Efficacy of Commercial Sanitizers Used in Food Processing Facilities for Inactivation of Listeria monocytogenes, E. Coli O157:H7, and Salmonella Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Growth of Enhanced Biofilms in Microplates
2.3. Washing Biofilms Generated in Microplates
2.4. Enzymatic Detachment of Adhered Cells from Microplates for Enumeration
2.5. Sanitizers Used in the Microplate Biofilm Assay
2.6. Microplate Biofilm Sanitizer Assay
2.7. Scanning Electron Microscopy (SEM) of Biofilms
2.8. Statistical Analysis
3. Results
3.1. Sanitizer Biofilm Microplate Assays vs. L. monocytogenes, Salmonella Montevideo, and E. coli O157:H7
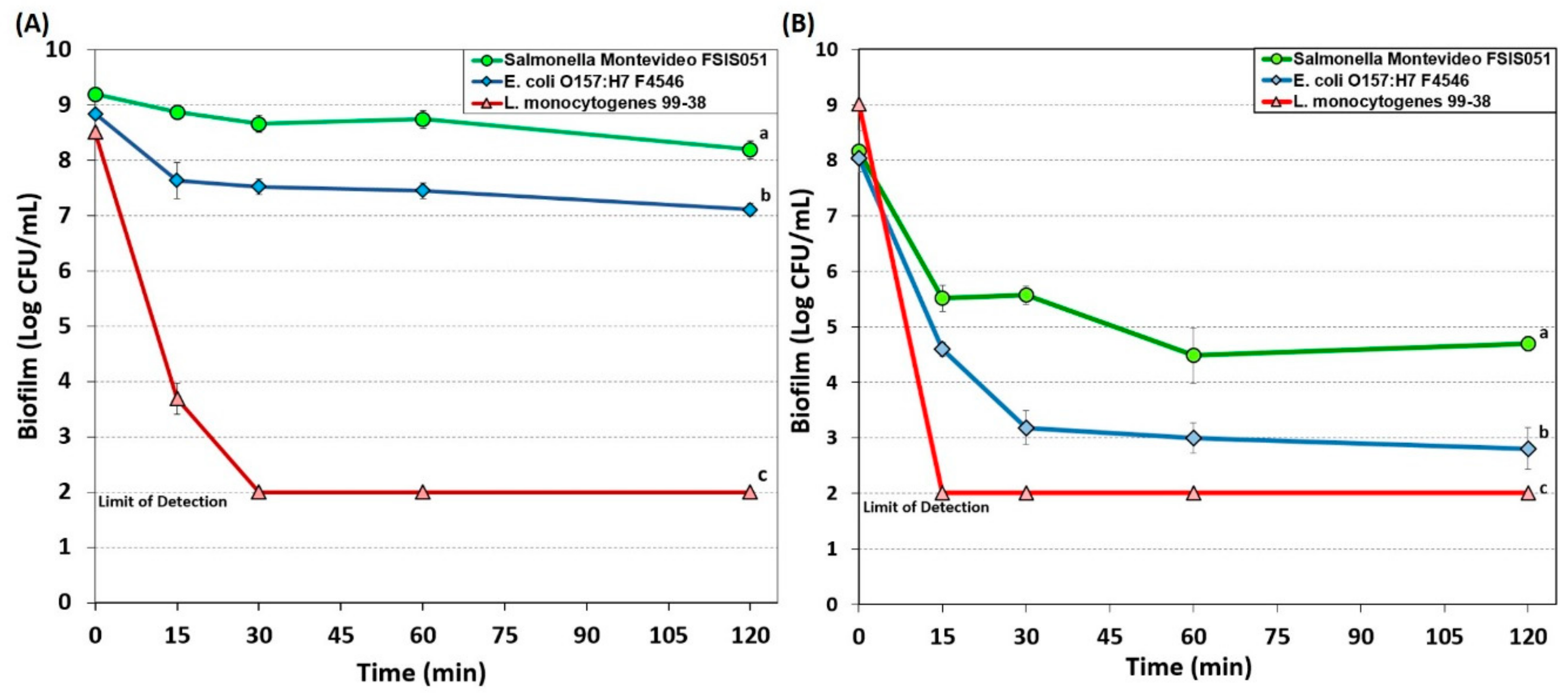
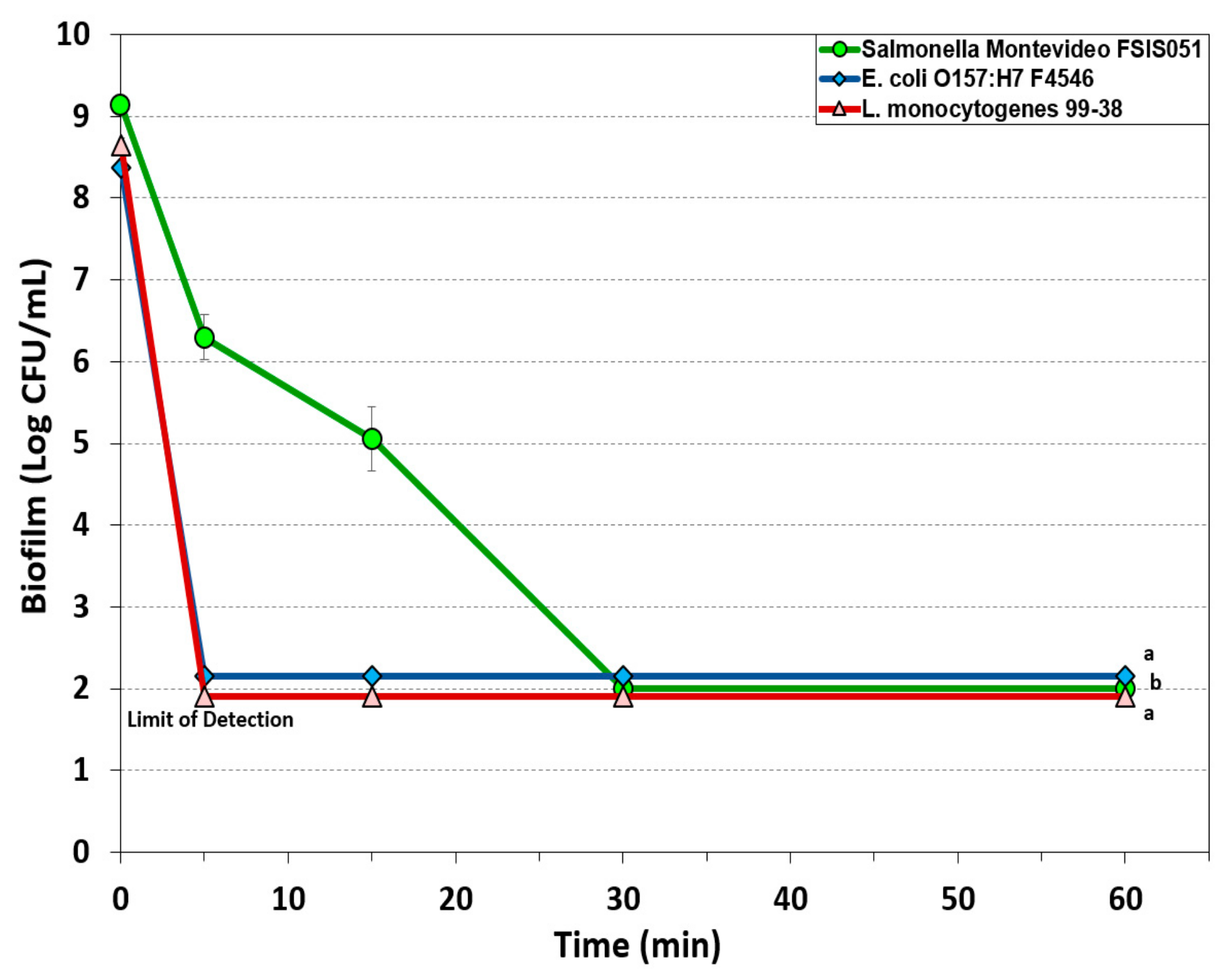
3.1.1. Hypochlorite-Based Sanitizer
3.1.2. Simple Quaternary Ammonium Chloride-Based Sanitizer
3.1.3. Peroxyacetic Acid-Based Sanitizer
3.1.4. New Generation Quaternary Ammonium Chloride-Based Sanitizers
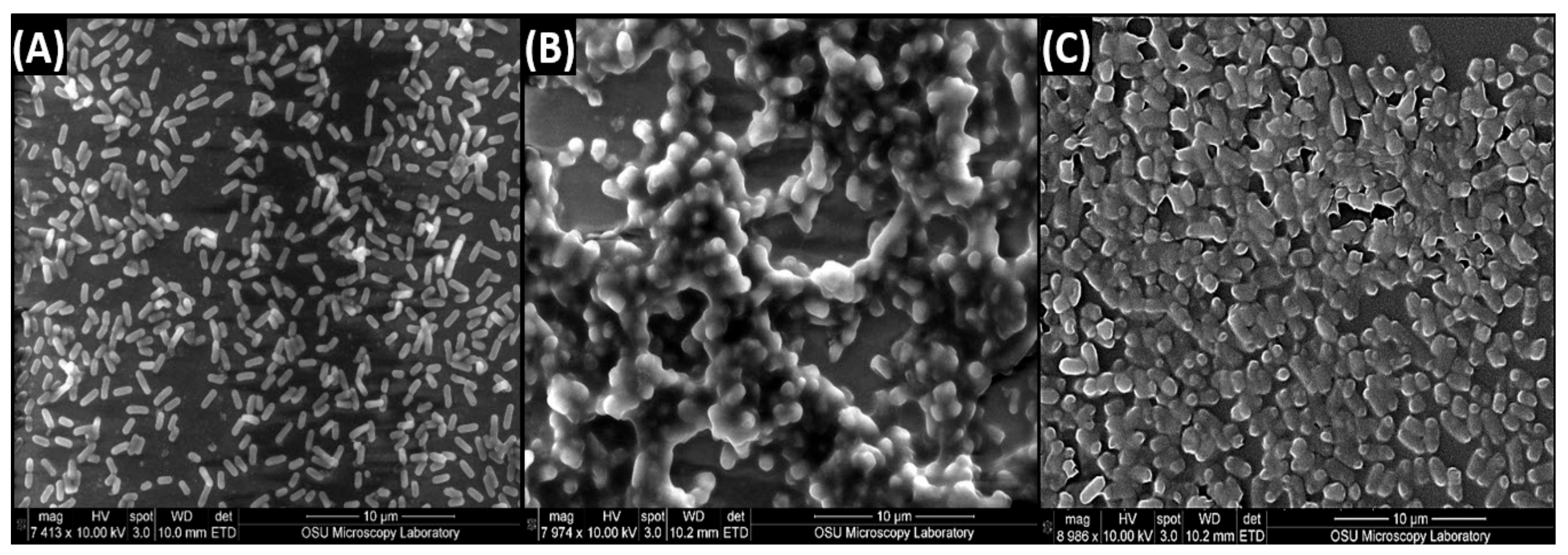
3.2. Scanning Electron Microscopy of Biofilms of L. monocytogenes 99-38, E. coli O157:H7 F4546, and S. Montevideo FSIS 051
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garvey, M. Food pollution: A comprehensive review of chemical and biological sources of food contamination and impact on human health. Nutrire 2019, 44, 1. [Google Scholar] [CrossRef]
- Jung, Y.; Jang, H.; Matthews, K.R. Effect of the food production chain from farm practices to vegetable processing on outbreak incidence. Microb. Biotech. 2014, 7, 517–527. [Google Scholar] [CrossRef]
- Schlegelová, J.; Babák, V.; Holasová, M.; Konstantinová, L.; Necidová, L.; Šišák, F.; Vlková, H.; Roubal, P.; Jaglic, Z. Microbial contamination after sanitation of food contact surfaces in dairy and meat processing plants. Czech. J. Food Sci. 2010, 28, 450–461. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Abraham, J. Chapter 2-Microbial Contamination, Prevention, and Early Detection in Food Industry. In Microbial Contamination and Food Degradation; Holban, A.M., Grumezescu, A.M., Eds.; Academic Press: London, UK, 2018; pp. 21–47. [Google Scholar]
- Dewanti, R.; Wong, A.C.L. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int. J. Food Microbiol. 1995, 26, 147–164. [Google Scholar] [CrossRef]
- Sharma, M.; Anand, S.K. Characterization of constitutive microflora of biofilms in dairy processing lines. Food Microbiol. 2002, 19, 627–636. [Google Scholar] [CrossRef]
- Srey, S.; Jahid, I.K.; Ha, S.-D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Khangholi, M.; Jamalli, A. The effects of sugars on the biofilm formation of Escherichia coli 185p on stainless steel and polyethylene terephthalate surfaces in a laboratory model. Jundishapur J. Microbiol. 2016, 9, e40137. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Gonzalo, D.; Pagán, R. Influence of environmental factors on bacterial biofilm formation in the food industry: A review. Postdoc J. 2015, 3, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Feng, G.; Moraru, C.I. Micro- and nanotopography sensitive bacterial attachment mechanisms: review. Front. Microbiol. 2019, 10, e191. [Google Scholar] [CrossRef] [Green Version]
- Grinberg, M.; Orevi, T.; Kashtan, N. Bacterial surface colonization, preferential attachment and fitness under periodic stress. PLoS Comput. Biol. 2019, 15, e1006815. [Google Scholar] [CrossRef]
- Hinsa, S.M.; Espinosa-Urgel, M.; Ramos, J.L.; O’toole, G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003, 49, 905–918. [Google Scholar] [CrossRef]
- Wagner, S.; Hauck, D.; Hoffmann, M.; Sommer, R.; Joachim, I.; Müller, R.; Imberty, A.; Varrot, A.; Titz, A. Covalent lectin inhibition and application in bacterial biofilm imaging. Angew. Chem. Int. Ed. 2017, 56, 16559–16564. [Google Scholar] [CrossRef] [Green Version]
- Beloin, C.; Roux, A.; Ghigo, J.M. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 249–289. [Google Scholar]
- Chmielewski, R.A.N.; Frank, J.F. Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32. [Google Scholar] [CrossRef]
- Sinde, E.; Carballo, J. Attachment of Salmonella spp. and Listeria monocytogenes to stainless steel, rubber and polytetrafluorethylene: The influence of free energy and the effect of commercial sanitizers. Food Microbiol. 2000, 17, 439–447. [Google Scholar] [CrossRef]
- Chia, T.W.R.; Goulter, R.M.; Mcmeekin, T.; Dykes, G.A.; Fegan, N. Attachment of different Salmonella serovars to materials commonly used in a poultry processing plant. Food Microbiol. 2009, 26, 853–859. [Google Scholar] [CrossRef]
- Storgards, E.; Simola, H.; Sjöberg, A.M.; Wirtanen, G. Hygiene of gasket materials used in food processing equipment Part 2: Aged materials. Food Bioprod. Process. 1999, 77, 146–155. [Google Scholar] [CrossRef]
- Gamble, R.; Muriana, P.M. Microplate fluorescence assay for measurement of the ability of strains of Listeria monocytogenes from meat and meat-processing plants to adhere to abiotic surfaces. Appl. Environ. Microbiol. 2007, 73, 5235–5244. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Zhu, X. Biofilm formation and food safety in food industries. Trends Foodsci. Technol. 2009, 20, 407–413. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef] [Green Version]
- Hood, S.K.; Zottola, E.A. Biofilms in food processing. Food Control 1995, 6, 9–18. [Google Scholar] [CrossRef]
- Kumar, C.G.; Anand, S.K. Significance of microbial biofilms in food industry: A review. Int. J. Food Microbiol. 1998, 42, 9–27. [Google Scholar] [CrossRef]
- Van Houdt, R.; Michiels, C.W. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 2005, 156, 626–633. [Google Scholar] [CrossRef]
- Bremer, P.J.; Monk, I.; Butler, R. Inactivation of Listeria monocytogenes/Flavobacterium spp. biofilms using chlorine: Impact of substrate, pH, time and concentration. Lett. Appl. Microbiol. 2002, 35, 321–325. [Google Scholar] [CrossRef]
- Kuda, T.; Yano, T.; Kuda, M.T. Resistances to benzalkonium chloride of bacteria dried with food elements on stainless steel surface. LWT-Food Sci. Technol. 2008, 41, 988–993. [Google Scholar] [CrossRef]
- Pan, Y.; Breidt, F., Jr.; Kathariou, S. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 2006, 72, 7711–7717. [Google Scholar] [CrossRef] [Green Version]
- Lajhar, S.A.; Brownlie, J.; Barlow, R. Characterization of biofilm-forming capacity and resistance to sanitizers of a range of E. coli O26 pathotypes from clinical cases and cattle in Australia. BMC Microbiol. 2018, 18, 41. [Google Scholar] [CrossRef] [Green Version]
- Simões, M.; Simões, L.C.; Vieira, M.J. A review of current and emergent biofilm control strategies. LWT-Food Sci. Technol. 2010, 43, 573–583. [Google Scholar] [CrossRef] [Green Version]
- Donaghy, J.A.; Jagadeesan, B.; Goodburn, K.; Grunwald, L.; Jensen, O.N.; Jespers, A.D.; Kanagachandran, K.; Lafforgue, H.; Seefelder, W.; Quentin, M.-C. Relationship of sanitizers, disinfectants, and cleaning agents with antimicrobial resistance. J. Food Prot. 2019, 82, 889–902. [Google Scholar] [CrossRef]
- Gupta, P.; Bhatia, M.; Gupta, P.; Omar, B.J. Emerging biocide resistance among multidrug-resistant bacteria: Myth or reality? A pilot study. J. Pharm. Bioallied Sci. 2018, 10, 96–101. [Google Scholar]
- Kampf, G. Biocidal agents used for disinfection can enhance antibiotic resistance in Gram-negative species. Antibiotics 2018, 7, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampf, G. Antibiotic resistance can be enhanced in Gram-positive species by some biocidal agents used for disinfection. Antibiotics 2019, 8, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simões, M.; Bennett, R.N.; Rosa, E.A.S. Understanding antimicrobial activities of phytochemicals against multidrug resistant bacteria and biofilms. Nat. Prod. Rep. 2009, 26, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Flint, S.; Bremer, P.; Brooks, J.; Palmer, J.; Sadiq, F.A.; Seale, B.; Teh, K.H.; Wu, S.; Md Zain, S.N. Bacterial fouling in dairy processing. Int. Dairy J. 2020, 101, 104593. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Wang, R. Biofilms and meat safety: A mini-review. J. Food Prot. 2019, 82, 120–127. [Google Scholar] [CrossRef]
- Aryal, M.; Pranatharthiharan, P.; Muriana, P.M. Optimization of a microplate assay for generating Listeria monocytogenes, E. coli O157:H7, and Salmonella biofilms and enzymatic recovery for enumeration. Foods 2019, 8, 541. [Google Scholar] [CrossRef] [Green Version]
- Kushwaha, K.; Muriana, P.M. Adherence characteristics of Listeria strains isolated from three ready-to-eat meat processing plants. J. Food Prot. 2009, 72, 2125–2131. [Google Scholar] [CrossRef]
- Wirtanen, G.; Salo, S. Disinfection in food processing—Efficacy testing of disinfectants. Rev. Environ. Sci. Bio. 2003, 2, 293–306. [Google Scholar] [CrossRef]
- Hoenicke, K.; Gatermann, R.; Hartig, L.; Mandix, M.; Otte, S. Formation of semicarbazide (SEM) in food by hypochlorite treatment: Is SEM a specific marker for nitrofurazone abuse? Food Addit. Contam. 2004, 21, 526–537. [Google Scholar] [CrossRef]
- Cunningham, H.M.; Lawrence, G.A. Effect of exposure of meat and poultry too chlorinated water on the retention of chlorinated compounds and water. J. Food Sci. 1977, 42, 1504–1509. [Google Scholar] [CrossRef]
- Marriott, N.G.; Gravani, R.B. Sanitizers. In Principles of Food Sanitation; Springer US: Boston, MA, USA, 2006; pp. 165–189. [Google Scholar]
- Veasey, S.; Muriana, P.M. Evaluation of electrolytically-generated hypochlorous acid (’Electrolyzed Water’) for sanitation of meat and meat-contact surfaces. Foods 2016, 5, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerba, C.P. Quaternary ammonium biocides: Efficacy in application. Appl. Environ. Microbiol. 2015, 81, 464–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laopaiboon, L.; Hall, S.J.; Smith, R.N. The effect of a quaternary ammonium biocide on the performance and characteristics of laboratory-scale rotating biological contactors. J. Appl. Microbiol. 2002, 93, 1051–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcbain, A.J.; Ledder, R.G.; Moore, L.E.; Catrenich, C.E.; Gilbert, P. Effects of quaternary-ammonium-based formulations on bacterial community dynamics and antimicrobial susceptibility. Appl. Environ. Microbiol. 2004, 70, 3449–3456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.; Yang, J. Germicidal efficacy of sanitizers on food-borne bacteria and effect of sanitizes in CIP and SIP simulation. Eur. Food Res. Technol. 2013, 237, 265–274. [Google Scholar] [CrossRef]
- Kitis, M. Disinfection of wastewater with peracetic acid: A review. Environ. Int. 2004, 30, 47–55. [Google Scholar] [CrossRef]
- Mcdonnell, G.; Russell, A.D. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [Green Version]
- Toté, K.; Horemans, T.; Berghe, D.V.; Maes, L.; Cos, P. Inhibitory effect of biocides on the viable masses and matrices of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2010, 76, 3135–3142. [Google Scholar] [CrossRef] [Green Version]
- Ukuku, D.O. Effect of hydrogen peroxide treatment on microbial quality and appearance of whole and fresh-cut melons contaminated with Salmonella spp. Int. J. Food Microbiol. 2004, 95, 137–146. [Google Scholar] [CrossRef]
- Ma, Z.; Bumunang, E.W.; Stanford, K.; Bie, X.; Niu, Y.D.; Mcallister, T.A. Biofilm formation by shiga toxin-producing Escherichia coli on stainless steel coupons as affected by temperature and incubation time. Microorganisms 2019, 7, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Beltrame, C.A.; Kubiak, G.B.; Lerin, L.A.; Rottava, I.; Mossi, A.J.; Oliveira, D.D.; Cansian, R.L.; Treichel, H.; Toniazzo, G. Influence of different sanitizers on food contaminant bacteria: Effect of exposure temperature, contact time, and product concentration. Food Sci. Technol. 2012, 32, 228–232. [Google Scholar] [CrossRef]
- Cabeça, T.K.; Pizzolitto, A.C.; Pizzolitto, E.L. Activity of disinfectants against foodborne pathogens in suspension and adhered to stainless steel surfaces. Braz. J. Microbiol. 2012, 43, 1112–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meesilp, N.; Mesil, N. Effect of microbial sanitizers for reducing biofilm formation of Staphylococcus aureus and Pseudomonas aeruginosa on stainless steel by cultivation with UHT milk. Food Sci. Biotechnol. 2019, 28, 289–296. [Google Scholar] [CrossRef] [PubMed]



| Trade Name | Active Ingredients | Use Level | Source |
|---|---|---|---|
| Bi-Quat | Dimethyl ethylbenzyl ammonium chloride (5.1%); Alkyl dimethyl benzyl ammonium chloride (5.1%); Ethanol (1.1%) | 200 ppm, | Birko Corp. |
| 1000 ppm | |||
| 10-Chlor | Sodium hypochlorite (<20%); Sodium hydroxide (<5%) | 200 ppm, | Birko Corp. |
| 1000 ppm | |||
| Sterilex solution | 1.Ultra Disinfectant Cleaner: Hydrogen peroxide (5.5%–7.2%), Alykl dimethyl ethyl benzyl ammonium chloride (2.5%–3.5%), Alkyl (C12,C14,C16) dimethyl benzyl ammonium chloride (2.5%–3.5%) | 5%, 10% | Sterilex Corp. |
| 2. Ultra Activator Solution: Sodium carbonate (4%–8%); Potassium carbonate (4%–8%); Tetrasodium ethylenediaminetetraacetate (3%–7%) | |||
| KC-610 | Peroxyacetic acid (5%–6%), Hydrogen peroxide (25%–58%), Acetic acid (5%–10%) | 500 ppm | Packers Chemical |
| Decon7 solution | 1.Quaternary ammonium chloride | 5%, 10% | Decon7 Systems |
| Benzyl-C12-C16 Alkyl Di-methyl Chlorides (5.5%–6.5%); | |||
| 2. Hydrogen peroxide (<8%); | |||
| 3. Accelerant: Diacetin (30%–60%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aryal, M.; Muriana, P.M. Efficacy of Commercial Sanitizers Used in Food Processing Facilities for Inactivation of Listeria monocytogenes, E. Coli O157:H7, and Salmonella Biofilms. Foods 2019, 8, 639. https://doi.org/10.3390/foods8120639
Aryal M, Muriana PM. Efficacy of Commercial Sanitizers Used in Food Processing Facilities for Inactivation of Listeria monocytogenes, E. Coli O157:H7, and Salmonella Biofilms. Foods. 2019; 8(12):639. https://doi.org/10.3390/foods8120639
Chicago/Turabian StyleAryal, Manish, and Peter M. Muriana. 2019. "Efficacy of Commercial Sanitizers Used in Food Processing Facilities for Inactivation of Listeria monocytogenes, E. Coli O157:H7, and Salmonella Biofilms" Foods 8, no. 12: 639. https://doi.org/10.3390/foods8120639
APA StyleAryal, M., & Muriana, P. M. (2019). Efficacy of Commercial Sanitizers Used in Food Processing Facilities for Inactivation of Listeria monocytogenes, E. Coli O157:H7, and Salmonella Biofilms. Foods, 8(12), 639. https://doi.org/10.3390/foods8120639






