Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis
Abstract
:1. Introduction
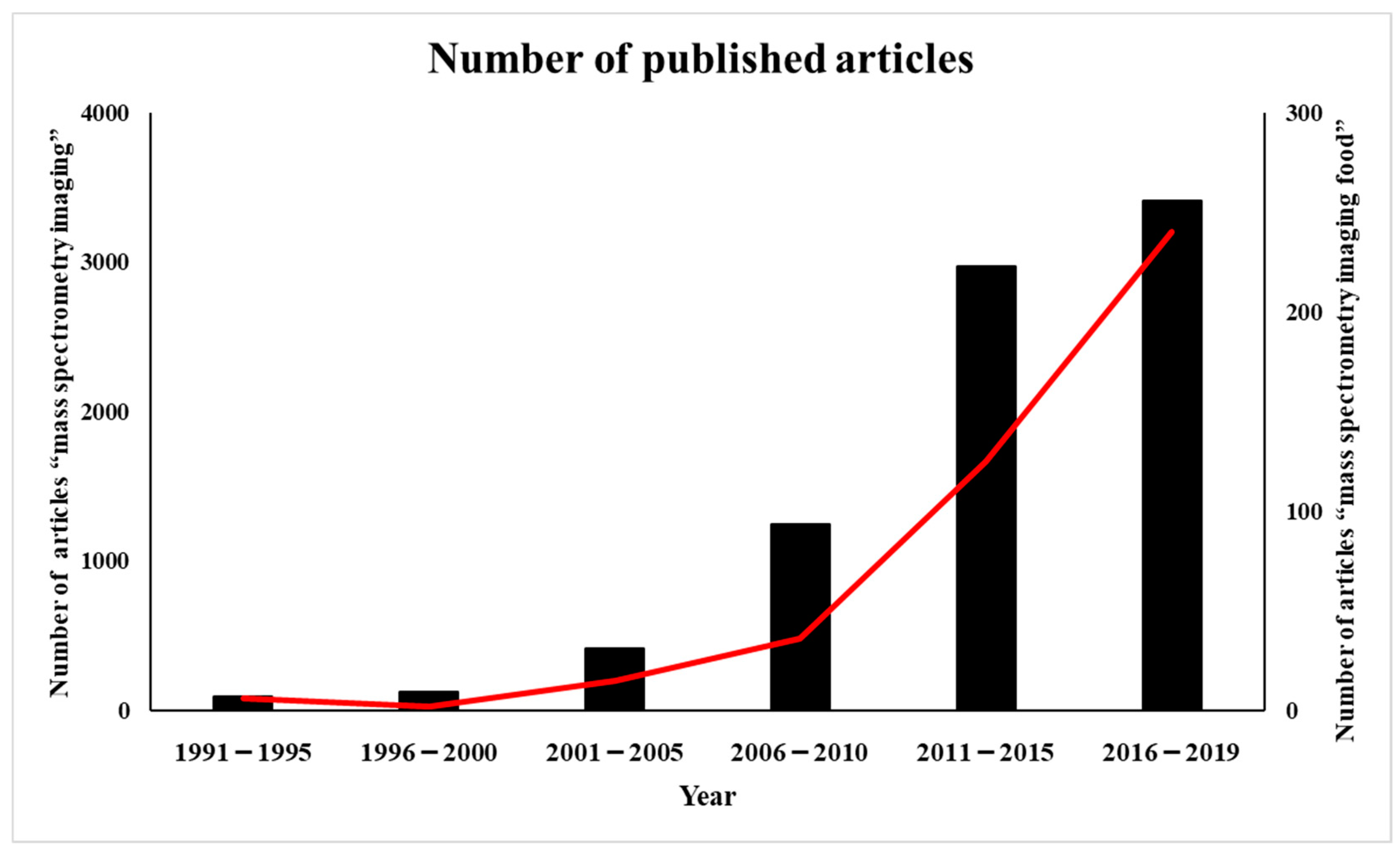
2. History of MALDI-MS Imaging Applications
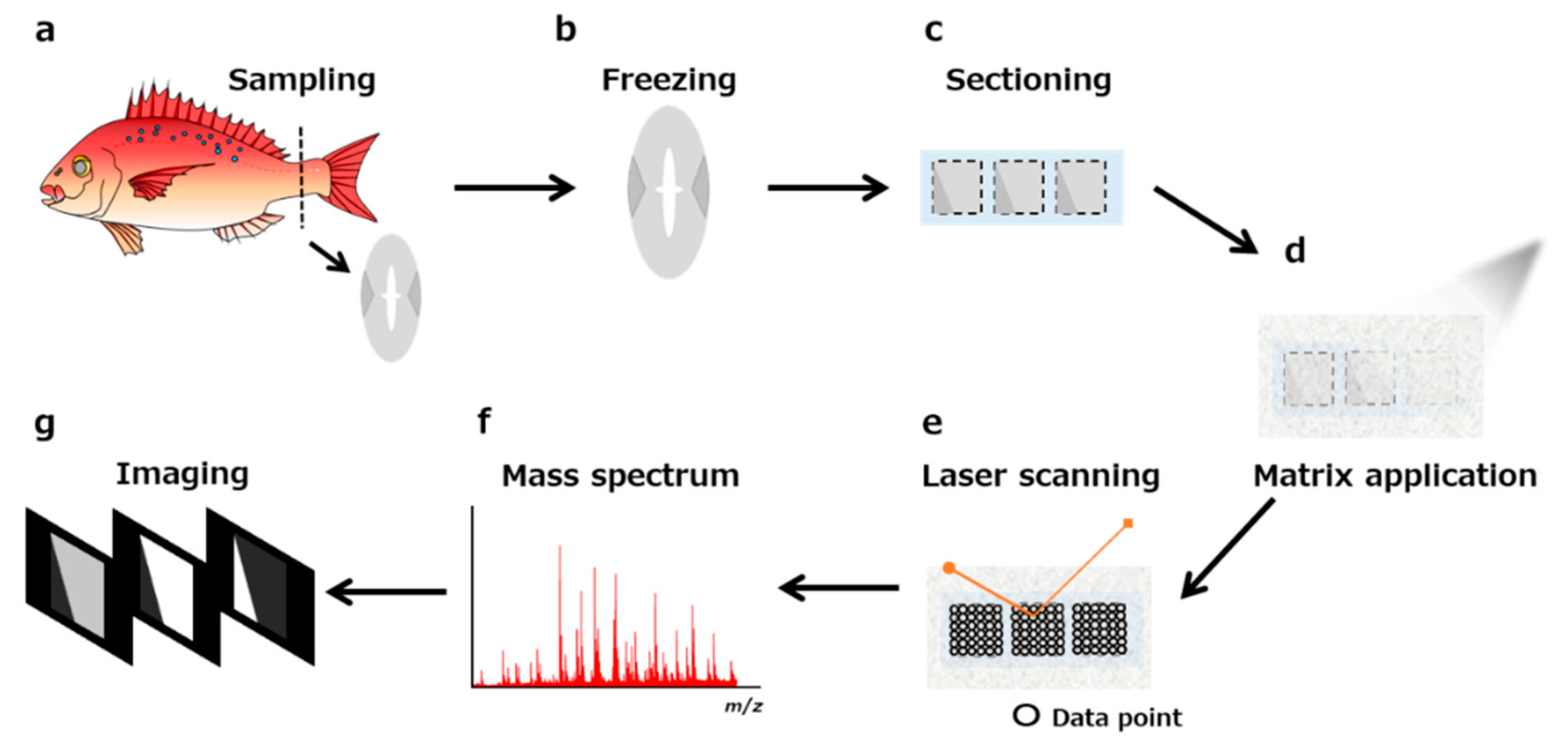
3. Sample Pretreatment for MALDI MS-Imaging
3.1. Sample Storage
3.2. Embedding
3.3. Sectioning
3.4. Sample Pretreatment (Washing, Digestion)
3.5. Derivatization for Minor Targets
3.6. Matrix Selection
3.7. Matrix Coating
4. Localization of Small Metabolites in Foods
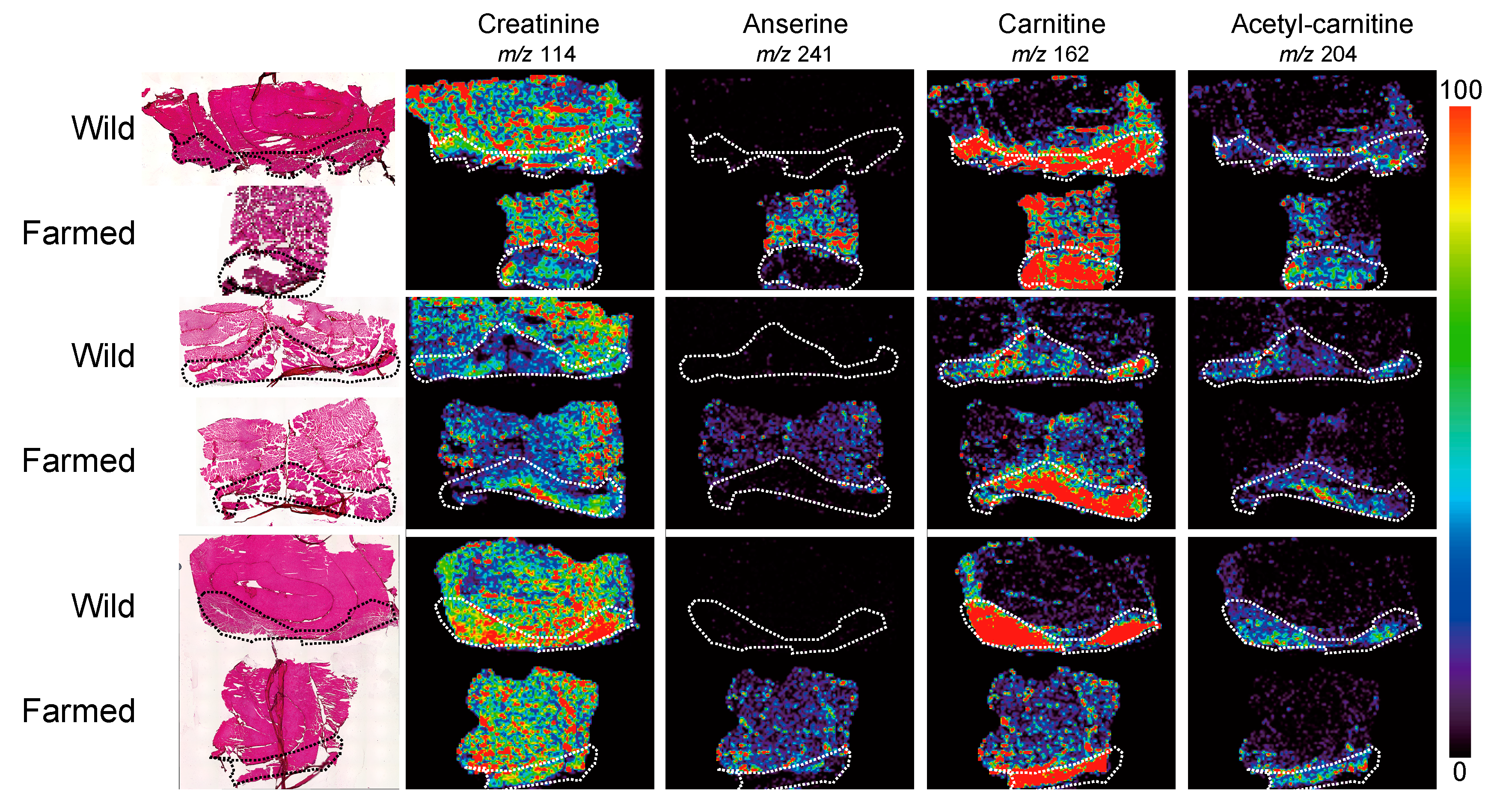
5. Breed Improvement with MALDI-MS Imaging-Based Localization Analysis
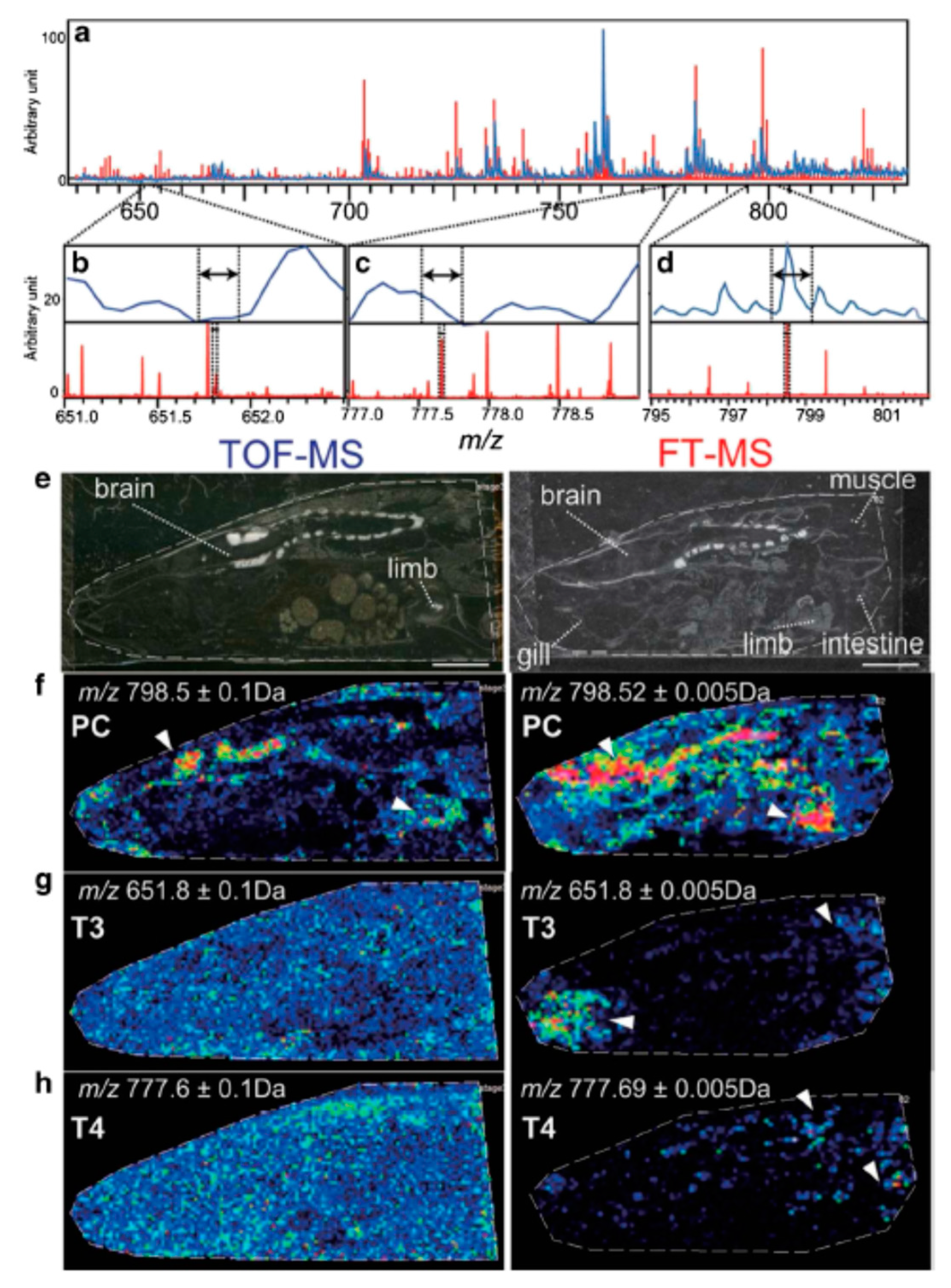
6. Recent Developments and Future Perspectives of Mass Spectrometry Imaging
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Christou, C.; Poulli, S.; Yiannopoulos, E.; Agapiou, A. GC-MS analysis of D-pinitol in carob: Syrup and fruit (flesh and seed). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1116, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Nimbalkar, M.S.; Pai, S.R.; Pawar, N.V.; Oulkar, D.; Dixit, G.B. Free amino acid profiling in grain amaranth using LC-MS/MS. Food Chem. 2012, 134, 2565–2569. [Google Scholar] [CrossRef] [PubMed]
- Zdunic, G.; Godevac, D.; Savikin, K.; Krivokuca, D.; Mihailovic, M.; Przic, Z.; Markovic, N. Grape seed polyphenols and fatty acids of autochthonous Prokupac vine variety from Serbia. Chem. Biodivers. 2019, 16, e1900053. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Martin, M.I.; Revilla, M.I.; Vivar-Quintana, A.M.; Betances Salcedo, E.V. Pesticide residues in propolis from Spain and Chile. An. approach using near infrared spectroscopy. Talanta 2017, 165, 533–539. [Google Scholar] [CrossRef]
- Ciampa, A.; Dell’Abate, M.T.; Masetti, O.; Valentini, M.; Sequi, P. Seasonal chemical–physical changes of PGI Pachino cherry tomatoes detected by magnetic resonance imaging (MRI). Food Chem. 2010, 122, 1253–1260. [Google Scholar] [CrossRef]
- Harada, T.; Yuba-Kubo, A.; Sugiura, Y.; Zaima, N.; Hayasaka, T.; Goto-Inoue, N.; Wakui, M.; Suematsu, M.; Takeshita, K.; Ogawa, K.; et al. Visualization of volatile substances in different organelles with an atmospheric-pressure mass microscope. Anal. Chem. 2009, 81, 9153–9157. [Google Scholar] [CrossRef]
- Burrell, M.; Earnshaw, C.; Clench, M. Imaging matrix assisted laser desorption ionization mass spectrometry: A technique to map plant metabolites within tissues at high spatial resolution. J. Exp. Bot. 2007, 58, 757–763. [Google Scholar] [CrossRef]
- Mullen, A.K.; Clench, M.R.; Crosland, S.; Sharples, K.R. Determination of agrochemical compounds in soya plants by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 2507–2516. [Google Scholar] [CrossRef]
- Enomoto, H.; Takeda, S.; Hatta, H.; Zaima, N. Tissue-specific distribution of sphingomyelin species in pork chop revealed by matrix-assisted laser desorption/ionization-imaging mass spectrometry. J. Food Sci. 2019, 84, 1758–1763. [Google Scholar] [CrossRef]
- Horn, P.J.; Silva, J.E.; Anderson, D.; Fuchs, J.; Borisjuk, L.; Nazarenus, T.J.; Shulaev, V.; Cahoon, E.B.; Chapman, K.D. Imaging heterogeneity of membrane and storage lipids in transgenic Camelina sativa seeds with altered fatty acid profiles. Plant. J. 2013, 76, 138–150. [Google Scholar]
- Fuchs, B.; Bischoff, A.; Suss, R.; Teuber, K.; Schurenberg, M.; Suckau, D.; Schiller, J. Phosphatidylcholines and -ethanolamines can be easily mistaken in phospholipid mixtures: A negative ion MALDI-TOF MS study with 9-aminoacridine as matrix and egg yolk as selected example. Anal. Bioanal. Chem. 2009, 395, 2479–2487. [Google Scholar] [CrossRef] [PubMed]
- Horn, P.J.; James, C.N.; Gidda, S.K.; Kilaru, A.; Dyer, J.M.; Mullen, R.T.; Ohlrogge, J.B.; Chapman, K.D. Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol. 2013, 162, 1926–1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chansela, P.; Goto-Inoue, N.; Zaima, N.; Sroyraya, M.; Sobhon, P.; Setou, M. Visualization of neuropeptides in paraffin-embedded tissue sections of the central nervous system in the decapod crustacean, Penaeus monodon, by imaging mass spectrometry. Peptides 2012, 34, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Groseclose, M.R.; Andersson, M.; Hardesty, W.M.; Caprioli, R.M. Identification of proteins directly from tissue: In situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom. 2007, 42, 254–262. [Google Scholar] [CrossRef]
- Rubakhin, S.S.; Li, L.; Moroz, T.P.; Sweedler, J.V. Characterization of the Aplysia californica cerebral ganglion F cluster. J Neurophysiol. 1999, 81, 1251–1260. [Google Scholar] [CrossRef] [Green Version]
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. [Google Scholar] [CrossRef]
- Stoeckli, M.; Chaurand, P.; Hallahan, D.E.; Caprioli, R.M. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 2001, 7, 493–496. [Google Scholar] [CrossRef]
- Aubagnac, J.L.; Enjalbal, C.; Drouot, C.; Combarieu, R.; Martinez, J. Imaging time-of-flight secondary ion mass spectrometry of solid-phase peptide syntheses. J. Mass Spectrom. 1999, 34, 749–754. [Google Scholar] [CrossRef]
- Trim, P.J.; Henson, C.M.; Avery, J.L.; McEwen, A.; Snel, M.F.; Claude, E.; Marshall, P.S.; West, A.; Princivalle, A.P.; Clench, M.R. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal. Chem. 2008, 80, 8628–8634. [Google Scholar] [CrossRef]
- Goodwin, R.J.; Scullion, P.; Macintyre, L.; Watson, D.G.; Pitt, A.R. Use of a solvent-free dry matrix coating for quantitative matrix-assisted laser desorption ionization imaging of 4-bromophenyl-1,4-diazabicyclo(3.2.2)nonane-4-carboxylate in rat brain and quantitative analysis of the drug from laser microdissected tissue regions. Anal. Chem. 2010, 82, 3868–3873. [Google Scholar]
- Chaurand, P.; Rahman, M.A.; Hunt, T.; Mobley, J.A.; Gu, G.; Latham, J.C.; Caprioli, R.M.; Kasper, S. Monitoring mouse prostate development by profiling and imaging mass spectrometry. Mol. Cell Proteom. 2008, 7, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, Y.; Goto-Inoue, N.; Moriyama, T.; Zaima, N. Significant advancement of mass spectrometry imaging for food chemistry. Food Chem. 2016, 210, 200–211. [Google Scholar] [CrossRef]
- Goto-Inoue, N.; Setou, M.; Zaima, N. Visualization of spatial distribution of gamma-aminobutyric acid in eggplant (Solanum melongena) by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Sci. 2010, 26, 821–825. [Google Scholar] [CrossRef] [Green Version]
- Enomoto, H.; Sato, K.; Miyamoto, K.; Ohtsuka, A.; Yamane, H. Distribution analysis of anthocyanins, sugars, and organic acids in strawberry fruits using matrix-assisted laser desorption/ionization-imaging mass spectrometry. J. Agric. Food Chem. 2018, 66, 4958–4965. [Google Scholar] [CrossRef]
- Taira, S.; Shimma, S.; Osaka, I.; Kaneko, D.; Ichiyanagi, Y.; Ikeda, R.; Konishi-Kawamura, Y.; Zhu, S.; Tsuneyama, K.; Komatsu, K. Mass spectrometry imaging of the capsaicin localization in the capsicum fruits. Int. J. Biotechnol. Wellness Ind. 2012, 1, 61–65. [Google Scholar] [CrossRef]
- Agar, N.Y.; Golby, A.J.; Ligon, K.L.; Norton, I.; Mohan, V.; Wiseman, J.M.; Tannenbaum, A.; Jolesz, F.A. Development of stereotactic mass spectrometry for brain tumor surgery. Neurosurgery 2011, 68, 280–289. [Google Scholar] [CrossRef] [Green Version]
- Eberlin, L.S.; Dill, A.L.; Costa, A.B.; Ifa, D.R.; Cheng, L.; Masterson, T.; Koch, M.; Ratliff, T.L.; Cooks, R.G. Cholesterol sulfate imaging in human prostate cancer tissue by desorption electrospray ionization mass spectrometry. Anal. Chem. 2010, 82, 3430–3434. [Google Scholar] [CrossRef] [Green Version]
- Masaki, N.; Ishizaki, I.; Hayasaka, T.; Fisher, G.L.; Sanada, N.; Yokota, H.; Setou, M. Three-Dimensional image of cleavage bodies in nuclei is configured using gas cluster ion beam with time-of-flight secondary ion. mass spectrometry. Sci. Rep. 2015, 5, 10000. [Google Scholar] [CrossRef] [Green Version]
- Takats, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef] [Green Version]
- Eberlin, L.S.; Ferreira, C.R.; Dill, A.L.; Ifa, D.R.; Cheng, L.; Cooks, R.G. Nondestructive, histologically compatible tissue imaging by desorption electrospray ionization mass spectrometry. ChemBioChem 2011, 12, 2129–2132. [Google Scholar] [CrossRef]
- Enomoto, H.; Sensu, T.; Sato, K.; Sato, F.; Paxton, T.; Yumoto, E.; Miyamoto, K.; Asahina, M.; Yokota, T.; Yamane, H. Visualisation of abscisic acid and 12-oxo-phytodienoic acid in immature Phaseolus vulgaris L. seeds using desorption electrospray ionisation-imaging mass spectrometry. Sci. Rep. 2017, 7, 42977. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Fu, X.; Zhou, H.; Rao, W.; Zeng, L.; Yang, Z. Visualized analysis of within-tissue spatial distribution of specialized metabolites in tea (Camellia sinensis) using desorption electrospray ionization imaging mass spectrometry. Food Chem. 2019, 292, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shrestha, B.; Vertes, A. Atmospheric pressure molecular imaging by infrared MALDI mass spectrometry. Anal. Chem. 2007, 79, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.; Warburton, K.; Seymour, M.; Clench, M.; Thomas-Oates, J. Localization of water-soluble carbohydrates in wheat stems using imaging matrix-assisted laser desorption ionization mass spectrometry. N. Phytol. 2007, 173, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Sroyraya, M.; Goto-Inoue, N.; Zaima, N.; Hayasaka, T.; Chansela, P.; Tanasawet, S.; Shrivas, K.; Sobhon, P.; Setou, M. Visualization of biomolecules in the eyestalk of the blue swimming crab, Portunus pelagicus, by imaging mass spectrometry using the atmospheric-pressure mass microscope. Surf. Interface Anal. 2010, 42, 1589–1592. [Google Scholar] [CrossRef]
- Zaima, N.; Goto-Inoue, N.; Hayasaka, T.; Setou, M. Application of imaging mass spectrometry for the analysis of Oryza sativa rice. Rapid Commun. Mass Spectrom. 2010, 24, 2723–2729. [Google Scholar] [CrossRef]
- Zaima, N.; Goto-Inoue, N.; Hayasaka, T.; Enomoto, H.; Setou, M. Authenticity assessment of beef origin by principal component analysis of matrix-assisted laser desorption/ionization mass spectrometric data. Anal. Bioanal. Chem. 2011, 400, 1865–1871. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Zaima, N.; Moriyama, T.; Kawamura, Y. Different localization patterns of anthocyanin species in the pericarp of black rice revealed by imaging mass spectrometry. PLoS ONE 2012, 7, e31285. [Google Scholar] [CrossRef]
- Hashizaki, R.; Komori, H.; Kazuma, K.; Konno, K.; Kawabata, K.; Kaneko, D.; Katano, H.; Taira, S. Localization analysis of natural toxin of Solanum tuberosum L. via mass spectrometric imaging. Int. J. Biotechnol. Well. Ind. 2016, 5, 1–5. [Google Scholar]
- Velickovic, D.; Saulnier, L.; Lhomme, M.; Damond, A.; Guillon, F.; Rogniaux, H. Mass Spectrometric Imaging of Wheat (Triticum spp.) and Barley (Hordeum vulgare L.) Cultivars: Distribution of major cell wall polysaccharides according to their main structural features. J. Agric. Food Chem. 2016, 64, 6249–6256. [Google Scholar] [CrossRef]
- Nakamura, J.; Morikawa-Ichinose, T.; Fujimura, Y.; Hayakawa, E.; Takahashi, K.; Ishii, T.; Miura, D.; Wariishi, H. Spatially resolved metabolic distribution for unraveling the physiological change and responses in tomato fruit using matrix-assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI). Anal. Bioanal. Chem. 2017, 409, 1697–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiono, K.; Hashizaki, R.; Nakanishi, T.; Sakai, T.; Yamamoto, T.; Ogata, K.; Harada, K.I.; Ohtani, H.; Katano, H.; Taira, S. Multi-imaging of cytokinin and abscisic acid on the roots of rice (Oryza sativa) using matrix-assisted laser desorption/ionization mass spectrometry. J. Agric. Food Chem. 2017, 65, 7624–7628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekelof, M.; McMurtrie, E.K.; Nazari, M.; Johanningsmeier, S.D.; Muddiman, D.C. Direct analysis of triterpenes from high.-salt fermented cucumbers using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI). J. Am. Soc. Mass Spectrom. 2017, 28, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Feenstra, A.D.; Alexander, L.E.; Song, Z.; Korte, A.R.; Yandeau-Nelson, M.D.; Nikolau, B.J.; Lee, Y.J. Spatial mapping and profiling of metabolite distributions during germination. Plant Physiol 2017, 174, 2532–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodfield, H.K.; Sturtevant, D.; Borisjuk, L.; Munz, E.; Guschina, I.A.; Chapman, K.; Harwood, J.L. Spatial and temporal mapping of key lipid species in Brassica napus seeds. Plant Physiol. 2017, 173, 1998–2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto-Inoue, N.; Sato, T.; Morisasa, M.; Igarashi, Y.; Mori, T. Characterization of Metabolite compositions in wild and farmed red sea bream (Pagrus major) using mass spectrometry imaging. J. Agric. Food Chem. 2019, 67, 7197–7203. [Google Scholar] [CrossRef] [PubMed]
- Fanuel, M.; Ropartz, D.; Guillon, F.; Saulnier, L.; Rogniaux, H. Distribution of cell wall hemicelluloses in the wheat grain endosperm: A 3D perspective. Planta 2018, 248, 1505–1513. [Google Scholar] [CrossRef]
- Resetar Maslov, D.; Svirkova, A.; Allmaier, G.; Marchetti-Deschamann, M.; Kraljevic Pavelic, S. Optimization of MALDI-TOF mass spectrometry imaging for the visualization and comparison of peptide distributions in dry-cured ham muscle fibers. Food Chem. 2019, 283, 275–286. [Google Scholar] [CrossRef]
- Horikawa, K.; Hirama, T.; Shimura, H.; Jitsuyama, Y.; Suzuki, T. Visualization of soluble carbohydrate distribution in apple fruit flesh utilizing MALDI-TOF MS imaging. Plant Sci. 2019, 278, 107–112. [Google Scholar] [CrossRef]
- Bednarz, H.; Roloff, N.; Niehaus, K. Mass spectrometry imaging of the spatial and temporal localization of alkaloids in Nightshades. J. Agric. Food Chem. 2019. [Google Scholar] [CrossRef]
- Jehl, B.; Bauer, R.; Dorge, A.; Rick, R. The use of propane/isopentane mixtures for rapid freezing of biological specimens. J. Microsc. 1981, 123, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Ly, A.; Longuespee, R.; Casadonte, R.; Wandernoth, P.; Schwamborn, K.; Bollwein, C.; Marsching, C.; Kriegsmann, K.; Hopf, C.; Weichert, W.; et al. Site-to-site reproducibility and spatial resolution in MALDI-MSI of peptides from formalin-fixed paraffin-embedded samples. Proteom. Clin. Appl. 2019, 13, e1800029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.A.; Reyzer, M.L.; Caprioli, R.M. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: Practical aspects of sample preparation. J. Mass Spectrom. 2003, 38, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Stoeckli, M.; Staab, D.; Schweitzer, A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int. J. Mass Spectrom. 2007, 260, 195–202. [Google Scholar] [CrossRef]
- Khatib-Shahidi, S.; Andersson, M.; Herman, J.L.; Gillespie, T.A.; Caprioli, R.M. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem. 2006, 78, 6448–6456. [Google Scholar] [CrossRef]
- Kawamoto, T. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch. Histol. Cytol. 2003, 66, 123–143. [Google Scholar] [CrossRef] [Green Version]
- Fujino, Y.; Minamizaki, T.; Yoshioka, H.; Okada, M.; Yoshiko, Y. Imaging and mapping of mouse bone using MALDI-imaging mass spectrometry. Bone Rep. 2016, 5, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Seeley, E.H.; Wilson, K.J.; Yankeelov, T.E.; Johnson, R.W.; Gore, J.C.; Caprioli, R.M.; Matrisian, L.M.; Sterling, J.A. Co-registration of multi-modality imaging allows for comprehensive analysis of tumor-induced bone disease. Bone 2014, 61, 208–216. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, R.; Hashimoto, K.; Toyooka, K.; Saito, K. Keeping the shape of plant tissue for visualizing metabolite features in segmentation and correlation analysis of imaging mass spectrometry in Asparagus officinalis. Metabolomics 2019, 15, 24. [Google Scholar] [CrossRef] [Green Version]
- Enthaler, B.; Bussmann, T.; Pruns, J.K.; Rapp, C.; Fischer, M.; Vietzke, J.P. Influence of various on-tissue washing procedures on the entire protein quantity and the quality of matrix-assisted laser desorption/ionization spectra. Rapid Commun. Mass Spectrom. 2013, 27, 878–884. [Google Scholar] [CrossRef]
- Shimma, S.; Furuta, M.; Ichimura, K.; Yoshida, Y.; Setou, M. Direct MS/MS analysis in mammalian tissue sections using MALDI-QIT-TOFMS and chemical inkjet technology. Surf. Interface Anal. 2006, 38, 1712–1714. [Google Scholar] [CrossRef]
- Heijs, B.; Carreira, R.J.; Tolner, E.A.; de Ru, A.H.; van den Maagdenberg, A.M.; van Veelen, P.A.; McDonnell, L.A. Comprehensive analysis of the mouse brain proteome sampled in mass spectrometry imaging. Anal. Chem. 2015, 87, 1867–1875. [Google Scholar] [CrossRef] [PubMed]
- Enthaler, B.; Trusch, M.; Fischer, M.; Rapp, C.; Pruns, J.K.; Vietzke, J.P. MALDI imaging in human skin tissue sections: Focus on various matrices and enzymes. Anal. Bioanal. Chem. 2013, 405, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Powers, T.W.; Jones, E.E.; Betesh, L.R.; Romano, P.R.; Gao, P.; Copland, J.A.; Mehta, A.S.; Drake, R.R. Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal. Chem. 2013, 85, 9799–9806. [Google Scholar] [CrossRef] [Green Version]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Adamczyk, B.; Tharmalingam, T.; Rudd, P.M. Glycans as cancer biomarkers. Biochim. Biophys. Acta 2012, 1820, 1347–1353. [Google Scholar] [CrossRef]
- Powers, T.W.; Neely, B.A.; Shao, Y.; Tang, H.; Troyer, D.A.; Mehta, A.S.; Haab, B.B.; Drake, R.R. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS ONE 2014, 9, e106255. [Google Scholar] [CrossRef]
- Bagley, M.C.; Stepanova, A.N.; Ekelof, M.; Alonso, J.M.; Muddiman, D.C. Development of a relative quantification method for IR-MALDESI mass spectrometry imaging of Arabidopsis root seedlings. Rapid Commun. Mass Spectrom. 2019. [Google Scholar] [CrossRef]
- Shariatgorji, M.; Nilsson, A.; Goodwin, R.J.; Kallback, P.; Schintu, N.; Zhang, X.; Crossman, A.R.; Bezard, E.; Svenningsson, P.; Andren, P.E. Direct targeted quantitative molecular imaging of neurotransmitters in brain tissue sections. Neuron 2014, 84, 697–707. [Google Scholar] [CrossRef] [Green Version]
- Karas, M.; Hillenkamp, F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988, 60, 2299–2301. [Google Scholar] [CrossRef]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T.; Matsuo, T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Chughtai, K.; Heeren, R.M. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 2010, 110, 3237–3277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaletas, B.K.; van der Wiel, I.M.; Stauber, J.; Lennard, J.D.; Guzel, C.; Kros, J.M.; Luider, T.M.; Heeren, R.M. Sample preparation issues for tissue imaging by imaging MS. Proteomics 2009, 9, 2622–2633. [Google Scholar] [CrossRef] [PubMed]
- Agar, N.Y.; Yang, H.W.; Carroll, R.S.; Black, P.M.; Agar, J.N. Matrix solution fixation: Histology-compatible tissue preparation for MALDI mass spectrometry imaging. Anal. Chem. 2007, 79, 7416–7423. [Google Scholar] [CrossRef]
- Gemperline, E.; Rawson, S.; Li, L. Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal. Chem. 2014, 86, 10030–10035. [Google Scholar] [CrossRef] [Green Version]
- Shimma, S.; Sugiura, Y. effective sample preparations in imaging mass spectrometry. Mass Spectrom. 2014, 3, S0029. [Google Scholar] [CrossRef] [Green Version]
- Goto-Inoue, N.; Sato, T.; Morisasa, M.; Kashiwagi, A.; Kashiwagi, K.; Sugiura, Y.; Sugiyama, E.; Suematsu, M.; Mori, T. Utilizing mass spectrometry imaging to map the thyroid hormones triiodothyronine and thyroxine in Xenopus tropicalis tadpoles. Anal. Bioanal. Chem. 2018, 410, 1333–1340. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Y.; Zhou, D.; Li, Z. Electric field-assisted matrix coating method enhances the detection of small molecule metabolites for mass spectrometry imaging. Anal. Chem. 2015, 87, 5860–5865. [Google Scholar] [CrossRef]
- Fowble, K.L.; Okuda, K.; Cody, R.B.; Musah, R.A. Spatial distributions of furan and 5-hydroxymethylfurfural in unroasted and roasted Coffea arabica beans. Food Res. Int. 2019, 119, 725–732. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Z.; Felder, M.A.; Yu, Q.; Shi, X.; Peng, Y.; Cao, Q.; Wang, B.; Puglielli, L.; Patankar, M.S.; et al. Mass spectrometry imaging of N-glycans from formalin-fixed paraffin-embedded tissue sections using a novel subatmospheric pressure ionization source. Anal. Chem. 2019, 91, 12942–12947. [Google Scholar] [CrossRef]
- Niehaus, M.; Soltwisch, J.; Belov, M.E.; Dreisewerd, K. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat. Methods 2019, 16, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Klein, O.; Strohschein, K.; Nebrich, G.; Fuchs, M.; Thiele, H.; Giavalisco, P.; Duda, G.N.; Winkler, T.; Kobarg, J.H.; Trede, D.; et al. Unraveling local tissue changes within severely injured skeletal muscles in response to MSC-based intervention using MALDI Imaging mass spectrometry. Sci. Rep. 2018, 8, 12677. [Google Scholar] [CrossRef] [PubMed]
- Paine, M.R.L.; Liu, J.; Huang, D.; Ellis, S.R.; Trede, D.; Kobarg, J.H.; Heeren, R.M.A.; Fernandez, F.M.; MacDonald, T.J. Three-dimensional mass spectrometry imaging identifies lipid markers of medulloblastoma metastasis. Sci. Rep. 2019, 9, 2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schramm, T.; Hester, Z.; Klinkert, I.; Both, J.P.; Heeren, R.M.A.; Brunelle, A.; Laprevote, O.; Desbenoit, N.; Robbe, M.F.; Stoeckli, M.; et al. ImzML—A common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteom. 2012, 75, 5106–5110. [Google Scholar] [CrossRef] [PubMed]
| Sample | Target Molecules | Sample Preparation Sample Type Thickness Embedding | Matrices | Reference |
|---|---|---|---|---|
| Soya leaf, stem | Mesotorione, azoxystrobin | Freeze-drying - - | CHCA | [8] |
| Strawberry fruit skin | Sucrose, fructose, glucose, citric acid | Fresh 0.2–0.5 mm with a sharp utility knife - | DHB | [33] |
| Wheat grain | Glucose-6-phosphate, sucrose | Frozen - Ice | CHCA | [7] |
| Wheat stem | Oligosaccharides | Freeze-drying 50 μm - | CHCA | [34] |
| Ginger rhizome (Zingiber officinale) | 6-gingerol, monoterpene | Fresh 0.2 mm - | - | [6] |
| Eggplant | GABA, nicotinic acid, arginine, 2-aminobenzoic acid, citric acid, saccharides | Frozen 14 μm - | DHB | [23] |
| Blue swimming crab (Portunus pelagicus) | Phospholipids, triacylglycerols | Frozen 14 μm 2% CMC | DHB | [35] |
| Rice seed | Phospholipids, α-tocopherol, arginine, ɤ-oryzanol, phytic acid | Frozen 8 μm with adhesive film (Kawamoto method) 2% CMC | DHB | [36] |
| Beef meat | Lipids | Frozen 8 μm - | DHB | [37] |
| Penaeus monodon | Neuropeptides | Frozen 5 μm Paraffin | CHCA | [13] |
| Capsicum annuum | Capsaicin | Frozen 70 μm - | CHCA | [25] |
| Black rice seed | Lysophosphatidylcholine, phosphatidylcholine, anthocyanins | Frozen 10 μm with adhesive film (Kawamoto method) 2% CMC | DHB | [38] |
| Camelina sativa seed transgenic | Lipids | Frozen 30–50 μm 10% gelatin | DHB | [10] |
| Potato (Solanum tuberosum L.) | α-solanine, α-chaconine | Frozen - - | CHCA | [39] |
| Wheat (Triticum aestivum L.) | Polysaccharides | Frozen 60 μm - | DHB | [40] |
| Tomato fruit (S. lycopersicum L.) | Organic acid, amino acid nucleotides, caffeic acid | Frozen 10 μm OCT compound | DHB, 9-AA | [41] |
| Rice (Oryza sativa L.) | Cytokinin, abscisic acid | Frozen 50 μm Ice | CHCA | [42] |
| Cucumber | Triterpenes | Frozen 50 μm - | - | [43] |
| Maize seed (Zea mays) | Triacylglycerols, amino acids | Frozen 10 μm - | DAN, DHB, 9-AA | [44] |
| Oilseed rape (Brassica napus) | Lipids | Frozen 30 μm - | DHB | [45] |
| Strawberry | Anthocyanins, sugars, organic acids | Frozen 80 μm 2% CMC | DHB | [24] |
| Red sea bream (Pagrus major) | Lipids | Frozen 15 μm - | DHB | [46] |
| Grain (Triticum aestivum L.) | Hemicelluloses | Frozen 80 μm - | DMA, DHB | [47] |
| Ham | Peptide | Frozen 12 μm - | CHCA | [48] |
| Apple | Soluble carbohydrate | Fresh 20 μm - | CHCA, DHB | [49] |
| Nightshades | Alkaloids | Frozen 40 μm Ice | DHB | [50] |
| Pork chop | Lipids | Frozen 10 μm - | CHCA, DHB | [9] |
| Matrices | Sample |
|---|---|
| 9-Aminoacridine (9-AA) | lipids, metabolites |
| Sinapinic acid (SA) | peptides, proteins |
| Nicotinic acid (NA) | nucleotide |
| 2,5-Dihydroxybenzoin acid (DHB) | lipids, glycopeptide, polymer |
| 3-Amino-4-hydroxybezoic acid (AHBA) | glycan |
| α-Cyano-4-hydroxycinnamic acid (CHCA) | peptides, proteins |
| 1,5-Diaminonapthalene (DAN) | lipids |
| t3-Indolacrylic acid (IAA) | polymer, aromatic |
| 2-(4-Hydroxyphenylazo)-benzoic acid (HABA) | polymer |
| 3-Aminoquinoline (3AQ) | glycan |
| Picolinic acid (PA) | nucleotide |
| Anthranilic acid (ANA) | nucleotide |
| 3-Hydroxypicolinic acid (3HPA) | nucleotide |
| 5-Chlorosalycilic acid (5CSA) | polymer |
| Dihydroxyacetone phosphate (DHAP) | lipids, glycan |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morisasa, M.; Sato, T.; Kimura, K.; Mori, T.; Goto-Inoue, N. Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis. Foods 2019, 8, 633. https://doi.org/10.3390/foods8120633
Morisasa M, Sato T, Kimura K, Mori T, Goto-Inoue N. Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis. Foods. 2019; 8(12):633. https://doi.org/10.3390/foods8120633
Chicago/Turabian StyleMorisasa, Mizuki, Tomohiko Sato, Keisuke Kimura, Tsukasa Mori, and Naoko Goto-Inoue. 2019. "Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis" Foods 8, no. 12: 633. https://doi.org/10.3390/foods8120633
APA StyleMorisasa, M., Sato, T., Kimura, K., Mori, T., & Goto-Inoue, N. (2019). Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis. Foods, 8(12), 633. https://doi.org/10.3390/foods8120633




