Non-Destructive Detection of Chilled Mutton Freshness Using a Dual-Branch Hierarchical Spectral Feature-Aware Network
Abstract
:1. Introduction
- (1)
- Utilizing multi-indicator integrated hyperspectral data for freshness evaluation, providing biologically and chemically relevant data support.
- (2)
- Establishing a generalized data processing strategy through optimized preprocessing and feature wavelength selection, supplemented by SHAP interpretability analyses.
- (3)
- Introducing the PBCA module, an advanced position-encoded bidirectional cross-attention mechanism, significantly enhancing the DBHSNet’s sensitivity to long-range dependencies and fine-grained details.
- (4)
- Developing the MSMHA module, a multi-scale enhanced multi-head attention approach, improving the DBHSNet’s capability to detect subtle freshness characteristics.
- (5)
- Proposing the HCM module, a hierarchical classifier with dynamically optimized loss functions, effectively mitigating misclassification caused by intermediate feature ambiguity and overlapping boundaries.
- (6)
- Designing a dual-branch spectral feature-aware network that integrates multiple modules to fully perceive spectral characteristics, achieving high-accuracy freshness detection of chilled mutton, and providing valuable insights and methodologies for future meat freshness research.
2. Materials and Methods
2.1. Preparation of Chilled Mutton Samples
2.2. Acquisition of Hyperspectral Images During Chilled Storage
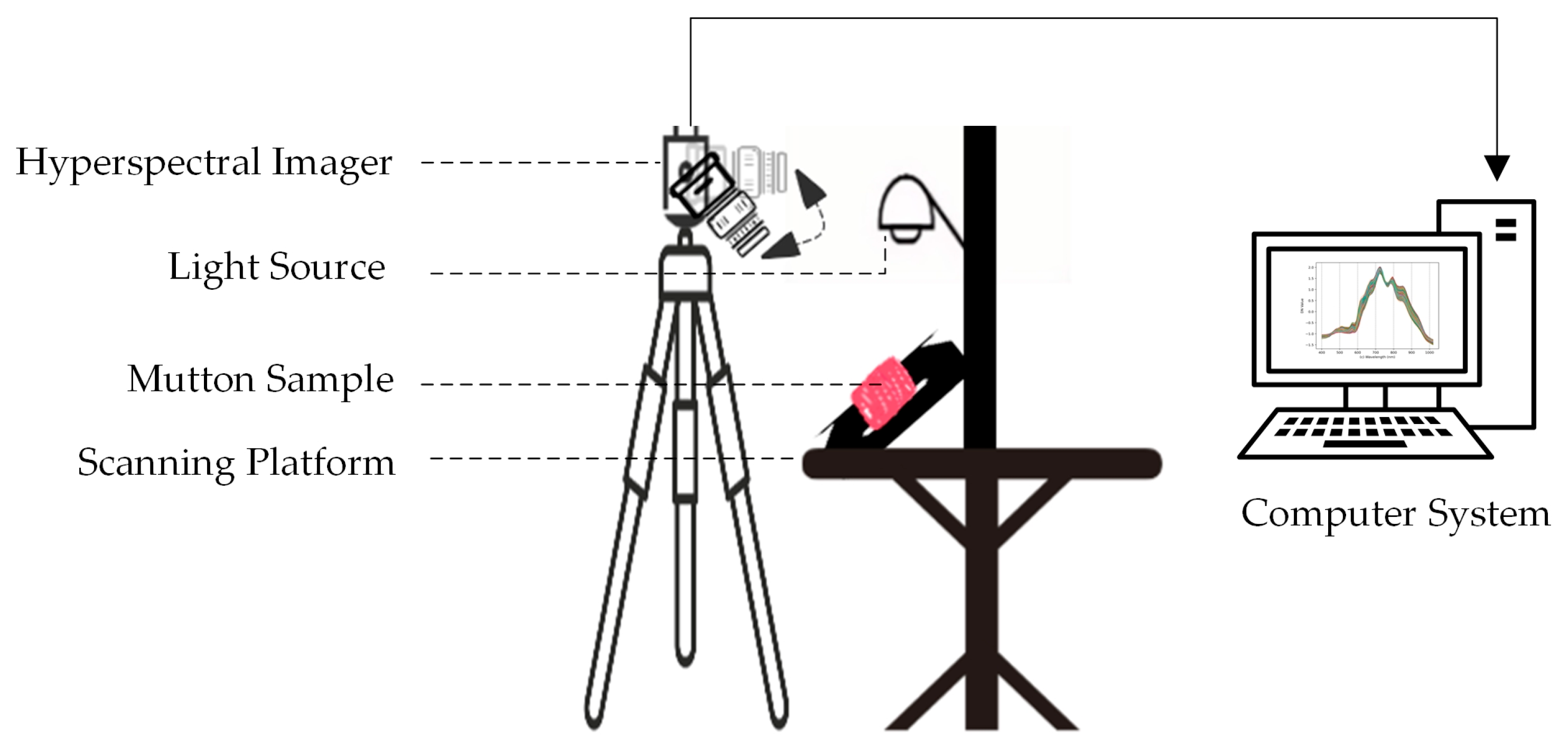
2.2.1. Hyperspectral Imaging Equipment
2.2.2. Hyperspectral Image Acquisition
2.2.3. Hyperspectral Data Extraction
2.3. Measurement of Freshness Indicators and Freshness Grading
2.4. Research Methods
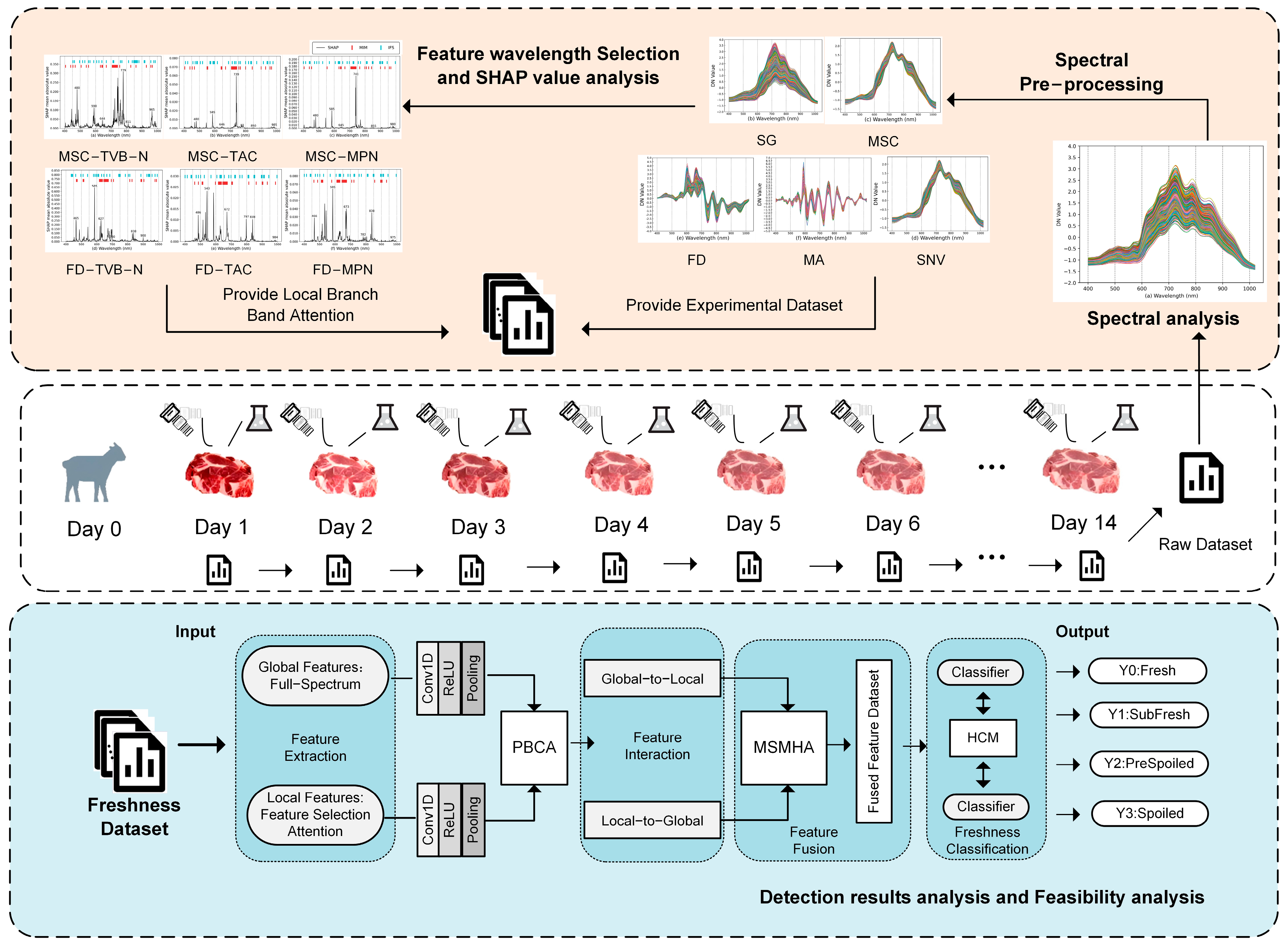
2.4.1. Research Framework
2.4.2. Preprocessing
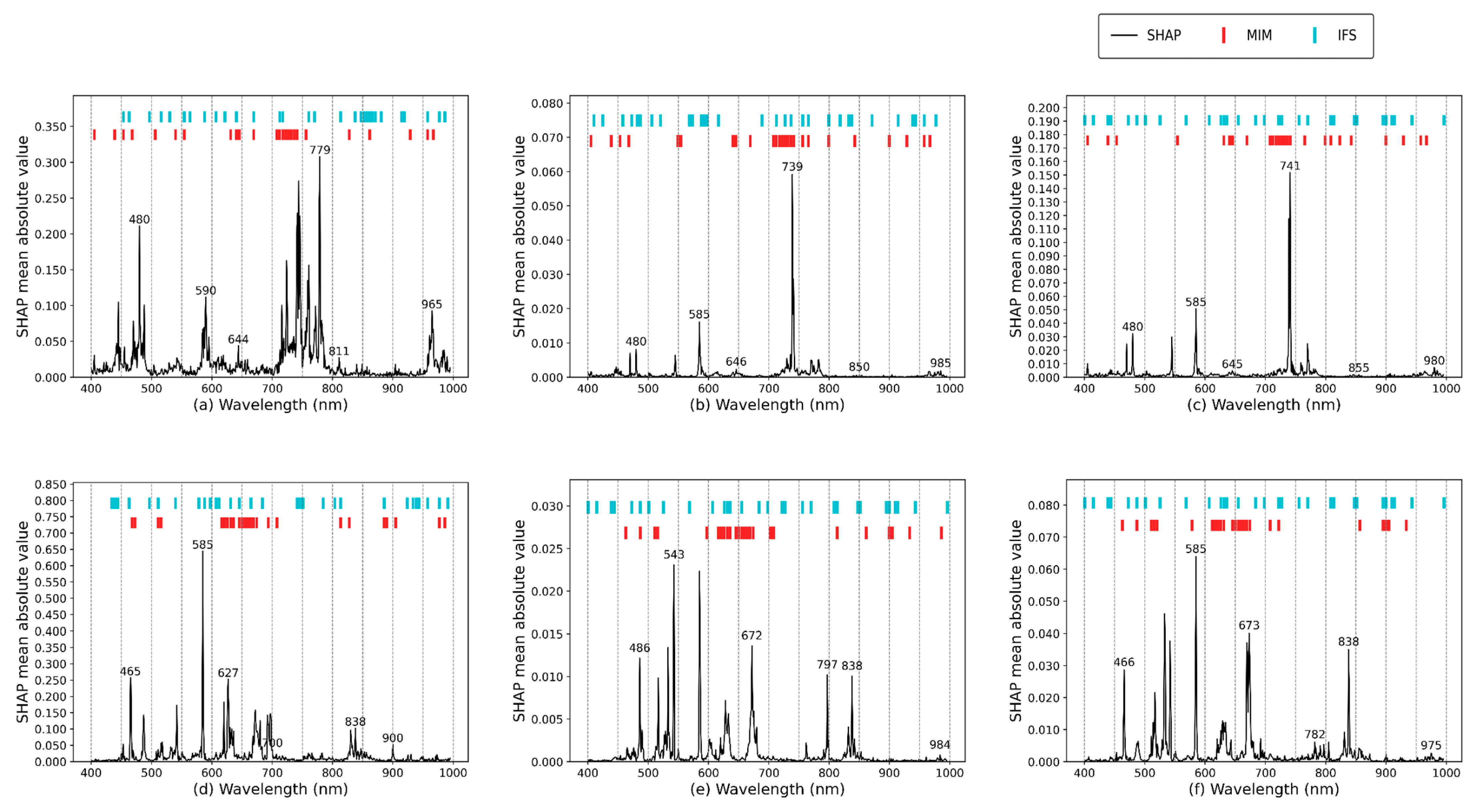
2.4.3. Feature Wavelength Selection
- MIM
- 2.
- IFS
- 3.
- SHAP value validation
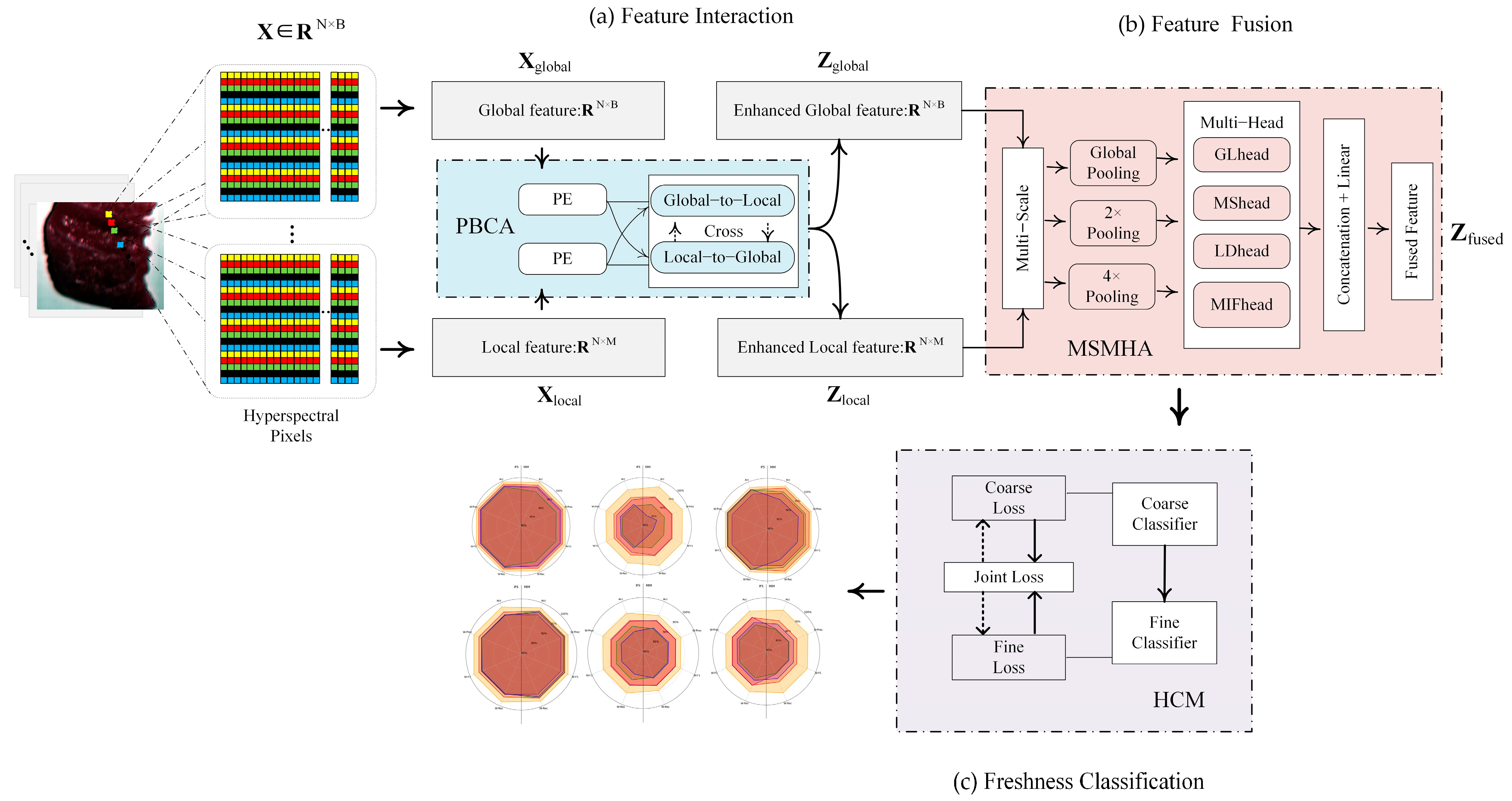
2.4.4. Dual-Branch Hierarchical Spectral Feature-Aware Network
- Feature interaction: Position-optimized bidirectional cross-attention (PBCA) module
- 2.
- Feature fusion: Multi-scale enhanced multi-head attention (MSMHA) module
- 3.
- Freshness classification: Hierarchical classification mechanism (HCM) module
2.4.5. Evaluation of Models
2.5. Implementation
3. Results and Discussion
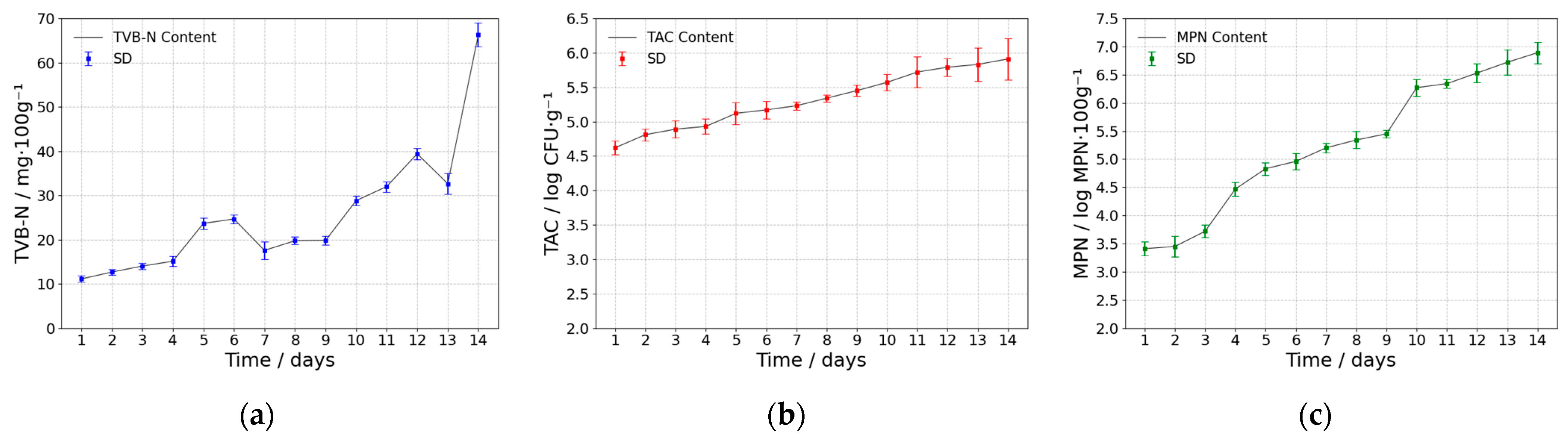
3.1. Analysis of Freshness Indicator Metrics
3.2. Data Preprocessing Results Analysis and Comparison
3.3. Analysis of Feature Selection
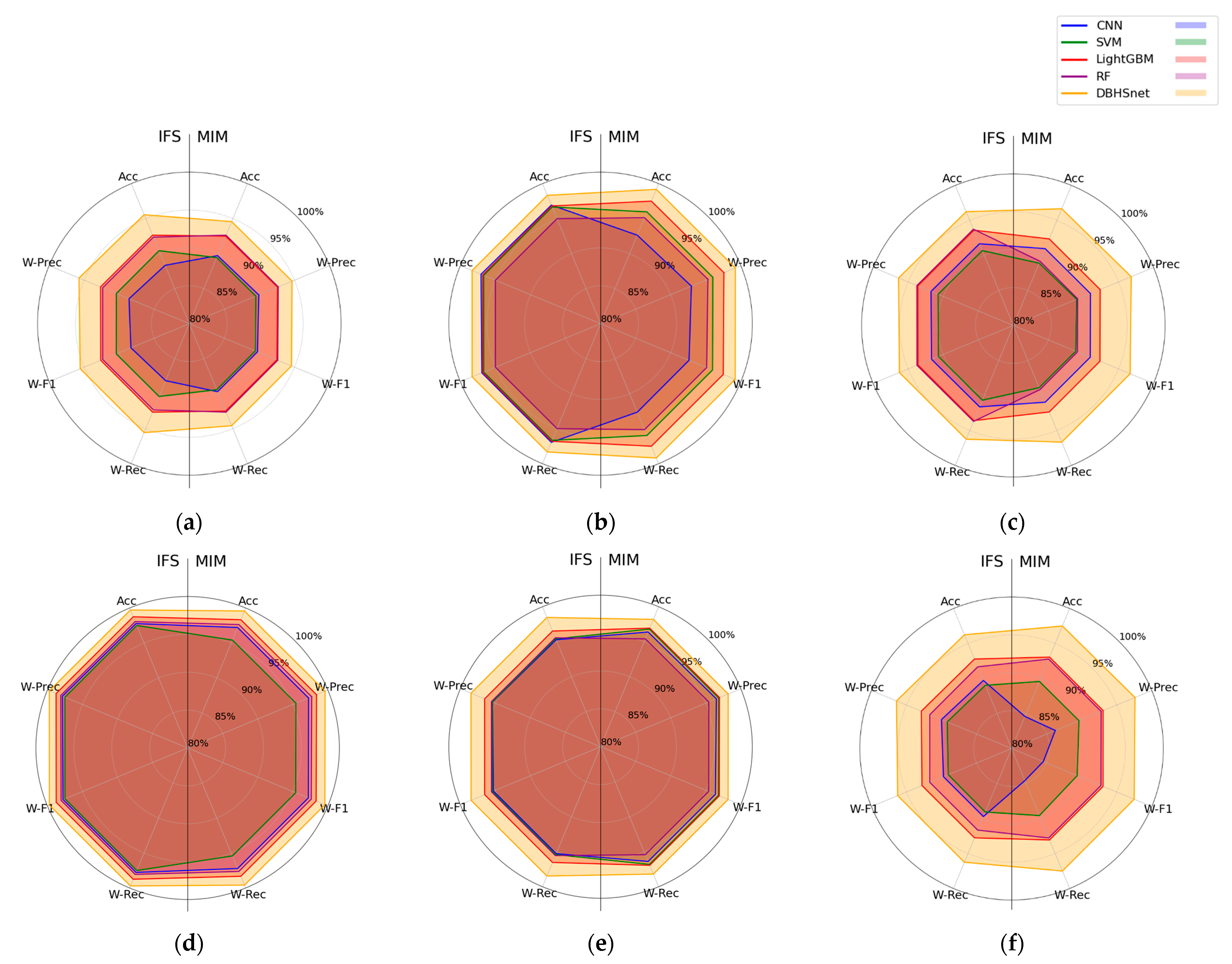
3.4. Analysis of Model Detection Results
3.4.1. Freshness Classification Results of Chilled Mutton
3.4.2. Ablation Study
3.4.3. Comparison of Classification Performance
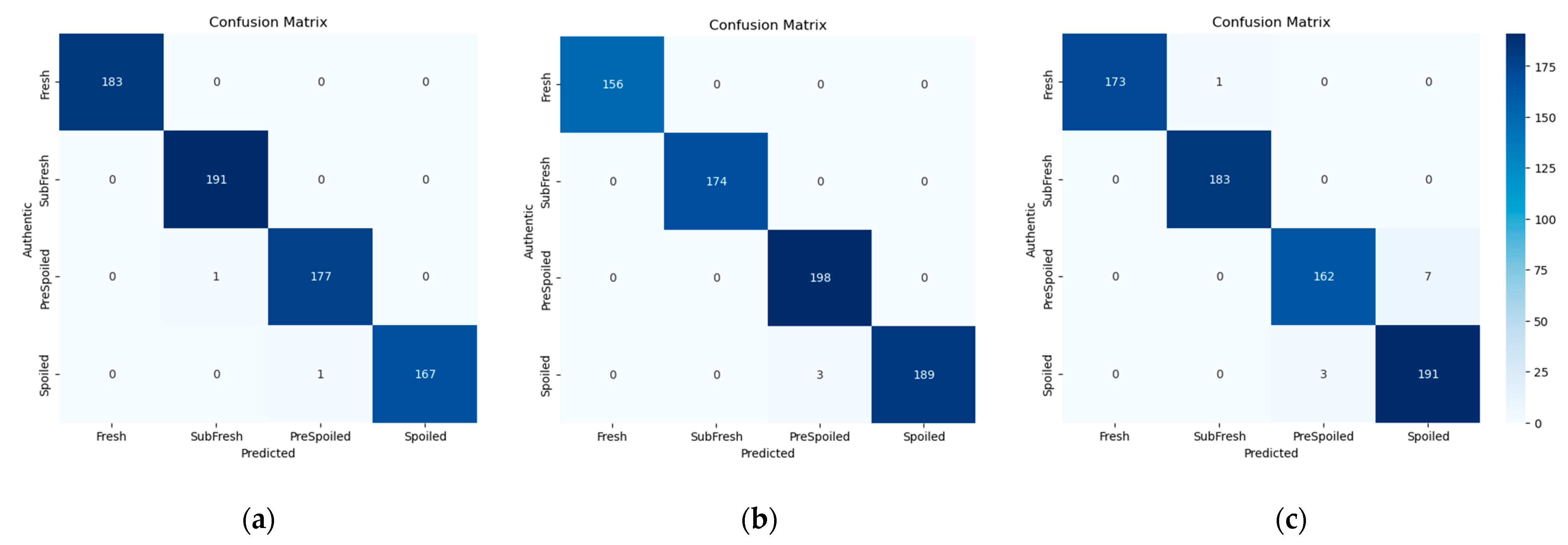
3.4.4. Confusion Matrix
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Junkuszew, A.; Nazar, P.; Milerski, M.; Margetin, M.; Brodzki, P.; Bazewicz, K. Chemical composition and fatty acid content in lamb and adult sheep meat. Arch. Anim. Breed. 2020, 63, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, S.; He, X.; Wu, L.; Guo, J. Combination of spectra and texture data of hyperspectral imaging for prediction and visualization of palmitic acid and oleic acid contents in lamb meat. Meat Sci. 2020, 169, 108194. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Liu, G.; He, J.; Luo, R.; Cheng, L.; Ma, C. Feature wavelength selection and model development for rapid determination of myoglobin content in nitrite-cured mutton using hyperspectral imaging. J. Food Eng. 2020, 287, 110090. [Google Scholar] [CrossRef]
- Jiang, X.; Xue, H.; Zhang, L.; Gao, X.; Wu, G.; Bai, J. Nondestructive detection of chilled mutton freshness based on multi-label information fusion and adaptive BP neural network. Comput. Electron. Agric. 2018, 155, 371–377. [Google Scholar] [CrossRef]
- Wei, J.; Qingyun, S.; Lin, S. Effect of irradiation treatment on the lipid composition and nutritional quality of goat meat. Food Chem. 2021, 351, 129295. [Google Scholar] [CrossRef]
- Kong, D.; Han, R.; Yuan, M.; Xi, Q.; Du, Q.; Li, P.; Yang, Y.; Rahman, S.M.E.; Wang, J. Slightly acidic electrolyzed water as a novel thawing media combined with ultrasound for improving thawed mutton quality, nutrients and microstructure. Food Chem. X 2023, 18, 100630. [Google Scholar] [CrossRef]
- Goudarzi, J.; Moshtaghi, H.; Shahbazi, Y.; Life, S. Kappa-carrageenan-poly (vinyl alcohol) electrospun fiber mats encapsulated with Prunus domestica anthocyanins and epigallocatechin gallate to monitor the freshness and enhance the shelf-life quality of minced beef meat. Food Packag. 2023, 35, 101017. [Google Scholar] [CrossRef]
- Cheng, L.; Liu, G.; He, J.; Wan, G.; Ban, J.; Yuan, R.; Fan, N. Development of a novel quantitative function between spectral value and metmyoglobin content in Tan mutton. Food Chem. 2021, 342, 128351. [Google Scholar] [CrossRef]
- Toomik, E.; Rood, L.; Bowman, J.P.; Kocharunchitt, C. Microbial spoilage mechanisms of vacuum-packed lamb meat: A review. Int. J. Food Microbiol. 2023, 387, 110056. [Google Scholar] [CrossRef]
- Zhao, H.; Feng, Y.; Chen, W.; Jia, G. Application of invasive weed optimization and least square support vector machine for prediction of beef adulteration with spoiled beef based on visible near-infrared (Vis-NIR) hyperspectral imaging. Meat Sci. 2019, 151, 75–81. [Google Scholar] [CrossRef]
- Zhou, Y.; Ai, Q.; Zhang, D. Changes in microflora on fresh mutton during chilled storage. Food Sci. 2015, 36, 242–245. [Google Scholar] [CrossRef]
- Burri, S.C.; Ekholm, A.; Bleive, U.; Püssa, T.; Jensen, M.; Hellström, J.; Mäkinen, S.; Korpinen, R.; Mattila, P.H.; Radenkovs, V. Lipid oxidation inhibition capacity of plant extracts and powders in a processed meat model system. Meat Sci. 2020, 162, 108033. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Gao, H.; Liu, Z.; Li, C.; Feng, X.; Chen, L. Ammonia/pH super-sensitive colorimetric labels based on gellan gum, sodium carboxymethyl cellulose, and dyes for monitoring freshness of lamb meat. Int. J. Biol. Macromol. 2024, 274, 133227. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Fan, S.; Liu, G.; He, J.; Wang, W.; Li, Y.; Cheng, L.; Ma, C.; Guo, M. Fusion of spectra and texture data of hyperspectral imaging for prediction of myoglobin content in nitrite-cured mutton. Food Control 2023, 144, 109332. [Google Scholar] [CrossRef]
- Samad, T.; Elliot, M.; Li, T.; Yves, B.Ń.; Yuqing, T.; Hui, H.; Yongkang, L. Novel intelligent films containing anthocyanin and phycocyanin for nondestructively tracing fish spoilage. Food Chem. 2023, 402, 134203. [Google Scholar] [CrossRef]
- Karambasti, P.R.; Shavisi, N. Development of guar gum-pectin nanofiber mats containing Papaver rhoeas petal anthocyanins and cellulose nanocrystals for real-time visual detection of lamb meat freshness. LWT 2024, 194, 115786. [Google Scholar] [CrossRef]
- Zou, Z.; Li, M.; Wang, Q.; Wu, Q.; Zhen, J.; Yuan, D.; Yin, S.; Zhou, M.; Cui, Q.; Xu, L. A non-destructive detection method of protein and TVB-N content changes in refrigerated and frozen-thawed salmon fillets using fluorescence hyperspectral technology. J. Food Compos. Anal. 2024, 133, 106435. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, W.; Zhang, H.; Liu, R.; Li, C. Discrimination among Fresh, Frozen–Stored and Frozen–Thawed Beef Cuts by Hyperspectral Imaging. Foods 2024, 13, 973. [Google Scholar] [CrossRef]
- Cheng, J.; Sun, J.; Shi, L.; Dai, C. An effective method fusing electronic nose and fluorescence hyperspectral imaging for the detection of pork freshness. Food Biosci. 2024, 59, 103880. [Google Scholar] [CrossRef]
- Zhuang, Q.; Peng, Y.; Nie, S.; Guo, Q.; Li, Y.; Zuo, J.; Chen, Y. Non-destructive detection of frozen pork freshness based on portable fluorescence spectroscopy. J. Food Compos. Anal. 2023, 118, 105175. [Google Scholar] [CrossRef]
- Yuan, R.; Liu, G.; He, J.; Ma, C.; Cheng, L.; Fan, N.; Ban, J.; Li, Y.; Sun, Y. Determination of metmyoglobin in cooked tan mutton using Vis/NIR hyperspectral imaging system. J. Food Sci. 2020, 85, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Dong, F.; Cui, J.; Hao, J.; Luo, R.; Wang, S.; Rodas-Gonzalez, A.; Liu, S. Fusion of spectral and textural data of hyperspectral imaging for glycine content prediction in beef using SFCN algorithms. Food Anal. Methods 2023, 16, 413–425. [Google Scholar] [CrossRef]
- Fan, N.; Ma, X.; Liu, G.; Ban, J.; Yuan, R.; Sun, Y. Rapid determination of TBARS content by hyperspectral imaging for evaluating lipid oxidation in mutton. J. Food Compos. Anal. 2021, 103, 104110. [Google Scholar] [CrossRef]
- He, W.; Huang, W.; Wang, Y.; Li, Z.; Blanka, T.; Zhang, X. A lamb freshness detection model using a flexible optoelectronic in-situ sensing system and multi-input multi-label causal ensemble learning. Food Chem. 2025, 471, 142803. [Google Scholar] [CrossRef]
- Kim, M.J.; Yu, W.H.; Song, D.J.; Chun, S.W.; Kim, M.S.; Lee, A.; Kim, G.; Shin, B.S.; Mo, C. Prediction of Soluble-Solid Content in Citrus Fruit Using Visible-Near-Infrared Hyperspectral Imaging Based on Effective-Wavelength Selection Algorithm. Sensors 2024, 24, 1512. [Google Scholar] [CrossRef]
- Huang, P.; Yuan, J.; Yang, P.; Xiao, F.; Zhao, Y. Nondestructive Detection of Sunflower Seed Vigor and Moisture Content Based on Hyperspectral Imaging and Chemometrics. Foods 2024, 13, 1320. [Google Scholar] [CrossRef]
- Binbin, F.; Rongguang, Z.; Dongyu, H.; Shichang, W.; Xiaomin, C.; Xuedong, Y. Evaluation of Mutton Adulteration under the Effect of Mutton Flavour Essence Using Hyperspectral Imaging Combined with Machine Learning and Sparrow Search Algorithm. Foods 2022, 11, 2278. [Google Scholar] [CrossRef]
- Zhong, L.; Guo, X.; Ding, M.; Ye, Y.; Jiang, Y.; Zhu, Q.; Li, J. SHAP values accurately explain the difference in modeling accuracy of convolution neural network between soil full-spectrum and feature-spectrum. Comput. Electron. Agric. 2024, 217, 108627. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, H.; Zhao, Y.; Shi, W.; Fu, H.; Zhao, Y.; Han, Z. A multi-verse optimizer-based CNN-BiLSTM pixel-level detection model for peanut aflatoxins. Food Chem. 2025, 463, 141393. [Google Scholar] [CrossRef]
- Cai, M.; Li, X.; Liang, J.; Liao, M.; Han, Y. An effective deep learning fusion method for predicting the TVB-N and TVC contents of chicken breasts using dual hyperspectral imaging systems. Food Chem. 2024, 456, 139847. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, Y.; Jiang, X.; Jin, S.; Li, S.; Xu, M. LGGFormer: A dual-branch local-guided global self-attention network for surface defect segmentation. Adv. Eng. Inform. 2025, 64, 103099. [Google Scholar] [CrossRef]
- Wan, B.; Wan, X.; Sun, Y.; Wang, T.; Lv, C.; Wang, S.; Yin, H.; Yan, C. ADNet: Anti-noise dual-branch network for road defect detection. Eng. Appl. Artif. Intell. 2024, 132, 107963. [Google Scholar] [CrossRef]
- Hao, X.; Wang, Z.; Zhang, Y.; Li, F.; Wang, M.; Li, J.; Mao, R. Detecting wheat yellow dwarf disease by employing a Dual-Branch multiscale model from UAV multispectral images. Comput. Electron. Agric. 2025, 230, 109898. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, F.; Su, H.; Hu, Y. Dual-branch multi-information aggregation network with transformer and convolution for polyp segmentation. Comput. Biol. Med. 2024, 168, 107760. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Yan, X.; Zhou, D.; Wang, C.; Zhang, W. IV-YOLO: A Lightweight Dual-Branch Object Detection Network. Sensors 2024, 24, 6181. [Google Scholar] [CrossRef]
- Sun, J.; Cheng, J.; Xu, M.; Yao, K. A method for freshness detection of pork using two-dimensional correlation spectroscopy images combined with dual-branch deep learning. J. Food Compos. Anal. 2024, 129, 106144. [Google Scholar] [CrossRef]
- Hou, Z.; Li, X.; Yang, C.; Ma, S.; Yu, W.; Wang, Y. Dual-branch network object detection algorithm based on dual-modality fusion of visible and infrared images. Multimed. Syst. 2024, 30, 333. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, X.; Qin, Y.; Yang, S.; Wen, X.; Cernava, T.; Migheli, Q.; Chen, X. Local and Global Feature-Aware Dual-Branch Networks for Plant Disease Recognition. Plant Phenomics 2024, 6, 0208. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Zhang, H.; Yang, S.; Lai, K.; Luan, D.; Yan, J. Development of a Novel Colorimetric pH Biosensor Based on A-Motif Structures for Rapid Food Freshness Monitoring and Spoilage Detection. Biosensors 2024, 14, 605. [Google Scholar] [CrossRef]
- GB 5009.228-2016; National Food Safety Standard: Determination of Volatile Basic Nitrogen in Food by Semi-micro Kjeldahl Method. National Health and Family Planning Commission of the People’s Republic of China: Beijing, China, 2016.
- GB 4789.2-2022; National Food Safety Standard: Microbiological Examination of Food—Determination of Total Bacterial Count. National Health Commission of the People’s Republic of China: Beijing, China, 2022.
- GB 4789.3-2016; National Food Safety Standard: Microbiological Examination of Food—Enumeration of Escherichia coli. National Health and Family Planning Commission of the People’s Republic of China: Beijing, China, 2016.
- Wentao, H.; Jie, X.; Xuepei, W.; Qinan, Z.; Mengjie, Z.; Xiaoshuan, Z. Improvement of non-destructive detection of lamb freshness based on dual-parameter flexible temperature-impedance sensor. Food Control 2023, 153, 109963. [Google Scholar] [CrossRef]
- Sooin, H.; HyeJin, K.; Seungah, L.; Jinwoo, C.; Aera, J.; Joonsung, B. Utilization of Electrical Impedance Spectroscopy and Image Classification for Non-Invasive Early Assessment of Meat Freshness. Sensors 2021, 21, 1001. [Google Scholar] [CrossRef] [PubMed]
- Qibin, Z.; Yankun, P.; Deyong, Y.; Yali, W.; Renhong, Z.; Kuanglin, C.; Qinghui, G. Detection of frozen pork freshness by fluorescence hyperspectral image. J. Food Eng. 2022, 316, 110840. [Google Scholar] [CrossRef]
- Jingjing, Z.; Guishan, L.; Yan, L.; Mei, G.; Fangning, P.; Han, W. Rapid identification of lamb freshness grades using visible and near-infrared spectroscopy (Vis-NIR). J. Food Compos. Anal. 2022, 111, 104590. [Google Scholar] [CrossRef]
- Huan, Y.; Seyoung, K.; Zhu, B.; Yingjun, K.; Ruilong, S.; Ting, D.; Lintao, Z.; Seung, K.J. Real-Time Fluorescence Screening Platform for Meat Freshness. Anal. Chem. 2022, 94, 15423–15432. [Google Scholar] [CrossRef]
- GB 2707-2016; National Food Safety Standard: Fresh (Frozen) Livestock and Poultry Products. China Food and Drug Administration: Beijing, China, 2016.
- Sandra, A.; Margarida, L.M. Cross-Classification Analysis of Food Products Based on Nutritional Quality and Degree of Processing. Nutrients 2023, 15, 3117. [Google Scholar] [CrossRef]
- Wang, T.; Chen, Y.; Huang, Y.; Zheng, C.; Liao, S.; Xiao, L.; Zhao, J. Prediction of the Quality of Anxi Tieguanyin Based on Hyperspectral Detection Technology. Foods 2024, 13, 4126. [Google Scholar] [CrossRef]
- Pu, H.; Xie, A.; Sun, D.-W.; Kamruzzaman, M.; Ma, J. Application of Wavelet Analysis to Spectral Data for Categorization of Lamb Muscles. Food Bioprocess. Technol. 2015, 8, 1–16. [Google Scholar] [CrossRef]
- Chang, X.; Liu, T.; Shi, B.; Zhang, G.; Xu, Y.; Zhang, J.; Zhang, P. An improved algorithm with particle swarm optimization-extreme gradient boosting to predict the contents of pyrolytic hydrocarbons in source rocks. J. Asian Earth Sci. 2024, 276, 106367. [Google Scholar] [CrossRef]
- Yu, F.; Guan, J.; Wu, H.; Wang, H.; Ma, B. Multi-population differential evolution approach for feature selection with mutual information ranking. Expert. Syst. Appl. 2025, 260, 125404. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Zheng, L.; Qin, Q.; Lee, W.S. Spectral features extraction for estimation of soil total nitrogen content based on modified ant colony optimization algorithm. Geoderma 2019, 333, 23–34. [Google Scholar] [CrossRef]
- Lu, Z.; Min, L.; Xinyi, Q.; Guangzhong, L. Succinylation Site Prediction Based on Protein Sequences Using the IFS-LightGBM (BO) Model. Comput. Math. Methods Med. 2020, 2020, 8858489. [Google Scholar] [CrossRef]
- Liang, Z.; Xi, G.; Zhe, X.; Meng, D. Soil properties: Their prediction and feature extraction from the LUCAS spectral library using deep convolutional neural networks. Geoderma 2021, 402, 115366. [Google Scholar] [CrossRef]
- Hamid, G.; Aliakbar, M.; Shahram, G.; Yougui, S.; Biswajeet, P. Interpretability of simple RNN and GRU deep learning models used to map land susceptibility to gully erosion. Sci. Total Environ. 2023, 904, 166960. [Google Scholar] [CrossRef]
- Alaiz-Rodriguez, R.; Parnell, A.C. A Machine Learning Approach for Lamb Meat Quality Assessment Using FTIR Spectra. IEEE Access 2020, 8, 52385–52394. [Google Scholar] [CrossRef]
- Pravin, A.; Deepa, C. Aggregate Plant Species Categorization with Deep CNN Based Positional Encoded Cross Attentional Mechanism. SN Comput. Sci. 2024, 5, 912. [Google Scholar] [CrossRef]
- Chen, M.; Li, H.; Peng, H.; Xiong, X.; Long, N. HPCDNet: Hybrid position coding and dual-frquency domain transform network for low-light image enhancement. Math. Biosci. Eng. MBE 2024, 21, 1917–1937. [Google Scholar] [CrossRef]
- Madani, M.; Behzadi, M.M.; Song, D.; Ilies, H.T.; Tarakanova, A. Improved inter-residue contact prediction via a hybrid generative model and dynamic loss function. Comput. Struct. Biotechnol. J. 2022, 20, 6138–6148. [Google Scholar] [CrossRef]
- Siyuan, K.; Qinglun, Z.; Hongru, W.; Yan, S. An efficient multiscale integrated attention method combined with hyperspectral system to identify the quality of rice with different storage periods and humidity. Comput. Electron. Agric. 2023, 213, 108259. [Google Scholar] [CrossRef]
- Harshula, T.; Biplab, B.; Mohan, B.K. Multi-head attention with CNN and wavelet for classification of hyperspectral image. Neural Comput. Appl. 2022, 35, 7595–7609. [Google Scholar] [CrossRef]
- Huaifei, S.; Yinghao, L.; Qingjiu, T.; Kaijian, X.; Junnan, J. A comparison of multiple classifier combinations using different voting-weights for remote sensing image classification. Int. J. Remote Sens. 2018, 39, 3705–3722. [Google Scholar] [CrossRef]
- Houman, K.; Mobin, G.N.; Abbas, S.; Mohammad, K.; Mokhtar, M. Feature Selection and Training Multilayer Perceptron Neural Networks Using Grasshopper Optimization Algorithm for Design Optimal Classifier of Big Data Sonar. J. Sens. 2022, 2022, 9620555. [Google Scholar] [CrossRef]
- de Abreu, D.P.; Losada, P.P.; Maroto, J.; Cruz, J.M. Lipid damage during frozen storage of Atlantic halibut (Hippoglossus hippoglossus) in active packaging film containing antioxidants. Food Chem. 2011, 126, 315–320. [Google Scholar] [CrossRef]
- Zhang, Z.; Chang, Z.; Huang, J.; Leng, G.; Xu, W.; Wang, Y.; Xie, Z.; Yang, J. Enhancing soil texture classification with multivariate scattering correction and residual neural networks using visible near-infrared spectra. J. Environ. Manag. 2024, 352, 120094. [Google Scholar] [CrossRef] [PubMed]
- Kucha, C.T.; Ngadi, M.O. Rapid assessment of pork freshness using miniaturized NIR spectroscopy. J. Food Meas. Charact. 2020, 14, 1105–1115. [Google Scholar] [CrossRef]
- Hasan, M.M.; Chaudhry, M.M.A.; Erkinbaev, C.; Paliwal, J.; Suman, S.P.; Rodas-Gonzalez, A. Application of Vis-NIR and SWIR spectroscopy for the segregation of bison muscles based on their color stability. Meat Sci. 2022, 188, 108774. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, M.; Zhang, F.; Xiong, Y.; Wang, X.; Ali, S.; Zhang, Y.; Fu, S. Hyperspectral imaging combined with GA-SVM for maize variety identification. Food Sci. Nutr. 2024, 12, 3177–3187. [Google Scholar] [CrossRef]
- Mahsa, H.; Abbas, M.; Reza, G. A Novel Scheme for Mapping of MVT-Type Pb–Zn Prospectivity: LightGBM, a Highly Efficient Gradient Boosting Decision Tree Machine Learning Algorithm. Nat. Resour. Res. 2023, 32, 2417–2438. [Google Scholar] [CrossRef]

| TVB-N/mg·100 g−1 | TAC/log CFU·g−1 | MPN/log MPN·100 g−1 | Label |
|---|---|---|---|
| (0, 15] | ≤6.0 | ≤5.0 | Fresh |
| (15, 25] | ≤6.0 | ≤5.0 | SubFresh |
| (15, 25] | ≤6.0 | >5.0 | PreSpoiled |
| >25 | >6.0 | >5.0 | Spoiled |
| Preprocessing Methods | Metric | MIM | IFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R² | RMSE | MAE | RPD | R² | RMSE | MAE | RPD | ||
| RAW | TVB-N | 0.6972 | 0.5621 | 0.3862 | 1.8171 | 0.5548 | 0.6815 | 0.4273 | 1.4988 |
| TAC | 0.7098 | 0.5538 | 0.3502 | 1.8563 | 0.7291 | 0.535 | 0.3358 | 1.9214 | |
| MPN | 0.7599 | 0.4974 | 0.3318 | 2.0409 | 0.8110 | 0.4413 | 0.2939 | 2.3004 | |
| MSC | TVB-N | 0.8399 | 0.4087 | 0.2948 | 2.4992 | 0.7939 | 0.4637 | 0.3066 | 2.2028 |
| TAC | 0.8891 | 0.3424 | 0.2353 | 3.0028 | 0.9019 | 0.3219 | 0.2214 | 3.1932 | |
| MPN | 0.8852 | 0.3439 | 0.2377 | 2.9520 | 0.8908 | 0.3355 | 0.2181 | 3.0257 | |
| MA | TVB-N | 0.5978 | 0.6477 | 0.4312 | 1.5768 | 0.6633 | 0.5926 | 0.3956 | 1.7233 |
| TAC | 0.7125 | 0.5512 | 0.3688 | 1.8650 | 0.7311 | 0.5331 | 0.3385 | 1.9283 | |
| MPN | 0.7697 | 0.4872 | 0.3297 | 2.0838 | 0.8114 | 0.4409 | 0.2944 | 2.3029 | |
| SNV | TVB-N | 0.8177 | 0.4361 | 0.3070 | 2.3421 | 0.8414 | 0.4067 | 0.2867 | 2.5110 |
| TAC | 0.8888 | 0.3428 | 0.2365 | 2.9985 | 0.8917 | 0.3383 | 0.2333 | 3.0389 | |
| MPN | 0.9001 | 0.3209 | 0.2204 | 3.1638 | 0.8900 | 0.3367 | 0.2188 | 3.0154 | |
| FD | TVB-N | 0.7586 | 0.5018 | 0.3239 | 2.0355 | 0.8757 | 0.3601 | 0.2563 | 2.8364 |
| TAC | 0.8336 | 0.4194 | 0.2459 | 2.4512 | 0.9261 | 0.2795 | 0.2114 | 3.6785 | |
| MPN | 0.9024 | 0.3172 | 0.2038 | 3.2009 | 0.9387 | 0.2514 | 0.1847 | 4.0378 | |
| SG | TVB-N | 0.5957 | 0.6494 | 0.4282 | 1.5727 | 0.6656 | 0.5906 | 0.3955 | 1.7293 |
| TAC | 0.7479 | 0.5161 | 0.3475 | 1.9917 | 0.6845 | 0.5774 | 0.3552 | 1.7804 | |
| MPN | 0.7584 | 0.4990 | 0.3479 | 2.0346 | 0.8121 | 0.4401 | 0.2931 | 2.3069 | |
| Model | Preprocessing | MIM | IFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy /% | Weighted Precision /% | Weighted Recall /% | Weighted F1 Score /% | Accuracy /% | Weighted Precision /% | Weighted Recall /% | Weighted F1 Score /% | ||
| CNN | Raw | 89.72 | 89.91 | 89.72 | 89.72 | 88.33 | 88.60 | 88.33 | 88.15 |
| MSC | 92.64 | 92.95 | 92.64 | 92.62 | 96.94 | 97.05 | 96.94 | 96.95 | |
| FD | 97.22 | 97.25 | 97.22 | 97.22 | 97.78 | 97.87 | 97.78 | 97.78 | |
| SG | 90.97 | 91.00 | 90.97 | 90.97 | 91.67 | 91.74 | 91.67 | 91.63 | |
| SNV | 96.39 | 96.52 | 96.39 | 96.39 | 95.28 | 95.42 | 95.28 | 95.27 | |
| MA | 89.91 | 89.27 | 89.58 | 89.44 | 89.72 | 90.00 | 89.72 | 89.72 | |
| SVM | Raw | 89.44 | 89.49 | 89.44 | 89.42 | 90.42 | 90.41 | 90.42 | 90.40 |
| MSC | 95.97 | 96.01 | 95.97 | 95.97 | 96.67 | 96.67 | 96.67 | 96.67 | |
| FD | 95.42 | 95.45 | 95.42 | 95.42 | 95.50 | 97.51 | 97.50 | 97.50 | |
| SG | 88.89 | 89.03 | 88.89 | 88.90 | 90.69 | 90.74 | 90.69 | 90.68 | |
| SNV | 96.81 | 96.83 | 96.81 | 96.81 | 95.42 | 95.44 | 95.42 | 95.41 | |
| MA | 89.58 | 89.61 | 89.58 | 89.57 | 89.03 | 89.18 | 89.03 | 89.07 | |
| LightGBM | Raw | 92.50 | 92.59 | 92.50 | 92.49 | 92.64 | 92.67 | 92.64 | 92.64 |
| MSC | 97.50 | 97.59 | 97.50 | 97.50 | 96.81 | 96.81 | 96.81 | 96.80 | |
| FD | 98.33 | 98.39 | 98.33 | 98.33 | 98.75 | 98.77 | 98.75 | 98.75 | |
| SG | 92.36 | 92.38 | 92.36 | 92.35 | 93.61 | 93.63 | 93.61 | 93.60 | |
| SNV | 96.94 | 96.97 | 96.94 | 96.94 | 96.53 | 96.53 | 96.53 | 96.53 | |
| MA | 93.06 | 93.06 | 93.06 | 93.05 | 92.78 | 92.88 | 92.78 | 92.76 | |
| RF | Raw | 92.64 | 92.69 | 92.64 | 92.62 | 92.36 | 92.36 | 92.36 | 92.33 |
| MSC | 95.14 | 95.31 | 95.14 | 95.13 | 95.00 | 95.00 | 95.00 | 94.99 | |
| FD | 97.64 | 97.70 | 97.64 | 97.64 | 98.06 | 98.11 | 98.06 | 98.06 | |
| SG | 89.17 | 89.17 | 89.17 | 89.14 | 93.75 | 93.76 | 93.75 | 93.71 | |
| SNV | 95.42 | 95.44 | 95.42 | 95.42 | 95.56 | 95.57 | 95.56 | 95.55 | |
| MA | 92.78 | 92.78 | 92.78 | 92.75 | 91.67 | 91.67 | 91.67 | 91.66 | |
| DBHSNet | Raw | 94.58 | 94.66 | 94.58 | 94.58 | 95.56 | 95.74 | 95.56 | 95.57 |
| MSC | 98.48 | 98.48 | 98.47 | 98.47 | 98.06 | 98.09 | 98.06 | 98.06 | |
| FD | 99.58 | 99.61 | 99.59 | 99.58 | 99.72 | 99.73 | 99.72 | 99.71 | |
| SG | 96.67 | 96.81 | 96.67 | 96.66 | 96.25 | 96.38 | 96.25 | 96.25 | |
| SNV | 98.19 | 98.20 | 98.19 | 98.19 | 98.33 | 98.34 | 98.33 | 98.33 | |
| MA | 97.50 | 97.60 | 97.50 | 97.49 | 96.25 | 96.42 | 96.25 | 96.25 | |
| Local data | PBCA | MSMHA | HCM | Accuracy /% | Weighted Precision /% | Weighted Recall /% | Weighted F1 Score /% |
|---|---|---|---|---|---|---|---|
| FD-MIM | ✗ | ✗ | ✗ | 86.53 | 86.81 | 86.53 | 86.44 |
| ✗ | ✗ | ✓ | 89.86 | 89.92 | 89.86 | 89.87 | |
| ✗ | ✓ | ✓ | 91.67 | 91.65 | 91.67 | 91.66 | |
| ✓ | ✗ | ✓ | 93.75 | 93.75 | 93.75 | 93.75 | |
| ✓ | ✓ | ✗ | 96.53 | 96.57 | 96.52 | 96.55 | |
| ✓ | ✓ | ✓ | 99.58 | 99.61 | 99.59 | 99.58 | |
| FD-IFS | ✗ | ✗ | ✗ | 87.92 | 88.06 | 87.92 | 87.91 |
| ✗ | ✗ | ✓ | 90.28 | 90.41 | 90.28 | 90.26 | |
| ✗ | ✓ | ✓ | 93.75 | 93.76 | 93.75 | 93.74 | |
| ✓ | ✗ | ✓ | 95.28 | 95.3 | 95.28 | 95.27 | |
| ✓ | ✓ | ✗ | 97.22 | 97.22 | 97.25 | 97.23 | |
| ✓ | ✓ | ✓ | 99.72 | 99.73 | 99.72 | 99.71 | |
| MSC-MIM | ✗ | ✗ | ✗ | 86.39 | 86.39 | 86.62 | 86.36 |
| ✗ | ✗ | ✓ | 89.31 | 89.28 | 89.31 | 89.20 | |
| ✗ | ✓ | ✓ | 92.92 | 92.91 | 92.92 | 92.91 | |
| ✓ | ✗ | ✓ | 94.17 | 94.30 | 94.17 | 94.18 | |
| ✓ | ✓ | ✗ | 96.25 | 96.26 | 96.25 | 96.25 | |
| ✓ | ✓ | ✓ | 98.48 | 98.48 | 98.47 | 98.47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
E, J.; Zhai, C.; Jiang, X.; Xu, Z.; Wudan, M.; Li, D. Non-Destructive Detection of Chilled Mutton Freshness Using a Dual-Branch Hierarchical Spectral Feature-Aware Network. Foods 2025, 14, 1379. https://doi.org/10.3390/foods14081379
E J, Zhai C, Jiang X, Xu Z, Wudan M, Li D. Non-Destructive Detection of Chilled Mutton Freshness Using a Dual-Branch Hierarchical Spectral Feature-Aware Network. Foods. 2025; 14(8):1379. https://doi.org/10.3390/foods14081379
Chicago/Turabian StyleE, Jixiang, Chengjun Zhai, Xinhua Jiang, Ziyang Xu, Muqiu Wudan, and Danyang Li. 2025. "Non-Destructive Detection of Chilled Mutton Freshness Using a Dual-Branch Hierarchical Spectral Feature-Aware Network" Foods 14, no. 8: 1379. https://doi.org/10.3390/foods14081379
APA StyleE, J., Zhai, C., Jiang, X., Xu, Z., Wudan, M., & Li, D. (2025). Non-Destructive Detection of Chilled Mutton Freshness Using a Dual-Branch Hierarchical Spectral Feature-Aware Network. Foods, 14(8), 1379. https://doi.org/10.3390/foods14081379






