Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention
Abstract
:1. Introduction
2. Materials and Methods
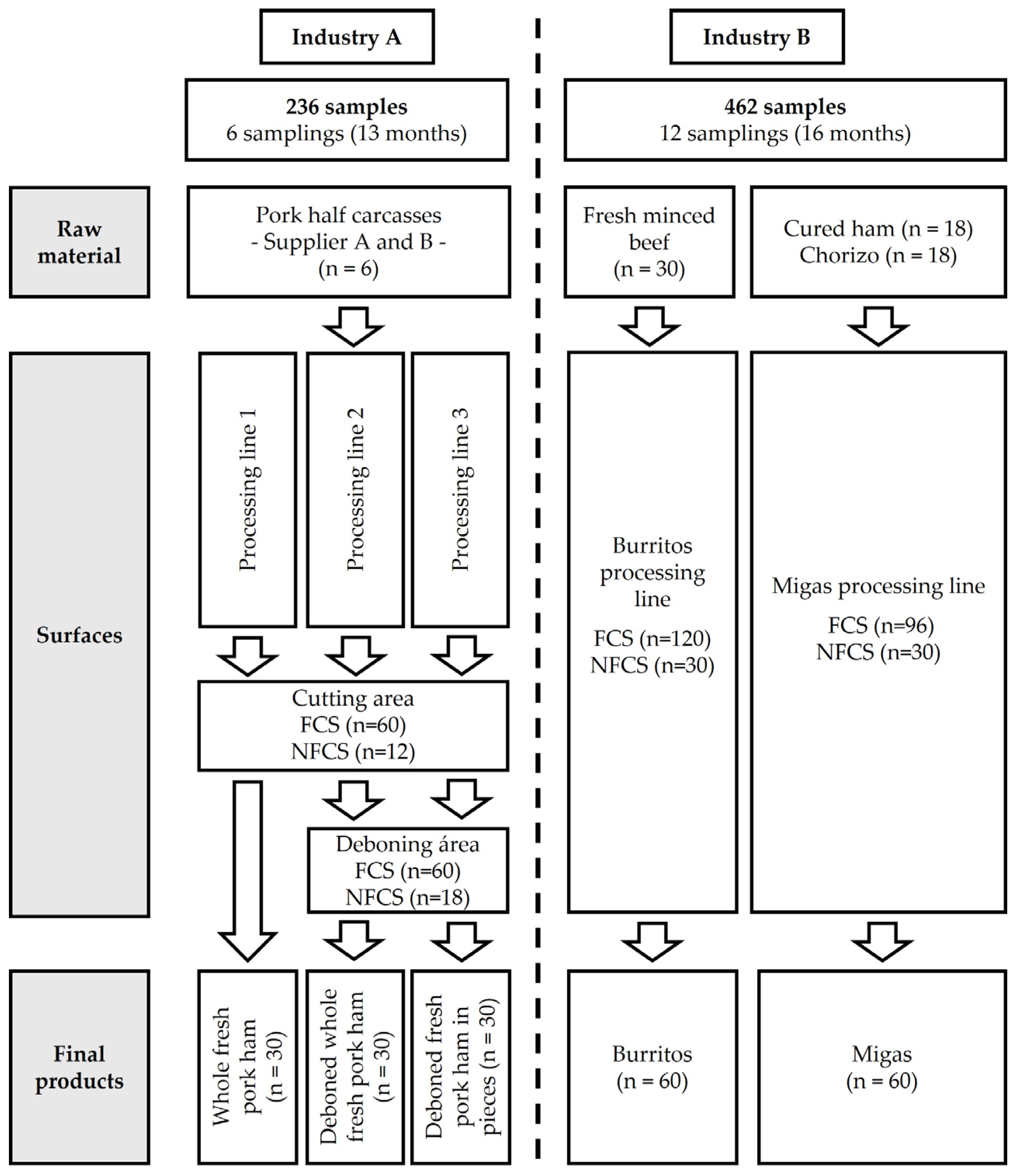
2.1. Study Design
2.2. Sampling Procedure
2.3. Detection of Listeria spp.
2.4. Isolation and Identification
2.5. L. monocytogenes Typing
2.5.1. Serotype Identification
2.5.2. Pulsed-Field Gel Electrophoresis Typing
2.6. Statistical Analysis
3. Results
3.1. Prevalence of Listeria spp. and L. monocytogenes
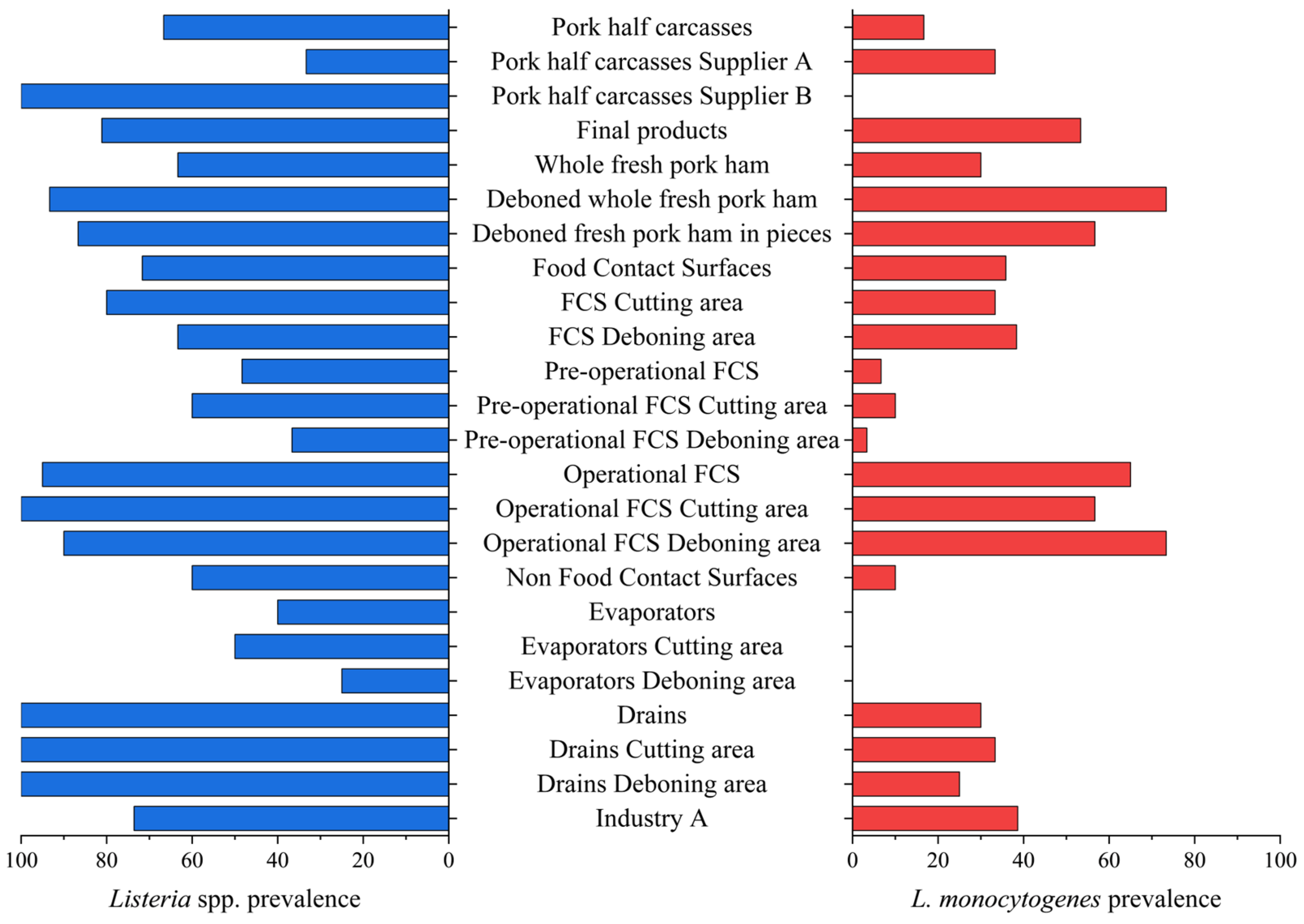
3.1.1. Industry A
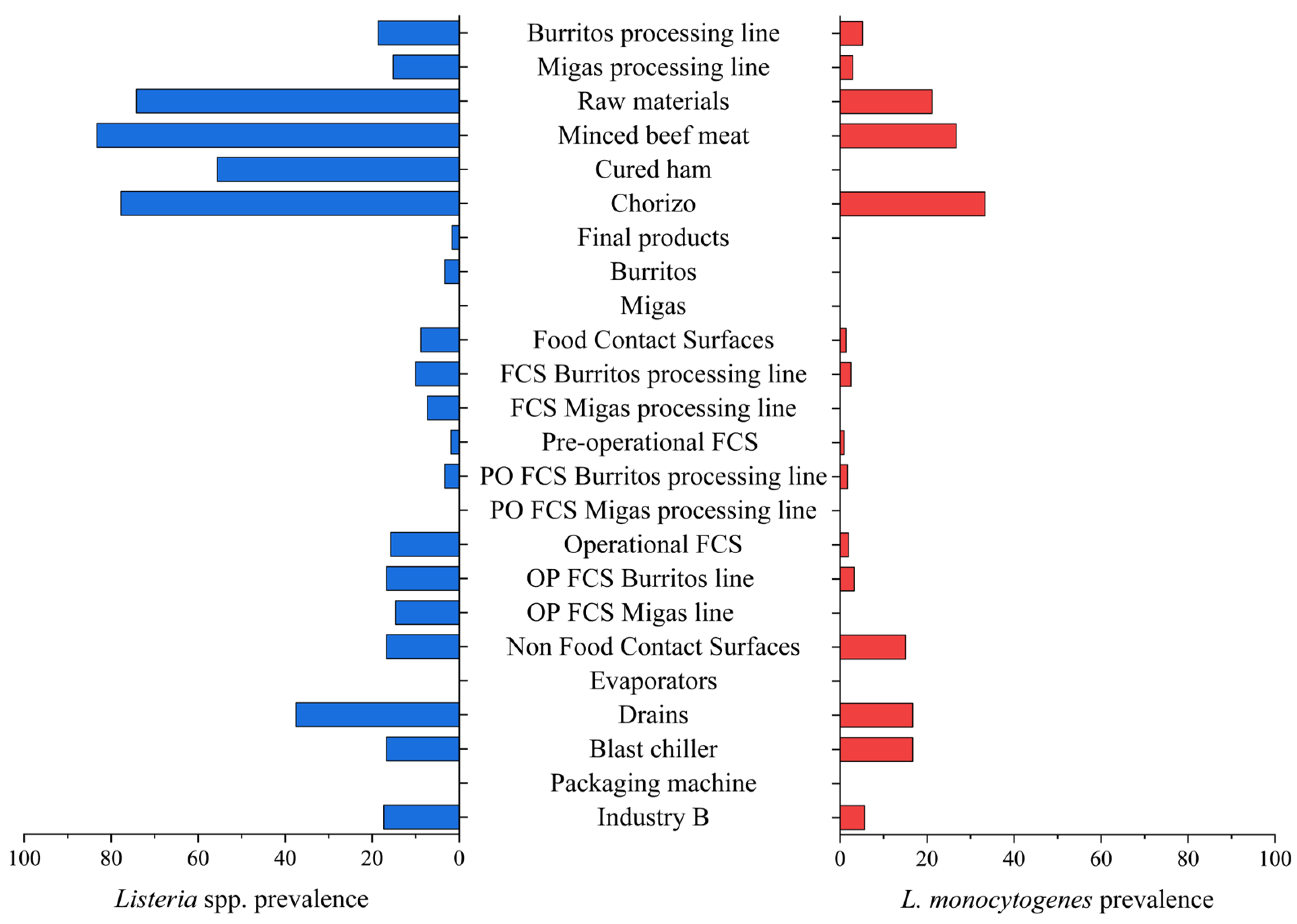
3.1.2. Industry B
3.2. Serotype Identification
3.2.1. Industry A
3.2.2. Industry B
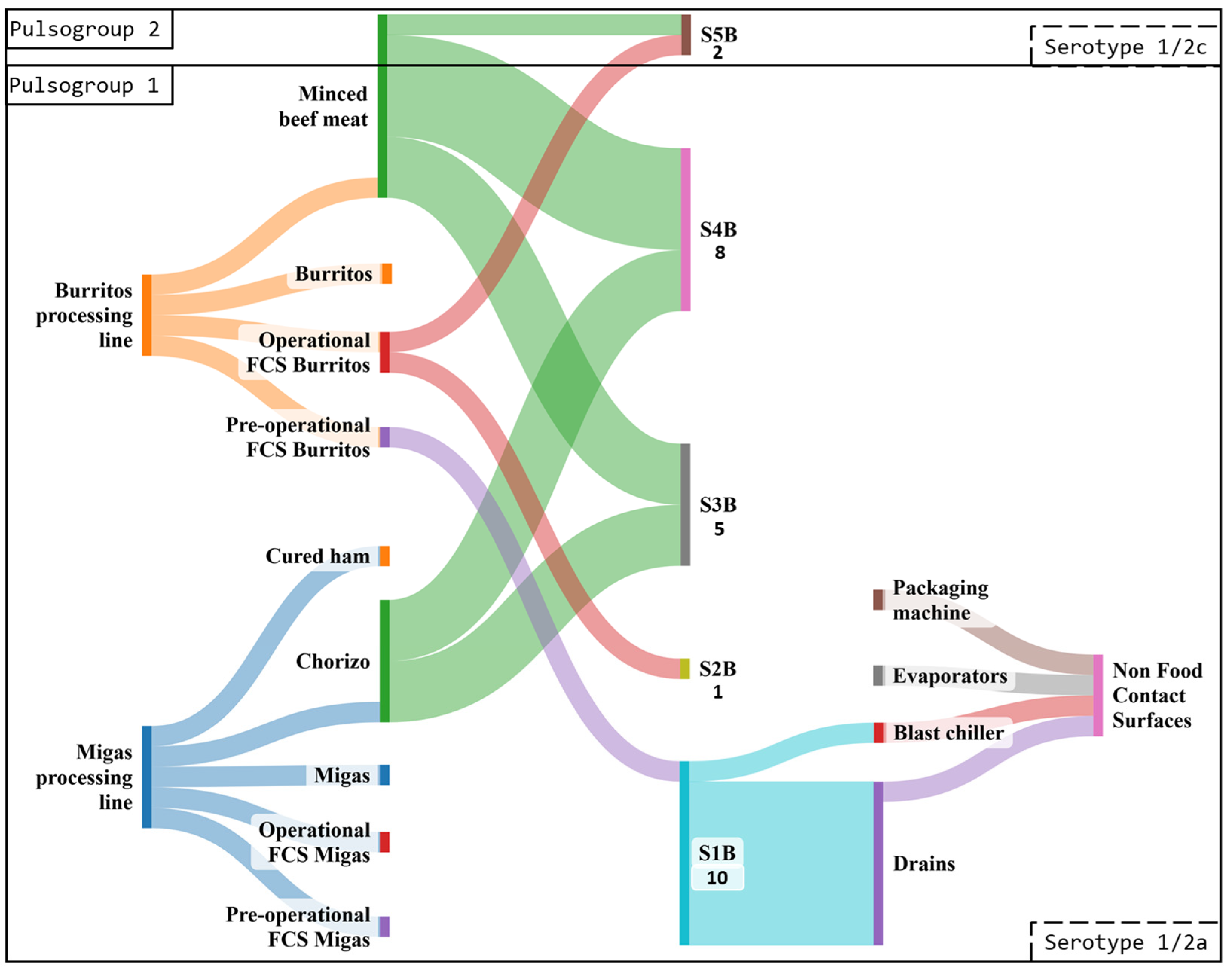
3.3. Pulsed-Field Gel Electrophoresis Typing
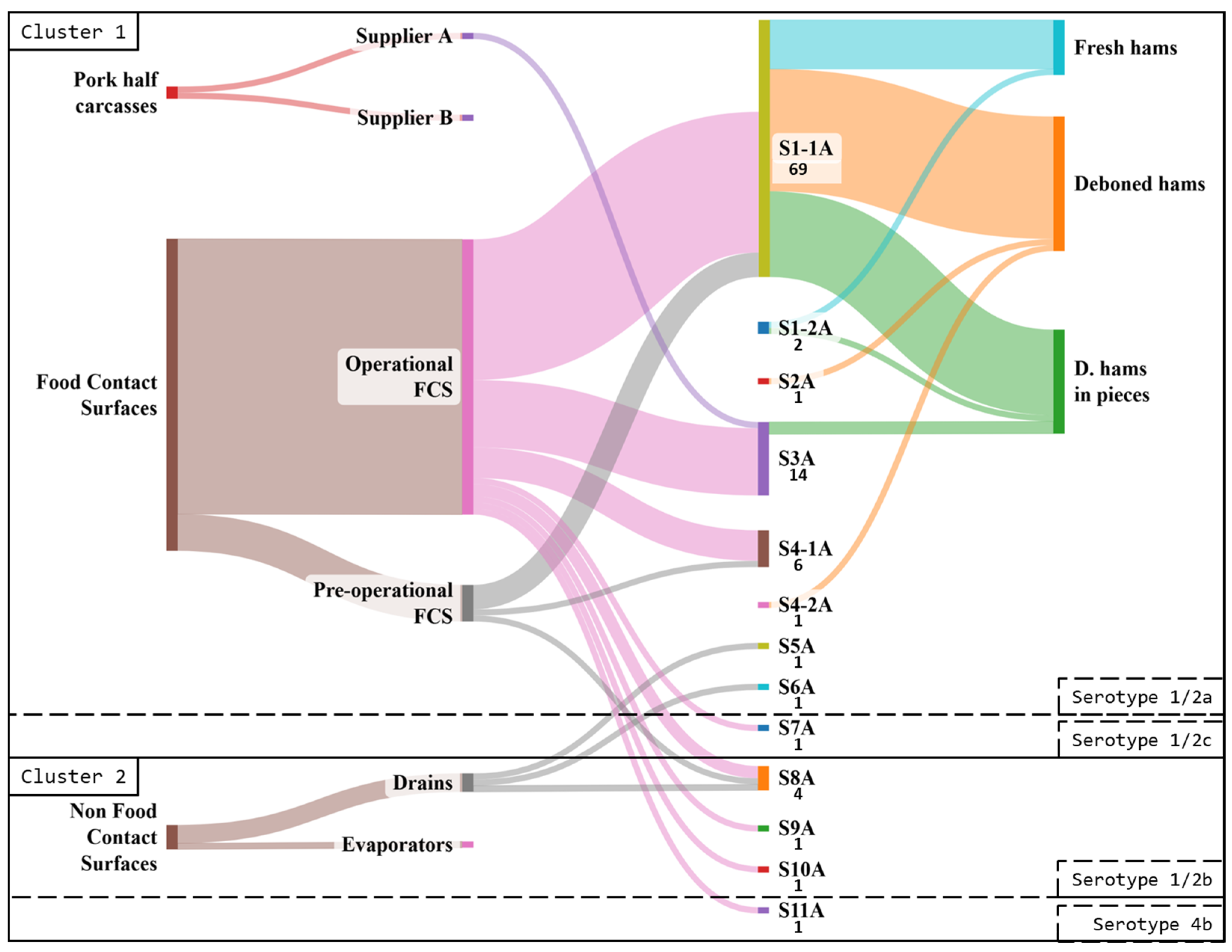
3.3.1. Industry A
3.3.2. Industry B
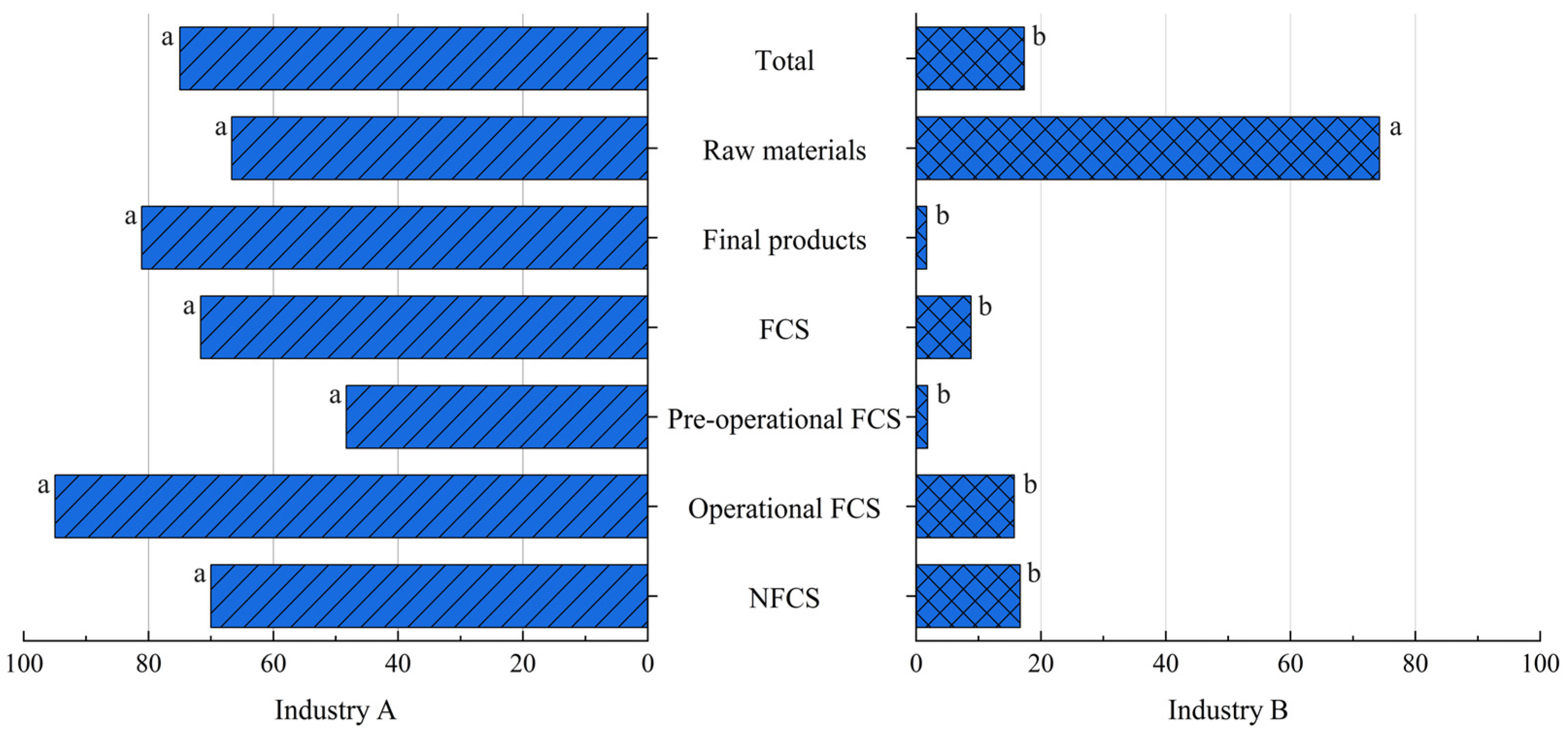
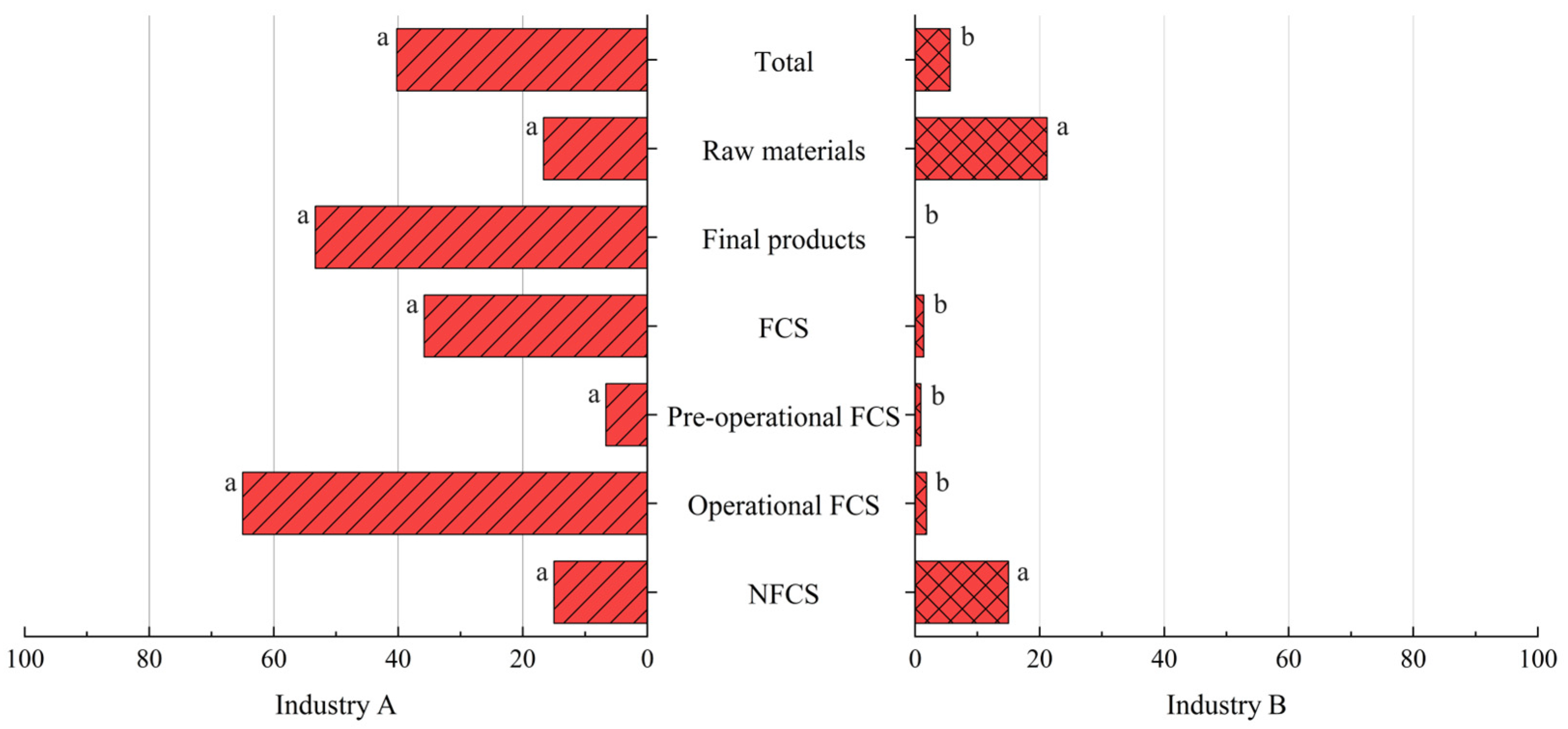
3.4. Comparison Between Industries A and B
3.4.1. Prevalence of Listeria spp. and L. monocytogenes
3.4.2. Pulsed-Field Gel Electrophoresis Typing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EU | European Union |
| RTE | Ready-To-Eat |
| GDP | Spanish Gross Domestic Product |
| C&D | Cleaning and Disinfection |
| FCS | Food Contact Surfaces |
| NFCS | Non-Food Contact Surfaces |
| OB | One Broth Listeria |
| OCLA | Oxoid Chromogenic Listeria Agar |
| TSA | Tryptone Soy Agar |
| TSB | Tryptone Soya Broth |
| PFGE | Pulsed-Field Gel Electrophoresis |
| FH | Whole Fresh Pork Hams |
| DFH | Deboned Whole Fresh Pork Hams |
| DFHP | Deboned Fresh Pork Hams In Pieces |
| OP | Operational |
| PO | Pre-Operational |
| PT | Pulsotype |
References
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2023 Zoonoses Report. EFSA J. 2024, 22, e9106. [Google Scholar]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed]
- Kérouanton, A.; Marault, M.; Petit, L.; Grout, J.; Dao, T.T.; Brisabois, A. Evaluation of a Multiplex PCR Assay as an Alternative Method for Listeria monocytogenes Serotyping. J. Microbiol. Methods 2010, 80, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, H.P.R.; Höhne, K. Chapter II Serotyping of Listeria monocytogenes and Related Species. In Methods in Microbiology; Bergan, T., Norris, J.R., Eds.; Academic Press: Cambridge, MA, USA, 1979; Volume 13, pp. 31–49. [Google Scholar]
- Yin, Y.; Yao, H.; Doijad, S.; Kong, S.; Shen, Y.; Cai, X.; Tan, W.; Wang, Y.; Feng, Y.; Ling, Z.; et al. A Hybrid Sub-Lineage of Listeria monocytogenes Comprising Hypervirulent Isolates. Nat. Commun. 2019, 10, 4283. [Google Scholar] [CrossRef]
- Borucki, M.K.; Call, D.R. Listeria monocytogenes Serotype Identification by PCR. J. Clin. Microbiol. 2003, 41, 5537–5540. [Google Scholar] [CrossRef]
- Bergis, H.; Bonanno, L.; Asséré, A.; Lombard, B. EURL Lm Technical Guidance Document on Challenge Tests and Durability Studies for Assessing Shelf-Life of Ready-to-Eat Foods Related to Listeria monocytogenes. EURL Lm 2021, 4, 196. [Google Scholar]
- Rugna, G.; Carra, E.; Bergamini, F.; Franzini, G.; Faccini, S.; Gattuso, A.; Morganti, M.; Baldi, D.; Naldi, S.; Serraino, A.; et al. Distribution, Virulence, Genotypic Characteristics and Antibiotic Resistance of Listeria monocytogenes Isolated over One-Year Monitoring from Two Pig Slaughterhouses and Processing Plants and Their Fresh Hams. Int. J. Food Microbiol. 2021, 336, 108912. [Google Scholar] [CrossRef]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in Biofilm Formation among Strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes Biofilms in the Wonderland of Food Industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef]
- Dygico, L.K.; Gahan, C.G.M.; Grogan, H.; Burgess, C.M. The Ability of Listeria monocytogenes to Form Biofilm on Surfaces Relevant to the Mushroom Production Environment. Int. J. Food Microbiol. 2020, 317, 108385. [Google Scholar] [CrossRef]
- Burdová, A.; Véghová, A.; Minarovičová, J.; Drahovská, H.; Kaclíková, E. The Relationship between Biofilm Phenotypes and Biofilm-Associated Genes in Food-Related Listeria monocytogenes Strains. Microorganisms 2024, 12, 1297. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA) Panel on Biological Hazards (BIOHAZ); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Listeria monocytogenes Contamination of Ready-to-Eat Foods and the Risk for Human Health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef] [PubMed]
- Tsaloumi, S.; Aspridou, Z.; Tsigarida, E.; Gaitis, F.; Garofalakis, G.; Barberis, K.; Tzoumanika, F.; Dandoulaki, M.; Skiadas, R.; Koutsoumanis, K. Quantitative Risk Assessment of Listeria monocytogenes in Ready-to-Eat (RTE) Cooked Meat Products Sliced at Retail Stores in Greece. Food Microbiol. 2021, 99, 103800. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2005, 338, 1–26. [Google Scholar]
- Asociación Nacional de Industrias de la Carne de España (ANICE) Memoria de Actividades. 2023. Available online: https://www.anice.es/industrias/memoria-anice/memoria-de-actividades-de-anice-2023_36573_201_48093_0_1_in.html (accessed on 18 July 2024).
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and Food Processing Environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- Osek, J.; Lachtara, B.; Wieczorek, K. Listeria monocytogenes—How This Pathogen Survives in Food-Production Environments? Front. Microbiol. 2022, 13, 866462. [Google Scholar] [CrossRef]
- Bonsaglia, E.C.R.; Silva, N.C.C.; Fernades Júnior, A.; Araújo Júnior, J.P.; Tsunemi, M.H.; Rall, V.L.M. Production of Biofilm by Listeria monocytogenes in Different Materials and Temperatures. Food Control 2014, 35, 386–391. [Google Scholar] [CrossRef]
- Conficoni, D.; Losasso, C.; Cortini, E.; Di Cesare, A.; Cibin, V.; Giaccone, V.; Corno, G.; Ricci, A. Resistance to Biocides in Listeria monocytogenes Collected in Meat-Processing Environments. Front. Microbiol. 2016, 7, 1627. [Google Scholar] [CrossRef]
- Ortiz, S.; López, V.; Villatoro, D.; López, P.; Dávila, J.C.; Martínez-Suárez, J.N.V. A 3-Year Surveillance of the Genetic Diversity and Persistence of Listeria monocytogenes in an Iberian Pig Slaughterhouse and Processing Plant. Foodborne Pathog. Dis. 2010, 7, 1177–1184. [Google Scholar] [CrossRef]
- Li, H.; Wang, P.; Lan, R.; Luo, L.; Cao, X.; Wang, Y.; Wang, Y.; Li, H.; Zhang, L.; Ji, S.; et al. Risk Factors and Level of Listeria monocytogenes Contamination of Raw Pork in Retail Markets in China. Front. Microbiol. 2018, 9, 1090. [Google Scholar] [CrossRef]
- Sereno, M.J.; Viana, C.; Pegoraro, K.; da Silva, D.A.L.; Yamatogi, R.S.; Nero, L.A.; Bersot, L. dos S. Distribution, Adhesion, Virulence and Antibiotic Resistance of Persistent Listeria monocytogenes in a Pig Slaughterhouse in Brazil. Food Microbiol. 2019, 84, 103234. [Google Scholar] [CrossRef] [PubMed]
- D’Arrigo, M.; Mateo-Vivaracho, L.; Guillamón, E.; Fernández-León, M.F.; Bravo, D.; Peirotén, Á.; Medina, M.; García-Lafuente, A. Characterization of Persistent Listeria monocytogenes Strains from Ten Dry-Cured Ham Processing Facilities. Food Microbiol. 2020, 92, 103581. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA) Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Persistence of Microbiological Hazards in Food and Feed Production and Processing Environments. EFSA J. 2024, 22, e8521. [Google Scholar] [CrossRef]
- Ortiz, S.; López-Alonso, V.; Rodríguez, P.; Martínez-Suárez, J.V. The Connection between Persistent, Disinfectant-Resistant Listeria monocytogenes Strains from Two Geographically Separate Iberian Pork Processing Plants: Evidence from Comparative Genome Analysis. Appl. Environ. Microbiol. 2016, 82, 308–317. [Google Scholar] [CrossRef]
- Pérez-Baltar, A.; Pérez-Boto, D.; Medina, M.; Montiel, R. Genomic Diversity and Characterization of Listeria monocytogenes from Dry-Cured Ham Processing Plants. Food Microbiol. 2021, 99, 103779. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission Regulation (EU) 2024/2895 of 20 November 2024 Amending Regulation (EC) No 2073/2005 as Regards Listeria monocytogenes. Off. J. Eur. Union 2024, 2895, 1–3. [Google Scholar]
- Larivière-Gauthier, G.; Letellier, A.; Kérouanton, A.; Bekal, S.; Quessy, S.; Fournaise, S.; Fravalo, P. Analysis of Listeria monocytogenes Strain Distribution in a Pork Slaughter and Cutting Plant in the Province of Quebec. J. Food Prot. 2014, 77, 2121–2128. [Google Scholar] [CrossRef]
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp. Part 1: Detection Method. International Organization for Standardization: Geneva, Switzerland, 2017.
- ISO 18593:2018; Microbiology of the Food Chain—Horizontal Methods for Surface Sampling. International Organization for Standardization: Geneva, Switzerland, 2018.
- Labrador, M.; Rota, M.C.; Pérez-Arquillué, C.; Herrera, A.; Bayarri, S. Comparative Evaluation of Impedanciometry Combined with Chromogenic Agars or RNA Hybridization and Real-Time PCR Methods for the Detection of L. monocytogenes in Dry-Cured Ham. Food Control 2018, 94, 108–115. [Google Scholar] [CrossRef]
- Labrador, M. Desarrollo de Métodos Rápidos Para el Análisis de Listeria monocytogenes y su Aplicación al Proceso de Evaluación del Riesgo en la Industria Cárnica. Ph.D. Thesis, Universidad de Zaragoza, Zaragoza, Spain, 2018. [Google Scholar]
- PulseNet. Standard Operating Procedure for Pulsenet PFGE of Listeria monocytogenes. Available online: https://www.pulsenetinternational.org/assets/PulseNet/uploads/pfge/PNL04_ListeriaPFGEProtocol.pdf (accessed on 18 September 2024).
- Graves, L.M.; Swaminathan, B. PulseNet Standardized Protocol for Subtyping Listeria monocytogenes by Macrorestriction and Pulsed-Field Gel Electrophoresis. Int. J. Food Microbiol. 2001, 65, 55–62. [Google Scholar] [CrossRef]
- Martin, P.; Jacquet, C.; Goulet, V.; Vaillant, V.; De Valk, H. Participants in the PulseNet Europe Feasibility Study Pulsed-Field Gel Electrophoresis of Listeria monocytogenes Strains: The PulseNet Europe Feasibility Study. Foodborne Pathog. Dis. 2006, 3, 303–308. [Google Scholar] [CrossRef]
- Meloni, D.; Consolati, S.G.; Mazza, R.; Mureddu, A.; Fois, F.; Piras, F.; Mazzette, R. Presence and Molecular Characterization of the Major Serovars of Listeria monocytogenes in Ten Sardinian Fermented Sausage Processing Plants. Meat Sci. 2014, 97, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Demaître, N.; Rasschaert, G.; De Zutter, L.; Geeraerd, A.; De Reu, K. Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: From Occasional Introduction to Plausible Persistence in Harborage Sites. Pathogens 2021, 10, 717. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical Index of the Discriminatory Ability of Typing Systems: An Application of Simpson’s Index of Diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed]
- Prencipe, V.A.; Rizzi, V.; Acciari, V.; Iannetti, L.; Giovannini, A.; Serraino, A.; Calderone, D.; Rossi, A.; Morelli, D.; Marino, L.; et al. Listeria monocytogenes Prevalence, Contamination Levels and Strains Characterization throughout the Parma Ham Processing Chain. Food Control 2012, 25, 150–158. [Google Scholar] [CrossRef]
- Sala, C.; Morar, A.; Tîrziu, E.; Nichita, I.; Imre, M.; Imre, K. Environmental Occurrence and Antibiotic Susceptibility Profile of Listeria monocytogenes at a Slaughterhouse Raw Processing Plant in Romania. J. Food Prot. 2016, 79, 1794–1797. [Google Scholar] [CrossRef]
- Thévenot, D.; Delignette-Muller, M.-L.; Christieans, S.; Leroy, S.; Kodjo, A.; Vernozy-Rozand, C. Serological and Molecular Ecology of Listeria monocytogenes Isolates Collected from 13 French Pork Meat Salting–Curing Plants and Their Products. Int. J. Food Microbiol. 2006, 112, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Belias, A.; Sullivan, G.; Wiedmann, M.; Ivanek, R. Factors That Contribute to Persistent Listeria in Food Processing Facilities and Relevant Interventions: A Rapid Review. Food Control 2022, 133, 108579. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Ye, L.; Wang, W.; Shi, L.; Yan, H.; Meng, H. Characterization of Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from a Pork Processing Plant and Its Respective Meat Markets in Southern China. Foodborne Pathog. Dis. 2016, 13, 262–268. [Google Scholar] [CrossRef]
- Knabel, S.J.; Reimer, A.; Verghese, B.; Lok, M.; Ziegler, J.; Farber, J.; Pagotto, F.; Graham, M.; Nadon, C.A.; Gilmour, M.W. Sequence Typing Confirms That a Predominant Listeria monocytogenes Clone Caused Human Listeriosis Cases and Outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 2012, 50, 1748–1751. [Google Scholar] [CrossRef]
- Arslan, S.; Baytur, S. Prevalence and Antimicrobial Resistance of Listeria Species and Subtyping and Virulence Factors of Listeria monocytogenesfrom Retail Meat. J. Food Saf. 2019, 39, e12578. [Google Scholar] [CrossRef]
- Paduro, C.; Montero, D.A.; Chamorro, N.; Carreño, L.J.; Vidal, M.; Vidal, R. Ten Years of Molecular Epidemiology Surveillance of Listeria monocytogenes in Chile 2008–2017. Food Microbiol. 2020, 85, 103280. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequences (5′–3′) | Product Size (pb) | Reference |
|---|---|---|---|
| lmo1118 | F: AGG GGT CTT AAA TCC TGG AA R: CGG CTT GTT CGG CAT ACT TA | 906 | [2] |
| lmo0737 | F: AGG GCT TCA AGG ACT TAC CC R: ACG ATT TCT GCT TGC CAT TC | 691 | [2] |
| ORF2110 | F: AGT GGA CAA TTG ATT GGT GAA R: CAT CCA TCC CTT ACT TTG GAC | 597 | [2] |
| ORF2819 | F: AGC AAA ATG CCA AAA CTC GT R: CAT CAC TAA AGC CTC CCA TTG | 471 | [2] |
| prs | F: GCT GAA GAG ATT GCG AAA GAA G R: CAA AGA AAC CTT GGA TTT GCG G | 370 | [2] |
| prfA | F: GAT ACA GAA ACA TCG GTT GGC R: GTG TAA TCT TGA TGC CAT CAG G | 274 | [3] |
| flaA | F: TTA CTA GAT CAA ACT GCT CC R: AAG AAA AGC CCC TCG TCC | 538 | [6] |
| Sample Category | No. of Samples | Listeria Species No. of Positive Samples (%) | |
|---|---|---|---|
| Listeria spp. | L. monocytogenes | ||
| Pork half carcasses | 6 | 4 (66.7) | 1 (16.7) |
| Pork half carcasses, Supplier A | 3 | 1 (33.3) | 1 (33.3) |
| Pork half carcasses, Supplier B | 3 | 3 (100.0) | 0 (0.0) |
| Final products | 90 | 73 (81.1) | 48 (53.3) |
| Whole fresh pork ham | 30 | 19 (63.3) a | 9 (30.0) a |
| Deboned whole fresh pork ham | 30 | 28 (93.3) b | 22 (73.3) b |
| Deboned fresh pork ham in pieces | 30 | 26 (86.7) ab | 17 (56.7) b |
| Total FCSs | 120 | 86 (71.7) | 43 (35.8) |
| FCSs cutting area | 60 | 48 (80.0) a | 20 (33.3) a |
| FCSs deboning area | 60 | 38 (63.3) b | 23 (38.3) a |
| Pre-operational FCSs | 60 | 29 (48.3) | 4 (6.7) |
| PO FCSs cutting area | 30 | 18 (60.0) a | 3 (10.0) a |
| PO FCSs deboning area | 30 | 11 (36.7) a | 1 (3.3) a |
| Operational FCSs | 60 | 57 (95.0) | 39 (65.0) |
| OP FCSs cutting area | 30 | 30 (100.0) a | 17 (56.7) a |
| OP FCSs deboning area | 30 | 27 (90.0) a | 22 (73.3) a |
| Total NFCSs | 20 | 14 (70.0) | 3 (15.0) |
| Evaporators | 10 | 4 (40.0) | 0 (0.0) |
| Evaporators cutting area | 6 | 3 (50.0) | 0 (0.0) |
| Evaporators deboning area | 4 | 1 (25.0) | 0 (0.0) |
| Drains | 10 | 10 (100.0) | 3 (30.0) |
| Drains cutting area | 6 | 6 (100.0) | 2 (33.3) |
| Drains deboning area | 4 | 4 (100.0) | 1 (25.0) |
| Industry A | 236 | 177 (75.0) | 95 (40.3) |
| Sample Category | No. of Samples | Listeria Species No. of Positive Samples (%) | |
|---|---|---|---|
| Listeria spp. | L. monocytogenes | ||
| Burritos processing line | 210 | 39 (18.6) a | 11 (5.2) a |
| Migas processing line | 192 | 31 (16.1) a | 6 (3.1) a |
| Raw materials | 66 | 49 (74.2) | 14 (21.2) |
| Minced beef meat | 30 | 25 (83.3) a | 8 (26.7) a |
| Cured ham | 18 | 10 (55.6) b | 0 (0.0) b |
| Chorizo | 18 | 14 (77.8) ab | 6 (33.3) a |
| Final products | 120 | 2 (1.7) | 0 (0.0) |
| Burritos | 60 | 2 (3.3) | 0 (0.0) |
| Migas | 60 | 0 (0.0) | 0 (0.0) |
| Total FCSs | 216 | 19 (8.8) | 3 (1.4) |
| FCSs Burritos processing line | 120 | 12 (10.0) a | 3 (2.5) a |
| FCSs Migas processing line | 96 | 7 (7.3) a | 0 (0.0) a |
| Pre-operational FCSs | 108 | 2 (1.9) | 1 (0.9) |
| PO FCSs Burritos processing line | 60 | 2 (3.3) a | 1 (1.7) a |
| PO FCSs Migas processing line | 48 | 0 (0.0) a | 0 (0.0) a |
| Operational FCSs | 108 | 17 (15.7) | 2 (1.9) |
| OP FCSs Burritos processing line | 60 | 10 (16.7) a | 2 (3.3) a |
| OP FCSs Migas processing line | 48 | 7 (14.6) a | 0 (0.0) a |
| Total NFCSs | 60 | 10 (16.7) | 9 (15.0) |
| Evaporators | 24 | 0 (0.0) | 0 (0.0) |
| Drains | 24 | 9 (37.5) | 8 (33.3) |
| Blast chiller | 6 | 1 (16.7) | 1 (16.7) |
| Packaging machine | 6 | 0 (0.0) | 0 (0.0) |
| Industry B | 462 | 80 (17.3) | 26 (5.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero de Castilla López, B.; Gómez Lozano, D.; Herrera Marteache, A.; Conchello Moreno, P.; Rota García, C. Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention. Foods 2025, 14, 1519. https://doi.org/10.3390/foods14091519
Romero de Castilla López B, Gómez Lozano D, Herrera Marteache A, Conchello Moreno P, Rota García C. Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention. Foods. 2025; 14(9):1519. https://doi.org/10.3390/foods14091519
Chicago/Turabian StyleRomero de Castilla López, Belén, Diego Gómez Lozano, Antonio Herrera Marteache, Pilar Conchello Moreno, and Carmen Rota García. 2025. "Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention" Foods 14, no. 9: 1519. https://doi.org/10.3390/foods14091519
APA StyleRomero de Castilla López, B., Gómez Lozano, D., Herrera Marteache, A., Conchello Moreno, P., & Rota García, C. (2025). Control of Persistent Listeria monocytogenes in the Meat Industry: From Detection to Prevention. Foods, 14(9), 1519. https://doi.org/10.3390/foods14091519





