 1998, 84, 315–327. [Google Scholar]
- Araque, I.; Gil, J.; Carreté, R.; Bordons, A.; Reguant, C. Detection of arc genes related with the ethyl carbamate precursors in wine lactic acid bacteria. J. Agric. Food Chem. 2009, 57, 1841–1847. [Google Scholar]
- Spano, G.; Chieppa, G.; Beneduce, L.; Massa, S. Expression analysis of putative arcA, arcB and arcC genes partially cloned from Lactobacillus plantarum isolated from wine. J. Appl. Microbiol. 2004, 96, 185–193. [Google Scholar]
- Tonon, T.; Bourdineaud, J.P.; Lonvaud-Funel, A. The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a CRP-like gene. Res. Microbiol. 2001, 152, 653–661. [Google Scholar]
- Divol, B.; Tonon, T.; Morichon, S.; Gindreau, E.; Lonvaud-Funel, A. Molecular characterization of Oenococcus oeni genes encoding proteins involved in arginine transport. J. Appl. Microbiol. 2003, 94, 738–746. [Google Scholar]
- Mira de Orduña, R.; Patchett, M.L.; Liu, S.Q.; Pilone, G.J. Growth and arginine metabolism of the wine lactic acid bacteria Lactobacillus buchneri and Oenococcus oeni at different pH values and arginine concentrations. Appl. Environ. Microbiol. 2001, 67, 1657–1662. [Google Scholar]
- Landete, J.M.; de las Rivas, B.; Marcobal, A.; Muñoz, R. Molecular methods for the detection of biogenic amine-producing bacteria on foods. Int. J. Food Microbiol. 2007, 117, 258–269. [Google Scholar] [PubMed]
- Lonvaud-Funel, A. Biogenic amines in wines: Role of lactic acid bacteria. FEMS Microbiol. Lett. 2001, 199, 9–13. [Google Scholar]
- Rosi, I.; Nannelli, F.; Giovani, G. Biogenic amine production by Oenococcus oeni during malolactic fermentation of wines obtained using different strains of Saccharomyces cerevisiae. LWT Food Sci. Technol. 2009, 42, 525–530. [Google Scholar]
- Coton, E.; Coton, M. Evidence of horizontal transfer as origin of strain to strain variation of the tyramine production trait in Lactobacillus brevis. Food Microbiol. 2009, 26, 52–57. [Google Scholar] [CrossRef]
- Henríquez-Aedo, K.; Durán, D.; Garcia, A.; Hengst, M.B.; Aranda, M. Identification of biogenic amines-producing lactic acid bacteria isolated from spontaneous malolactic fermentation of chilean red wines. LWT Food Sci. Technol. 2016, 68, 183–189. [Google Scholar]
- Wang, P.; Li, A.; Sun, H.; Dong, M.; Wei, M.; Fan, M. Selection and characterization of Oenococcus oeni strains for use as new malolactic fermentation starter cultures. Ann. Microbiol. 2016, 66, 1285–1292. [Google Scholar]
- Chen, Y.; Lei, X.; Jiang, J.; Qin, Y.; Jiang, L.; Liu, Y.L. Microbial diversity on grape epidermis and wine volatile aroma in spontaneous fermentation comprehensively driven by geography, subregion, and variety. Int. J. Food Microbiol. 2023, 404, 110315. [Google Scholar]
- Yu, D.; Shi, K.; Wen, X.; Xie, F.; Wang, T.; Liu, S.; He, L. Evidence of the genetic diversity and clonal population structure of Oenococcus oeni strains isolated from different wine-making regions of China. J. Microbiol. 2018, 56, 556–564. [Google Scholar]
- Li, Y.; Wang, Y.; Fan, L.; Wang, F.; Liu, X.; Zhang, H.; Zhou, J. Assessment of β-D-glucosidase activity and bgl gene expression of Oenococcus oeni SD-2a. PLoS ONE 2020, 15, e0240484. [Google Scholar]
- Li, Y.; Ma, Y.; Huang, K.; Zhang, H. Identification and Localization of β-D-Glucosidase from Two Typical Oenococcus oeni Strains. Pol. J. Microbiol. 2016, 65, 209–213. [Google Scholar]
- Liu, L.; Peng, S.; Song, W.; Zhao, H.; Li, H.; Wang, H. Genomic Analysis of an Excellent Wine-Making Strain Oenococcus oeni SD-2a. Pol. J. Microbiol. 2022, 71, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wang, T.; Li, Y.Y.; Li, J.; Zhang, Y.; Wang, Y.; Wang, H.; Li, H. Antioxidant properties of wine lactic acid bacteria: Oenococcus oeni. Appl. Microbiol. Biotechnol. 2015, 99, 5189–5202. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yang, X.; Meng, Q.; Zhao, L.; Yuan, Y.; Chi, W.; He, L.; Shi, K.; Liu, S. Integrative multiomics analysis of the acid stress response of Oenococcus oeni mutants at different growth stages. Food Microbiol. 2022, 102, 103905. [Google Scholar] [CrossRef] [PubMed]
- GB/T 15038-2006; Analytical Methods of Wine and Fruit Wine. China Standard Publishing House: Beijing, China, 2006.
- Du, Q.; Ye, D.; Zang, X.; Nan, H.; Liu, Y. Effect of low temperature on the shaping of yeast-derived metabolite compositions during wine fermentation. Food Res. Int. 2022, 162 Pt A, 112016. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, D.; Liu, H.; Shen, J.; Zhang, J.; He, L.; Li, J.; Zhou, P.; Guan, X.; Liu, S.; et al. Impact of indigenous Oenococcus oeni and Lactiplantibacillus plantarum species co-culture on Cabernet Sauvignon wine malolactic fermentation: Kinetic parameters, color and aroma. Food Chem. X 2024, 22, 101369. [Google Scholar] [CrossRef]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; La Hens, D.V.; Hollmann, A.; Delfederico, L.; Caballero, A.; Tymczyszyn, E.E.; Semorile, L. Comparative vinification assays with selected Patagonian strains of Oenococcus oeni and Lactobacillus plantarum. LWT Food Sci. Technol. 2017, 77, 348–355. [Google Scholar] [CrossRef]
- Selli, S.; Cabaroglu, T.; Canbas, A.; Erten, H.; Nurgel, C.; Lepoutre, J.P.; Gunata, Z. Volatile composition of red wine from cv. Kalecik Karasι grown in central Anatolia. Food Chem. 2004, 85, 207–213. [Google Scholar] [CrossRef]
- Díez, J.; Domínguez, C.; Guillén, D.A.; Veas, R.; Barroso, C.G. Optimisation of stir bar sorptive extraction for the analysis of volatile phenols in wines. J. Chromatogr. A 2004, 1025, 263–267. [Google Scholar] [CrossRef]
- Styger, G.; Prior, B.; Bauer, F.F. Wine flavor and aroma. J. Ind. Microbiol. Biotechnol. 2011, 38, 1145–1159. [Google Scholar] [CrossRef]
- Dein, M.; Kerley, T.; Munafo, J.P., Jr. Characterization of Odorants in a 10-Year-Old Riesling Wine. J. Agric. Food Chem. 2021, 69, 11372–11381. [Google Scholar] [CrossRef]
- Cao, W.; Shu, N.; Wen, J.; Yang, Y.; Jin, Y.; Lu, W. Characterization of the Key Aroma Volatile Compounds in Nine Different Grape Varieties Wine by Headspace Gas Chromatography-Ion Mobility Spectrometry (HS-GC-IMS), Odor Activity Values (OAV) and Sensory Analysis. Foods 2022, 11, 2767. [Google Scholar] [CrossRef] [PubMed]
- Sumby, K.M.; Grbin, P.R.; Jiranek, V. Microbial modulation of aromatic esters in wine: Current knowledge and future prospects. Food Chem. 2010, 121, 1–16. [Google Scholar]
- Diez-Ozaeta, I.; Lavilla, M.; Amárita, F. Wine aroma profile modification by Oenococcus oeni strains from Rioja Alavesa region: Selection of potential malolactic starters. Int. J. Food Microbiol. 2021, 356, 109324. [Google Scholar]
- Jiang, B.; Xi, Z.; Luo, M.; Zhang, Z. Comparison on aroma compounds in Cabernet Sauvignon and Merlot wines from four wine grape-growing regions in China. Food Res. Int. 2013, 51, 482–489. [Google Scholar]


| Simulated Wine Number No. | pH | Ethanol (%(v/v)) |
|---|---|---|
| 1 | 3.8 | 10 |
| 2 | 3.5 | 10 |
| 3 | 3.2 | 10 |
| 4 | 3.8 | 12 |
| 5 | 3.5 | 12 |
| 6 | 3.2 | 12 |
| 7 | 3.8 | 14 |
| 8 | 3.5 | 14 |
| 9 | 3.2 | 14 |
| Sample | Alcohol Degree %(v/v) | Residual Sugar g/L | Total Acid g/L | Volatile Acid g/L | pH | Color Intensity |
|---|---|---|---|---|---|---|
| Before MLF | 13.43 ± 0.2 ab | 3.4 ± 0.1 b | 7.16 ± 0.04 a | 0.30 ± 0.01 h | 3.49 ± 0.04 c | 9.68 ± 0.07 a |
| Spontaneous MLF | 13.63 ± 0.14 a | 2.3 ± 0.2 e | 4.93 ± 0.03 e | 0.57 ± 0.02 a | 3.75 ± 0.1 a | 7.01 ± 0.01 h |
| SD-2a | 13.45 ± 0.03 ab | 2.6 ± 0.12 d | 5.24 ± 0.04 d | 0.50 ± 0.02 bc | 3.74 ± 0.01 a | 7.16 ± 0.04 fg |
| 3-31 | 13.47 ± 0.11 ab | 2.0 ± 0.1 f | 5.26 ± 0.02 cd | 0.44 ± 0.03 de | 3.64 ± 0.03 ab | 7.31 ± 0.04 e |
| 7-04 | 13.63 ± 0.01 a | 2.0 ± 0.1 f | 5.19 ± 0.04 d | 0.42 ± 0.03 ef | 3.68 ± 0.1 ab | 7.47 ± 0.17 d |
| 9-10 | 13.36 ± 0.02 b | 1.6 ± 0.1 g | 5.20 ± 0.1 d | 0.52 ± 0.02 b | 3.71 ± 0.04 ab | 7.55 ± 0.05 d |
| 9-13 | 13.44 ± 0.1 ab | 1.6 ± 0.1 g | 5.22 ± 0.06 d | 0.45 ± 0.02 de | 3.69 ± 0.1 ab | 7.87 ± 0.05 b |
| 9-50 | 13.47 ± 0.12 ab | 1.9 ± 0.1 f | 5.34 ± 0.04 bc | 0.47 ± 0.04 cd | 3.74 ± 0.04 a | 7.19 ± 0.04 ef |
| 9-51 | 13.40 ± 0.06 b | 3.2 ± 0.1 c | 5.35 ± 0.05 b | 0.39 ± 0.01 fg | 3.66 ± 0.02 ab | 7.04 ± 0.01 gh |
| 8-17 | 13.38 ± 0.08 b | 3.7 ± 0.1 a | 5.23 ± 0.02 d | 0.39 ± 0.02 g | 3.61 ± 0.03 b | 7.79 ± 0.09 c |
| RT | Compounds | Aroma Concentration (μg/L) | Thresholds | OAV | Descriptor | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SD-2a | 3-31 | 7-04 | 9-10 | 9-13 | 9-50 | 9-51 | 8-17 | |||||
| 6.41 | Ethyl acetate (mg/L) | 47.78 ± 6.03 bc | 57.87 ± 4.67 a | 45.33 ± 0.37 c | 53.13 ± 3.52 ab | 46.24 ± 0.61 bc | 52.96 ± 0.25 ab | 49.65 ± 0.02 bc | 49.87 ± 0.80 bc | 7500 | >1 | Fruity, Sweet |
| 9.64 | Isobutyl acetate | 53.98 ± 8.23 abc | 61.04 ± 2.65 a | 51.79 ± 1.12 bc | 60.32 ± 3.35 ab | 50.73 ± 3.40 c | 59.28 ± 2.08 abc | 57.55 ± 0.61 abc | 50.85 ± 0.42 c | 30 | >1 | Banana |
| 10.37 | Ethyl butyrate | 313.7 ± 43.33 bcd | 380.5 ± 23.99 a | 300.82 ± 3.2 cd | 355.64 ± 6.43 ab | 293.13 ± 11.75 d | 346.05 ± 0.13 abc | 324.45 ± 1.29 bcd | 324.61 ± 31.29 cd | 20 | >1 | Strawberry |
| 11.38 | Ethyl isovalerate | 7.03 ± 2.82 a | 8.14 ± 3.07 a | 5.29 ± 0.19 a | 0.00 | 7.13 ± 0.63 a | 6.13 ± 0.16 a | 5.95 ± 0.09 a | 0.00 | 3 | >1 | Strawberry, Sweet |
| 13.32 | Isoamyl acetate | 684.14 ± 95.72 bc | 708.86 ± 30 abc | 624.39 ± 59.96 c | 804.56 ± 0.8 a | 726.38 ± 39.66 abc | 686.85 ± 0.24 abc | 774.09 ± 31.23 ab | 686.12 ± 41.70 abc | 30 | >1 | Fresh, Banana |
| 17.59 | Ethyl hexanoate | 269.13 ± 56.02 d | 388.54 ± 46.84 a | 283.64 ± 3.64 cd | 367.92 ± 14.12 ab | 305.78 ± 16.36 bcd | 341.45 ± 8.56 abc | 338.70 ± 1.06 abc | 318.01 ± 4.89 bcd | 14 | >1 | Green apple, Strawberry |
| 19.04 | Hexyl acetate | 2.15 ± 0.36 bc | 2.45 ± 0.3 bc | 2.35 ± 0.02 bc | 3.25 ± 0.1 a | 2.20 ± 0.22 bc | 2.85 ± 0.69 ab | 3.32 ± 0.35 a | 1.97 c | 1500 | <0.1 | Pear |
| 21.3 | Ethyl oenanthate | 2.94 ± 0.43 bc | 4.04 ± 0.63 a | 3.07 ± 0.03 bc | 3.63 ± 0.18 ab | 2.94 ± 0.01 bc | 3.14 ± 0.14 bc | 2.93 ± 0.3 bc | 2.86 ± 0.01 c | 220 | <0.1 | Pineapple |
| 23.38 | Methyl caprylate | 5.18 ± 0.62 bc | 6.66 ± 0.83 a | 5.00 ± 0.10 bc | 5.68 ± 0.07 b | 4.24 ± 0.27 c | 5.06 ± 0.32 bc | 4.57 ± 0.08 c | 4.50 ± 0.13 c | 100–400 | <0.1 | Sweet, orange |
| 25.02 | Ethyl octanoate | 340.54 ± 38.76 c | 436.27 ± 47.95 a | 352.56 ± 12.69 bc | 407.07 ± 23.05 ab | 306.14 ± 17.15 c | 345.85 ± 13.77 c | 331.49 ± 7.7 c | 317.24 ± 5.39 c | 5 | >1 | Pineapple Pear, Floral |
| 31.81 | Ethyl decanoate | 102.34 ± 8.11 cd | 116.77 ± 5.63 ab | 99.93 ± 0.39 c | 119.94 ± 7.53 a | 99.82 ± 2.66 c | 117.68 ± 9.16 ab | 108.38 ± 5.62 abc | 83.36 ± 6.17 d | 200 | 0.1~1 | Fruity, Comfort |
| 32.45 | Isoamyl caprylate | 6.17 ± 0.3 b | 7.41 ± 0.42 ab | 5.99 ± 0.06 b | 8.36 ± 1.56 a | 6.40 ± 0.3 b | 7.03 ± 0.83 ab | 8.29 ± 0.86 a | 6.73 ± 0.50 ab | 125 | <0.1 | Sweet, Cheese |
| 33.47 | Ethyl 9-decanoate | 5.35 ± 0.4 bc | 7.05 ± 1.25 a | 5.46 ± 0.34 bc | 6.07 ± 0.27 ab | 4.38 ± 0.03 c | 5.48 ± 0.46 bc | 5.95 ± 0.56 ab | 5.00 ± 0.26 bc | 100 | <0.1 | Fruity |
| 34.88 | Ethyl undecanoate | 4.21 ± 0.78 abc | 4.78 ± 0.45 ab | 4.12 ± 0.63 abc | 5.57 ± 1.32 a | 3.80 ± 0.54 bc | 4.51 abc | 5.13 ± 0.24 ab | 3.08 ± 0.04 c | n.f | n.f | Coconut |
| 37.08 | Phenethyl acetate | 12.03 ± 1.49 b | 14.35 ± 0.86 a | 11.55 ± 0.2 b | 14.39 ± 1.11 a | 11.29 ± 0.67 b | 11.36 ± 0.29 b | 13.35 ± 0.08 ab | 11.35 ± 1.45 b | 250 | <0.1 | Rose, Sweet |
| 16.62 | Isoamyl alcohol (mg/L) | 189.40 ± 22.16 cd | 235.101 ± 7.64 a | 180.77 ± 0.12 d | 213.13 ± 1.54 b | 176.93 ± 1.22 d | 202.8 ± 0.43 bc | 187.78 ± 2.68 cd | 206.30 ± 3.12 bc | 30,000 | >1 | Mellow |
| 29.02 | Octanol | 188.89 ± 18.6 a | 185.06 ± 7.25 a | 140.23 ± 1.67 b | 186.12 ± 16.08 a | 142.76 ± 0.66 b | 155.03 ± 0.17 b | 154.76 ± 3.79 b | 159.75 ± 6.33 b | 900 | 0.1~1 | Rose, Organic, sweet |
| 32.31 | Nonanol | 59.86 ± 7.36 cd | 73.07 ± 4.27 ab | 70.28 ± 1.49 bc | 82.31 ± 10.02 a | 60.57 ± 0.41 cd | 65.42 ± 0.7 bc | 71.44 ± 0.75 ab | 57.21 ± 1.91 d | 600 | 0.1~1 | Orange, Strawberry |
| 35.32 | Decanol | 139.40 ± 13.81 a | 78.85 ± 8.12 bc | 57.11 ± 1.37 d | 80.82 ± 9.98 b | 63.10 ± 0.24 cd | 66.19 ± 0.23 bcd | 71.74 ± 0.86 bcd | 59.97 ± 0.8 d | 400 | 0.1~1 | Sweet floral |
| 29.38 | Isobutyric acid | 3145.64 ± 174.68 ab | 3390.17 ± 186.02 ab | 2711.15 ± 27.26 b | 3737.4 ± 811.43 a | 2751.98 ± 120.88 b | 2917.50 ± 38.89 ab | 2668.17 ± 142.97 b | 3444.28 ± 441.84 ab | 8100 | 0.1~1 | Chemical |
| 32.67 | β-Methylvaleric acid | 2359.02 ± 164.93 bc | 2833.93 ± 113.59 a | 2256.06 ± 28.49 c | 2746.89 ± 195.95 a | 2150.49 ± 36.93 c | 2368.88 ± 31.91 bc | 2598.79 ± 142.07 ab | 2346.25 ± 102.96 bc | n.f | n.f | Mint |
| 41.88 | Octanoic acid | 1604.25 ± 203.46 bc | 1848.35 ± 118.11 a | 1490.10 ± 46.35 bc | 1693.97 ± 92.80 ab | 1438.23 ± 34.10 c | 1627.50 ± 8.44 abc | 1558.51 ± 1.71 bc | 1471.46 ± 28.9 bc | 8100 | 0.1~1 | Cheese |
| 46.28 | Decanoic acid | 156.69 ± 28.47 a | 151.29 ± 12.5 a | 132.77 ± 6.32 a | 130.14 ± 11.46 ab | 122.58 ± 17.54 ab | 133.62 ± 9.99 a | 128.96 ± 12.24 ab | 96.09 ± 0.85 b | 1000 | 0.1~1 | Fatty |
| Total (mg/L) | 246.65 | 303.69 | 234.71 | 277.09 | 231.72 | 265.04 | 246.66 | 265.62 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Hong, X.; Xu, Z.; Liu, S.; Shi, K. Bioprospecting Indigenous Oenococcus oeni Strains from Chinese Wine Regions: Multivariate Screening for Stress Tolerance and Aromatic Competence. Foods 2025, 14, 1207. https://doi.org/10.3390/foods14071207
Zhu Y, Hong X, Xu Z, Liu S, Shi K. Bioprospecting Indigenous Oenococcus oeni Strains from Chinese Wine Regions: Multivariate Screening for Stress Tolerance and Aromatic Competence. Foods. 2025; 14(7):1207. https://doi.org/10.3390/foods14071207
Chicago/Turabian StyleZhu, Yongzhang, Xiaoqing Hong, Zhenghua Xu, Shuwen Liu, and Kan Shi. 2025. "Bioprospecting Indigenous Oenococcus oeni Strains from Chinese Wine Regions: Multivariate Screening for Stress Tolerance and Aromatic Competence" Foods 14, no. 7: 1207. https://doi.org/10.3390/foods14071207
APA StyleZhu, Y., Hong, X., Xu, Z., Liu, S., & Shi, K. (2025). Bioprospecting Indigenous Oenococcus oeni Strains from Chinese Wine Regions: Multivariate Screening for Stress Tolerance and Aromatic Competence. Foods, 14(7), 1207. https://doi.org/10.3390/foods14071207







