Transcriptomic Identification of Core Regulatory Genes for Higher Alcohol Production in Saccharomyces cerevisiae at Different Sugar Concentrations in Wine Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Fermentation Conditions
2.3. Determination of the Basic Physicochemical Parameters
2.4. Transcriptome Sequencing
2.5. Transcriptome Analysis
2.6. Plasmid Construction and Recombinant Strain Screening
2.7. Statistical Analysis
3. Results
3.1. Fermentation Performance at Three Sugar Concentrations
3.2. Effect of Sugar Concentration on the Higher Alcohol Production by S. cerevisiae
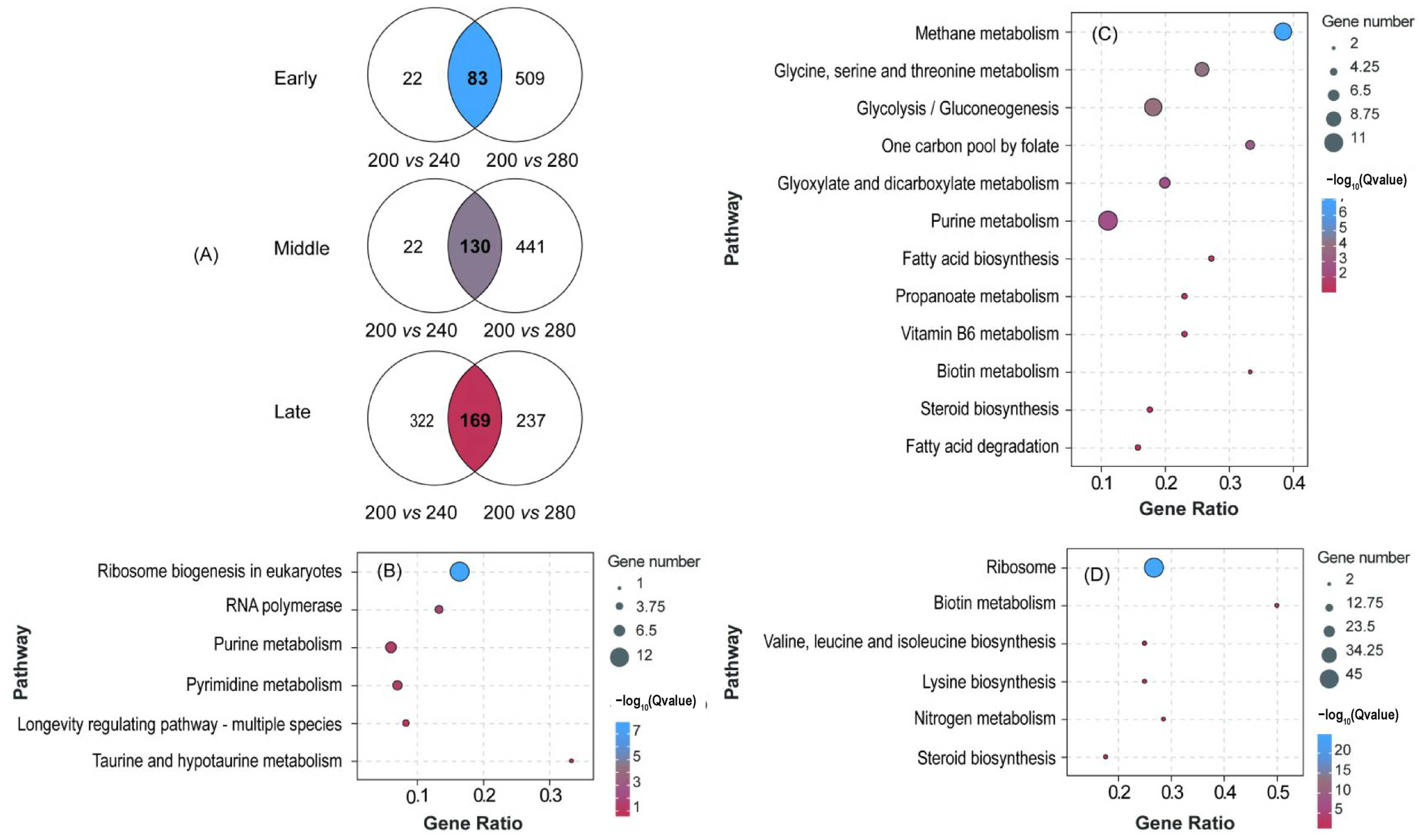
3.3. Transcriptional Response to Sugar Concentration
3.4. Genes Related to Higher Alcohol Metabolism
3.5. Venn Diagram Identification of Key Gene
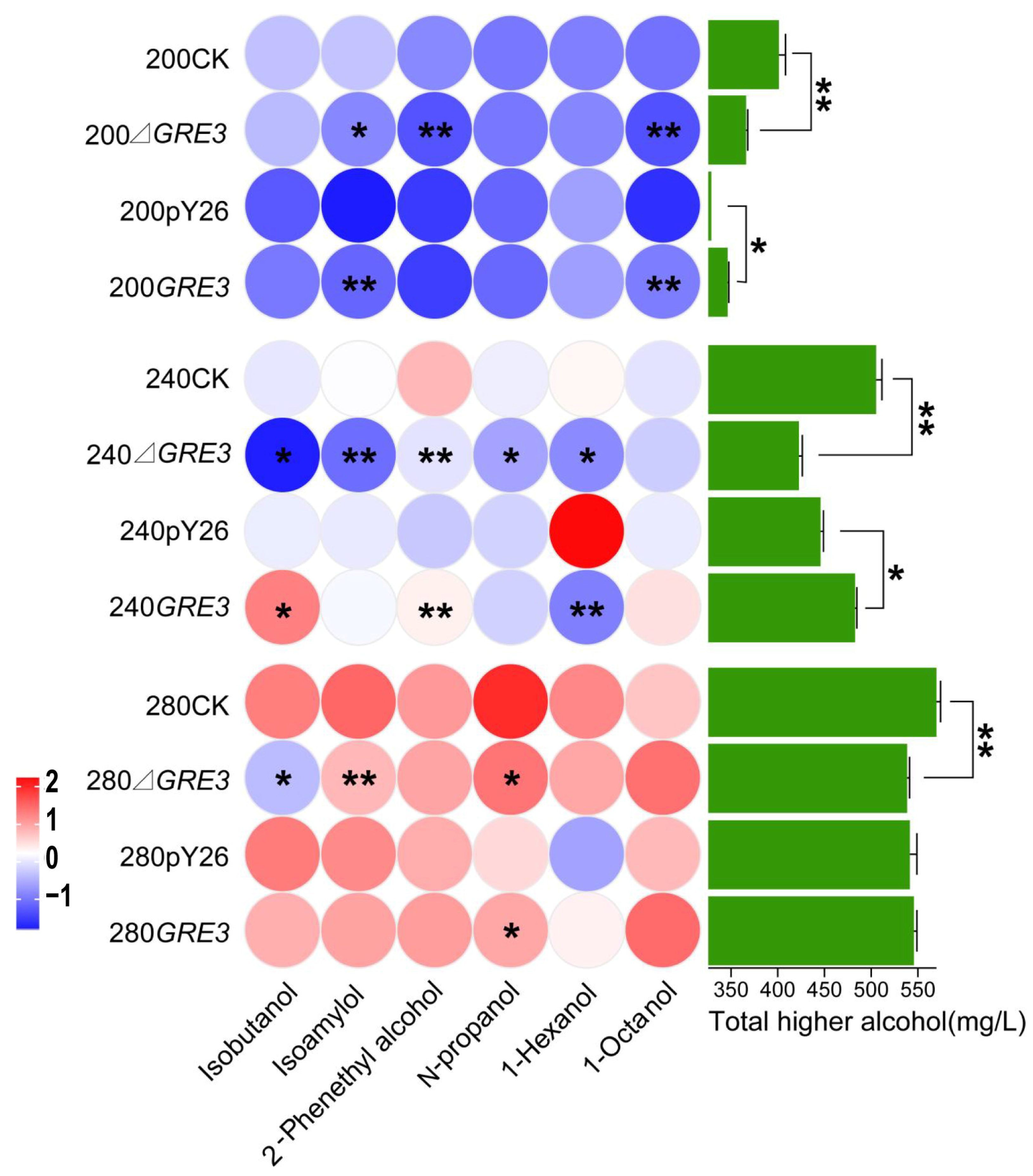
3.6. Production of Higher Alcohols in GRE3 Recombinant Strains
4. Discussion
4.1. Effects of High Sugar on Higher Alcohols and Other Metabolites of S. cerevisiae
4.2. Transcriptome Explanation of the Effect of High Sugar on Production of Higher Alcohols by S. cerevisiae
4.3. GRE3 Is Involved in the Production of Higher Alcohols
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Behringer, K.I.; Kapeluch, J.; Fischer, A.; Hellwig, M. Metabolization of free oxidized aromatic amino acids by Saccharomyces cerevisiae. J. Agric. Food Chem. 2024, 72, 5766–5776. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, M.; Meng, N.; Li, H.; Sun, J.; Sun, B. Influence of nitrogen status on fermentation performances of non-Saccharomyces yeasts: A review. Food Sci. Hum. Wellness 2024, 13, 556–567. [Google Scholar] [CrossRef]
- Roberts, R.; Khomenko, I.; Eyres, G.T.; Bremer, P.; Silcock, P.; Betta, E.; Biasioli, F. Online monitoring of higher alcohols and esters throughout beer fermentation by commercial Saccharomyces cerevisiae and Saccharomyces pastorianus yeast. J. Mass. Spectrom. 2023, 58, e4959. [Google Scholar] [CrossRef]
- Mandlaa; Ren, Y.; Qiao, M.; Yang, Y.; Ren, G.; Chen, Z.; Sun, Z. Profiling the potential producers of higher alcohol in different Daqu (a starter of Baijiu) by amplifying the key enzyme gene. Cogent Food Agric. 2024, 10, 2306014. [Google Scholar] [CrossRef]
- Wang, X.; Dang, C.; Liu, Y.; Ge, X.; Suo, R.; Ma, Q.; Wang, J. Effect of indigenous Saccharomyces cerevisiae strains on microbial community successions and volatile compounds changes during Longyan wine fermentation. Food Biosci. 2024, 57, 103595. [Google Scholar] [CrossRef]
- Prusova, B.; Humaj, J.; Sochor, J.; Baron, M. Formation, losses, preservation and recovery of aroma compounds in the winemaking process. Fermentation 2022, 8, 93. [Google Scholar] [CrossRef]
- Cordente, A.G.; Espinase Nandorfy, D.; Solomon, M.; Schulkin, A.; Kolouchova, R.; Francis, I.L.; Schmidt, S.A. Aromatic higher alcohols in wine: Implication on aroma and palate attributes during chardonnay aging. Molecules 2021, 26, 4979. [Google Scholar] [CrossRef]
- Huang, D.; Zhong, Y.; Liu, Y.L.; Song, Y.Y.; Zhao, X.X.; Qin, Y. Reducing higher alcohols by integrating indigenous Saccharomyces cerevisiae, nitrogen compensation, and chaptalization methods during fermentation of kiwifruit wine. LWT 2023, 184, 115059. [Google Scholar] [CrossRef]
- Kokkinakis, M.; Tsakiris, I.; Tzatzarakis, M.; Vakonaki, E.; Alegakis, A.; Papachristou, S.; Karzi, V.; Kokkinaki, A.; Goumenou, M.; Kallionakis, M.; et al. Carcinogenic, ethanol, acetaldehyde and noncarcinogenic higher alcohols, esters, and methanol compounds found in traditional alcoholic beverages. A risk assessment approach. Toxicol. Rep. 2020, 7, 1057–1065. [Google Scholar] [CrossRef]
- Coulibaly, W.H.; Bouatenin, K.M.J.P.; Boli, Z.B.I.A.; Camara, F.; Sanogo, Y.M.; Akissi, D.M.; Kouame, H.K.; Rigou, P.; Djameh, C.; Djè, K.M. Volatile compounds of traditional sorghum beer (tchapalo) produced in Côte d’Ivoire, comparison between wild yeasts and pure culture of Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2021, 37, 75. [Google Scholar] [CrossRef]
- Wang, Y.P.; Sun, Z.G.; Zhang, C.Y.; Zhang, Q.Z.; Guo, X.W.; Xiao, D.G. Comparative transcriptome analysis reveals the key regulatory genes for higher alcohol formation by yeast at different alpha-amino nitrogen concentrations. Food Microbiol. 2021, 95, 103713. [Google Scholar] [CrossRef] [PubMed]
- Massera, A.; Assof, M.; Sari, S.; Ciklic, I.; Mercado, L.; Jofré, V.; Combina, M. Effect of low temperature fermentation on the yeast-derived volatile aroma composition and sensory profile in Merlot wines. LWT 2021, 142, 111069. [Google Scholar] [CrossRef]
- Ntuli, R.G.; Saltman, Y.; Ponangi, R.; Jeffery, D.W.; Bindon, K.; Wilkinson, K.L. Impact of fermentation temperature and grape solids content on the chemical composition and sensory profiles of Cabernet Sauvignon wines made from flash détente treated must fermented off-skins. Food Chem. 2022, 369, 130861. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, L.; Wang, Y.; Wang, X.; Xiao, D. Higher alcohols metabolism by Saccharomyces cerevisiae: A mini review. Chin. J. Biotechnol. 2021, 37, 429–447. [Google Scholar]
- Choi, Y.J.; Lee, J.; Jang, Y.S.; Lee, S.Y. Metabolic engineering of microorganisms for the production of higher alcohols. MBio 2014, 5, 10–1128. [Google Scholar] [CrossRef]
- Hranilovic, A.; Gambetta, J.M.; Jeffery, D.W.; Grbin, P.R.; Jiranek, V. Lower-alcohol wines produced by Metschnikowia pulcherrima and Saccharomyces cerevisiae co-fermentations: The effect of sequential inoculation timing. Int. J. Food Microbiol. 2020, 329, 108651. [Google Scholar] [CrossRef]
- Mateo, J.J.; Jimenez, M.; Pastor, A.; Huerta, T. Influence of the inoculation time of high sugar content must on the formation of wine aroma. World J. Microbiol. Biotechnol. 1998, 14, 357–363. [Google Scholar] [CrossRef]
- Marullo, P.; Trujillo, M.; Viannais, R.; Hercman, L.; Guillaumie, S.; Colonna-Ceccaldi, B.; Albertin, W.; Barbe, J.C. Metabolic, organoleptic and transcriptomic impact of Saccharomyces cerevisiae genes involved in the biosynthesis of linear and substituted esters. Int. J. Mol. Sci. 2021, 22, 4026. [Google Scholar] [CrossRef]
- Jin, D.F.; Gu, B.T.; Xiong, D.E.; Huang, G.C.; Huang, X.P.; Liu, L.; Xiao, J.A. Transcriptomic analysis of Saccharomyces cerevisiae under the stress of 2-phenylethanol. Curr. Microbiol. 2018, 75, 1068–1076. [Google Scholar] [CrossRef]
- Ma, D.N.; Yuan, L.; Mao, J.Q.; Liu, T.T.; Zhao, Y.Z.; Han, X.; Ji, Z.W.; Liu, S.P.; Mao, J. Optimizing Huangjiu fermentation for enhanced aroma: Insights into Saccharomyces cerevisiae jiangnan1# strain. J. Food Compos. Anal. 2025, 139, 107051. [Google Scholar]
- Drozdova, P.; Gurkov, A.; Saranchina, A.; Vlasevskaya, A.; Zolotovskaya, E.; Indosova, E.; Timofeyev, M.; Borvinskaya, E. Transcriptional response of Saccharomyces cerevisiae to lactic acid enantiomers. Appl. Microbiol. Biotechnol. 2024, 108, 121. [Google Scholar] [CrossRef] [PubMed]
- Spiropoulos, A.; Tanaka, J.; Flerianos, I.; Bisson, L.F. Characterization of hydrogen sulfide formation in commercial and natural wine isolates of Saccharomyces. Am. J. Enol. Vitic. 2000, 51, 233–248. [Google Scholar] [CrossRef]
- Modesti, M.; Brizzolara, S.; Forniti, R.; Ceccantoni, B.; Bellincontro, A.; Catelli, C.; Mencarelli, F.; Tonutti, P. Postharvest ozone fumigation of grapes (cv Sangiovese) differently affects volatile organic compounds and polyphenol profiles of berries and wine. Aust. J. Grape Wine Res. 2023, 2023, 8244309. [Google Scholar] [CrossRef]
- Moreno-Beltrán, M.; Gore-Lloyd, D.; Chuck, C.; Henk, D. Variation among Metschnikowia pulcherrima Isolates for genetic modification and homologous recombination. Microorganisms 2021, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- de Andrade Silva, C.A.; Oka, M.L.; da Silva, P.G.P.; Honma, J.M.; Leite, R.S.R.; Fonseca, G.G. Physiological evaluation of yeast strains under anaerobic conditions using glucose.; fructose.; or sucrose as the carbon source. J. Biosci. Bioeng. 2024, 137, 420–428. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Querol, A.; Barrio, E. Application of a substrate inhibition model to estimate the effect of fructose concentration on the growth of diverse Saccharomyces cerevisiae strains. J. Ind. Microbiol. Biotechnol. 2009, 36, 663–669. [Google Scholar] [CrossRef]
- Añón, A.; López, J.F.; Hernando, D.; Orriols, I.; Revilla, E.; Losada, M.M. Effect of five enological practices and of the general phenolic composition on fermentation-related aroma compounds in Mencia young red wines. Food Chem. 2014, 148, 268–275. [Google Scholar] [CrossRef]
- Duncan, J.D.; Setati, M.E.; Divol, B. Redox cofactor metabolism in Saccharomyces cerevisiae and its impact on the production of alcoholic fermentation end-products. Food Res. Int. 2023, 163, 112276. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Liu, Y.; Li, H.; Yang, Q.; Wu, Q.; Chen, S.; Tang, J.; Xu, Y. Modeling and regulation of higher alcohol production through the combined effects of the C/N ratio and microbial interaction. J. Agric. Food Chem. 2019, 67, 10694–10701. [Google Scholar] [CrossRef]
- Xie, D.; Lei, Y.; Sun, Y.; Li, X.; Zheng, J. Regulation of fructose levels on carbon flow and metabolites in yeast during food fermentation. Food Sci. Technol. Int. 2025, 31, 69–82. [Google Scholar] [CrossRef]
- Bambina, P.; Vitaggio, C.; Pollon, M.; Gargano, M.; Martinico, A.; Celotti, E.; Cinquanta, L.; Corona, O. Fermentation performance of Saccharomyces cerevisiae yeast strain with high alcoholigenous power. Eur. Food Res. Technol. 2024, 250, 425–440. [Google Scholar] [CrossRef]
- Csoma, H.; Kállai, Z.; Czentye, K.; Sipiczki, M. Starmerella lactis-condensi, a yeast that has adapted to the conditions in the oenological environment. Int. J. Food Microbiol. 2023, 401, 110282. [Google Scholar] [CrossRef]
- Lee, S.W.; Oh, M.K. Improved production of N-acetylglucosamine in Saccharomyces cerevisiae by reducing glycolytic flux. Biotechnol. Bioeng. 2016, 113, 2524–2528. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Andalis, A.A.; Fink, G.R.; Guarente, L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Biol. 2002, 22, 8056–8066. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, J.; Fan, Z.H.; Li, F.; Liu, H.Q.; Zhu, W.B. MAL62 overexpression enhances uridine diphosphoglucose-dependent trehalose synthesis and glycerol metabolism for cryoprotection of baker’s yeast in lean dough. Microb. Cell Factories 2020, 19, 196. [Google Scholar] [CrossRef] [PubMed]
- Chidi, B.S.; Bauer, F.; Rossouw, D. Organic acid metabolism and the impact of fermentation practices on wine acidity—A review. S. Afr. J. Enol. Vitic. 2018, 39, 1–15. [Google Scholar] [CrossRef]
- Lencioni, L.; Taccari, M.; Ciani, M.; Domizio, P. Zygotorulaspora florentina and Starmerella bacillaris in multistarter fermentation with Saccharomyces cerevisiae to reduce volatile acidity of high sugar musts. Aust. J. Grape Wine Res. 2018, 24, 368–372. [Google Scholar] [CrossRef]
- Volschenk, H.; Van Vuuren, H.J.J.; Viljoen-Bloom, M. Malic acid in wine: Origin, function and metabolism during vinification. S. Afr. J. Enol. Vitic. 2006, 27, 123–136. [Google Scholar] [CrossRef]
- Pugh, T.A.; Maurer, J.M.; Pringle, A.T. The impact of wort nitrogen limitation on yeast fermentation performance and diacetyl. Discussion. Tech. Q.-Master Brew. Assoc. Am. 1997, 34, 185–189. [Google Scholar]
- Koonthongkaew, J.; Ploysongsri, N.; Toyokawa, Y.; Ruangpornvisuti, V.; Takagi, H. Improvement of fusel alcohol production by engineering of the yeast branched-chain amino acid aminotransaminase. Appl. Environ. Microbiol. 2022, 88, e00557-22. [Google Scholar] [CrossRef]
- Wang, Z.; Bai, X.; Guo, X.; He, X. Regulation of crucial enzymes and transcription factors on 2-phenylethanol biosynthesis via Ehrlich pathway in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2017, 44, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Eden, A.; Van Nedervelde, L.; Drukker, M.; Benvenisty, N.; Debourg, A. Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl. Microbiol. Biotechnol. 2001, 55, 296–300. [Google Scholar] [CrossRef]
- Lilly, M.; Bauer, F.F.; Styger, G.; Lambrechts, M.G.; Pretorius, I.S. The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res. 2006, 6, 726–743. [Google Scholar] [CrossRef] [PubMed]
- Iraqui, I.; Vissers, S.; Cartiaux, M.; Urrestarazu, A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 1998, 257, 238–248. [Google Scholar] [CrossRef]
- Li, W.; Chen, S.J.; Wang, J.H.; Zhang, C.Y.; Shi, Y.; Guo, X.W.; Chen, Y.F.; Xiao, D.G. Genetic engineering to alter carbon flux for various higher alcohol productions by Saccharomyces cerevisiae for Chinese Baijiu fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Holloway, P.; Subden, R.E. The nucleotide sequence and initial characterization of pyruvate decarboxylase from the yeast Hanseniaspora uvarum. Yeast 1994, 10, 1581–1589. [Google Scholar] [CrossRef]
- Schifferdecker, A.J.; Siurkus, J.; Andersen, M.R.; Joerck-Ramberg, D.; Ling, Z.; Zhou, N.; Blevins, J.E.; Sibirny, A.A.; Piškur, J.; Ishchuk, O.P. Erratum to: Alcohol dehydrogenase gene ADH3 activates glucose alcoholic fermentation in genetically engineered Dekkera bruxellensis yeast. Appl. Microbiol. Biotechnol. 2016, 100, 3233. [Google Scholar] [CrossRef]
- Yang, D.D.; de Billerbeck, G.M.; Zhang, J.J.; Rosenzweig, F.; Francois, J.M. Deciphering the origin.; evolution.; and physiological function of the subtelomeric aryl-Alcohol dehydrogenase gene family in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2018, 84, e01553-17. [Google Scholar] [CrossRef]
- Zheng, N.; Jiang, S.; He, Y.; Chen, Y.; Zhang, C.; Guo, X.; Ma, L.; Xiao, D. Production of low-alcohol Huangjiu with improved acidity and reduced levels of higher alcohols by fermentation with scarless ALD6 overexpression yeast. Food Chem. 2020, 321, 126691. [Google Scholar] [CrossRef]
- Aguilera, J.; Prieto, J. The Saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr. Genet. 2001, 39, 273–283. [Google Scholar] [CrossRef]
- Khattab, S.M.R.; Kodaki, T. Efficient bioethanol production by overexpression of endogenous Saccharomyces cerevisiae xylulokinase and NADPH-dependent aldose reductase with mutated strictly NADP+-dependent Pichia stipitis xylitol dehydrogenase. Process Biochem. 2014, 49, 1838–1842. [Google Scholar] [CrossRef]
- Masuda, C.A.; Previato, J.O.; Miranda, M.N.; Assis, L.J.; Penha, L.L.; Mendonça-Previato, L.; Montero-Lomelí, M. Overexpression of the aldose reductase GRE3 suppresses lithium-induced galactose toxicity in Saccharomyces cerevisiae. FEMS Yeast Res. 2008, 8, 1245–1253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Albillos-Arenal, S.; Minebois, R.; Querol, A.; Barrio, E. Understanding the role of GRE3 in the erythritol biosynthesis pathway in Saccharomyces uvarum and its implication in osmoregulation and redox homeostasis. Microb. Biotechnol. 2023, 16, 1858–1871. [Google Scholar] [CrossRef] [PubMed]
- Minebois, R.; Pérez-Torrado, R.; Querol, A. A time course metabolism comparison among Saccharomyces cerevisiae, S. uvarum and S. kudriavzevii species in wine fermentation. Food Microbiol. 2020, 90, 103484. [Google Scholar] [CrossRef]
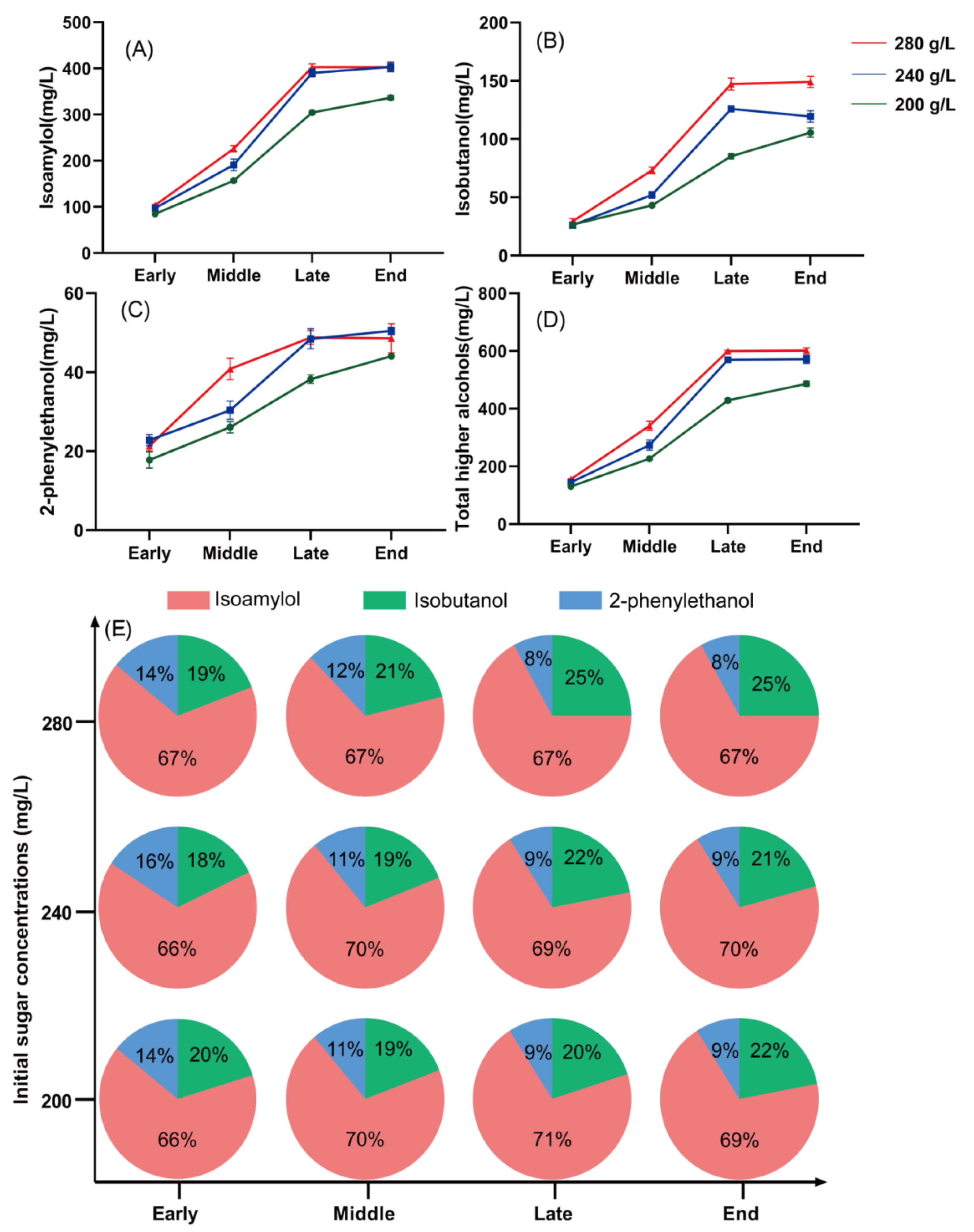


| Metabolite | Early | Middle | Late | End | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 g/L | 240 g/L | 280 g/L | 200 g/L | 240 g/L | 280 g/L | 200 g/L | 240 g/L | 280 g/L | 200 g/L | 240 g/L | 280 g/L | |
| Residual sugar (g/L) | 151.50 ± 3.02 a | 188.11 ± 2.33 b | 227.02 ± 7.45 c | 104.11 ± 5.29 a | 124.11 ± 6.35 b | 164.71 ± 6.10 c | 11.89 ± 1.99 a | 22.88 ± 3.04 b | 39.88 ± 6.98 c | 1.73 ± 0.49 a | 2.58 ± 0.33 b | 1.73 ± 1.06 a |
| Ethanol (% v/v) | 2.63 ± 0.03 a | 2.77 ± 0.12 a | 2.63 ± 0.30 a | 5.07 ± 0.23 a | 6.20 ± 0.36 b | 4.83 ± 2.47 ab | 10.47 ± 0.12 a | 12.67 ± 0.40 b | 11.80 ± 2.00 ab | 11.23 ± 0.29 a | 13.97 ± 0.29 b | 15.10 ± 0.14 c |
| Higher alcohol (mg/L) | 130.01 ± 7.56 c | 144.70 ± 3.26 b | 155.55 ± 1.97 a | 227.22 ± 4.70 c | 273.89 ± 17.62 b | 340.80 ± 15.92 a | 428.75 ± 6.63 c | 569.51 ± 6.92 b | 599.50 ± 2.68 a | 486.60 ± 8.90 c | 571.62 ± 14.31 b | 601.92 ± 9.31 a |
| Total acid (g/L) | 9.94 ± 0.9 a | 10.41 ± 0.93 ab | 11.78 ± 0.59 b | 8.06 ± 0.55 a | 8.40 ± 0.82 ab | 9.54 ± 0.38 b | 6.89 ± 0.42 a | 7.91 ± 0.17 a | 7.50 ± 0.89 a | 6.03 ± 0.12 a | 6.95 ± 0.05 b | 6.90 ± 0.09 b |
| pH | 3.22 ± 0.04 a | 3.25 ± 0.01 a | 3.25 ± 0.01 a | 3.13 ± 0.01 a | 3.20 ± 0.03 b | 3.15 ± 0.01 a | 3.07 ± 0.06 a | 3.14 ± 0.01 a | 3.14 ± 0.03 a | 3.17 ± 0.02 a | 3.30 ± 0.02 c | 3.24 ± 0.02 b |
| Glycerol (g/L) | 1.73 ± 0.12 a | 2.07 ± 0.12 b | 1.73 ± 0.12 a | 2.73 ± 0.42 a | 3.33 ± 0.31 b | 4.47 ± 0.23 c | 4.00 ± 0.20 a | 4.93 ± 0.12 b | 5.93 ± 0.76 c | 4.33 ± 0.31 a | 5.33 ± 0.12 b | 6.20 ± 0.72 c |
| Citrate (g/L) | 0.7 ± 0.06 a | 0.66 ± 0.05 a | 0.70 ± 0.06 a | 0.70 ± 0.04 a | 0.68 ± 0.04 a | 0.68 ± 0.04 a | 0.43 ± 0.02 a | 0.45 ± 0.02 a | 0.43 ± 0.05 a | 0.45 ± 0.06 a | 0.40 ± 0.04 a | 0.39 ± 0.00 a |
| Succinate (g/L) | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.75 ± 0.02 a | 0.82 ± 0.20 a | 0.69 ± 0.24 a | 0.61 ± 0.04 a | 0.69 ± 0.02 a | 0.63 ± 0.03 a |
| Lactate (g/L) | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.17 ± 0.02 a | 0.18 ± 0.03 a | 0.18 ± 0.02 a | 0.20 ± 0.02 a | 0.17 ± 0.01 | 0.20 ± 0.02 a |
| Malate (g/L) | 7.02 ± 0.57 a | 7.64 ± 0.67 ab | 8.18 ± 0.05 b | 5.61 ± 0.26 a | 6.31 ± 0.88 ab | 6.88 ± 0.35 b | 3.00 ± 0.43 a | 3.79 ± 0.05 b | 4.07 ± 0.30 b | 2.39 ± 0.05 a | 2.51 ± 0.03 a | 2.53 ± 0.11 a |
| Pyruvate (g/L) | 0.07 ± 0.02 a | 0.09 ± 0.01 a | 0.08 ± 0.03 a | 0.11 ± 0.02 a | 0.10 ± 0.02 a | 0.12 ± 0.01 a | 0.10 ± 0.01 a | 0.12 ± 0.00 a | 0.10 ± 0.02 a | 0.07 ± 0.01 a | 0.06 ± 0.00 a | 0.05 ± 0.01 b |
| Acetate (g/L) | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.36 ± 0.16 a | 0.70 ± 0.15 a | 0.52 ± 0.17 a | 0.50 ± 0.08 a | 0.72 ± 0.09 b | 0.86 ± 0.40 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Ren, X.; Wang, Y.; Hao, D.; Liang, Y.; Qin, Y. Transcriptomic Identification of Core Regulatory Genes for Higher Alcohol Production in Saccharomyces cerevisiae at Different Sugar Concentrations in Wine Fermentation. Foods 2025, 14, 1476. https://doi.org/10.3390/foods14091476
Chen L, Ren X, Wang Y, Hao D, Liang Y, Qin Y. Transcriptomic Identification of Core Regulatory Genes for Higher Alcohol Production in Saccharomyces cerevisiae at Different Sugar Concentrations in Wine Fermentation. Foods. 2025; 14(9):1476. https://doi.org/10.3390/foods14091476
Chicago/Turabian StyleChen, Lu, Xiaona Ren, Yanan Wang, Dongshu Hao, Yanying Liang, and Yi Qin. 2025. "Transcriptomic Identification of Core Regulatory Genes for Higher Alcohol Production in Saccharomyces cerevisiae at Different Sugar Concentrations in Wine Fermentation" Foods 14, no. 9: 1476. https://doi.org/10.3390/foods14091476
APA StyleChen, L., Ren, X., Wang, Y., Hao, D., Liang, Y., & Qin, Y. (2025). Transcriptomic Identification of Core Regulatory Genes for Higher Alcohol Production in Saccharomyces cerevisiae at Different Sugar Concentrations in Wine Fermentation. Foods, 14(9), 1476. https://doi.org/10.3390/foods14091476







