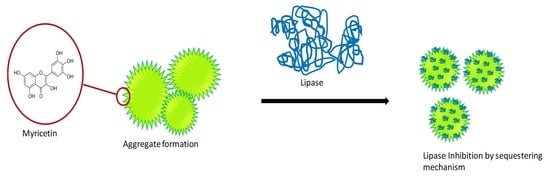
Interaction between Myricetin Aggregates and Lipase under Simplified Intestinal Conditions †
Abstract
:1. Introduction
2. Methodology
2.1. Chemical Compounds
2.2. Sample Preparation
2.3. Aggregate Formation
2.4. Myricetin–Lipase Interaction
2.4.1. Sample Treatment
2.4.2. AF4 Instrumentation
2.4.3. AF4 Method Parameters
2.4.4. AF4 Data Processing
2.5. Statistical Analysis
2.6. Molecular Dynamic Simulations
Calculation of the Hydrodynamic Radius from the Molecular Structure
3. Results and Discussions
3.1. Aggregates Formation
3.2. Myricetin—Lipase Interaction
3.2.1. Sequestering of Lipase
3.2.2. Aggregate Characterization
3.2.3. Molecular Dynamic Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yuda, N.; Tanaka, M.; Suzuki, M.; Asano, Y.; Ochi, H.; Iwatsuki, K. Polyphenols Extracted from Black Tea (Camellia sinensis) Residue by Hot-Compressed Water and Their Inhibitory Effect on Pancreatic Lipasein vitro. J. Food Sci. 2012, 77, H254–H261. [Google Scholar] [CrossRef]
- Yao, L.H.; Jiang, Y.; Shi, J.; Datta, N.; Singanusong, R.; Chen, S.S. Flavonoids in Food and Their Health Benefits. Plant Foods Hum. Nutr. 2004, 59, 113–122. [Google Scholar] [CrossRef]
- Semwal, D.; Semwal, R.B.; Combrinck, S.; Viljoen, A.M. Myricetin: A Dietary Molecule with Diverse Biological Activities. Nutrition 2016, 8, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, T.K.; Jung, I.; Kim, M.E.; Bae, S.K.; Lee, J.S. Anti-cancer activity of myricetin against human papillary thyroid cancer cells involves mitochondrial dysfunction–mediated apoptosis. Biomed. Pharmacother. 2017, 91, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-J.; Park, J.-H.Y.; Choi, H.-N.; Kim, J.-I. α-glucosidase inhibitory activities of myricetin in animal models of diabetes mellitus. Food Sci. Biotechnol. 2015, 24, 1897–1900. [Google Scholar] [CrossRef]
- Tan, Y.; Chang, S.K.; Zhang, Y. Comparison of α-amylase, α-glucosidase and lipase inhibitory activity of the phenolic substances in two black legumes of different genera. Food Chem. 2017, 214, 259–268. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.M.; Lee, S.S.; Chung, B.Y.; Cho, J.-Y.; Lee, I.C.; Ahn, S.R.; Jang, S.J.; Kim, T.H. Pancreatic Lipase Inhibition by C-Glycosidic Flavones Isolated from Eremochloa ophiuroides. Molecule 2010, 15, 8251–8259. [Google Scholar] [CrossRef] [Green Version]
- Guerciolini, R. Mode of action of orlistat. Int. J. Obes. Relat. Metab. Disord. J. Int. Assoc. Stud. Obes. 1997, 21, 12–23. [Google Scholar]
- Martinez-Gonzalez, A.I.; Alvarez-Parrilla, E.; Díaz-Sánchez, Á.G.; De La Rosa, L.A.; Núñez-Gastélum, J.A.; Vázquez-Flores, A.A.; González-Aguilar, G.A. In Vitro Inhibition of Pancreatic Lipase by Polyphenols: A Kinetic, Fluorescence Spectroscopy and Molecular Docking Study. Food Technol. Biotechnol. 2017, 55, 519–530. [Google Scholar] [CrossRef]
- Glisan, S.L.; Grove, K.A.; Yennawar, N.H.; Lambert, J.D. Inhibition of pancreatic lipase by black tea theaflavins: Comparative enzymology and in silico modeling studies. Food Chem. 2016, 216, 296–300. [Google Scholar] [CrossRef] [Green Version]
- Birner-Grünberger, R.; Scholze, H.; Faber, K.; Hermetter, A.; Birner-Gruenberger, R. Identification of various lipolytic enzymes in crude porcine pancreatic lipase preparations using covalent fluorescent inhibitors. Biotechnol. Bioeng. 2003, 85, 147–154. [Google Scholar] [CrossRef] [PubMed]
- McGovern, S.L.; Caselli, E.; Grigorieff, N.; Shoichet, B.K. A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. J. Med. Chem. 2002, 45, 1712–1722. [Google Scholar] [CrossRef] [PubMed]
- McGovern, S.L.; Helfand, B.T.; Feng, B.; Shoichet, B.K. A Specific Mechanism of Nonspecific Inhibition. J. Med. Chem. 2003, 46, 4265–4272. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, J.A.; Pérez-Jiménez, J.; Neveu, V.; Medina-Remón, A.; M’Hiri, N.; García-Lobato, P.; Manach, C.; Knox, C.; Eisner, R.; Wishart, D.S.; et al. Phenol-Explorer 3.0: A major update of the Phenol-Explorer database to incorporate data on the effects of food processing on polyphenol content. Database 2013, 2013, bat070. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Xie, Y.; Yao, Y.; Li, G.; Yuan, X.; Shen, H. A Novel Strategy for Pharmaceutical Cocrystal Generation Without Knowledge of Stoichiometric Ratio: Myricetin Cocrystals and a Ternary Phase Diagram. Pharm. Res. 2014, 32, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Lin, G.; Xie, Y.; Duan, J.; Ma, P.; Li, G.; Ji, G. Quantitative Determination of Myricetin in Rat Plasma by Ultra Performance Liquid Chromatography Tandem Mass Spectrometry and its Absolute Bioavailability. Drug Res. 2013, 64, 516–522. [Google Scholar] [CrossRef]
- Qian, J.; Meng, H.; Xin, L.; Xia, M.; Shen, H.; Li, G.; Xie, Y. Self-nanoemulsifying drug delivery systems of myricetin: Formulation development, characterization, and in vitro and in vivo evaluation. Colloids Surf. B Biointerfaces 2017, 160, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.-F.; Guo, J.; Qian, D.; Duan, J.-A.; Shu, Y. Absorption of flavonoids from Abelmoschus manihot extract by in situ intestinal perfusion. Yao Xue Xue Bao = Acta Pharm. Sin. 2011, 46, 454–459. [Google Scholar]
- Wahlund, K.-G.; Nilsson, L. Flow FFF–Basics and Key Applications. In Field-Flow Fractionation in Biopolymer Analysis; Springer-Verlag Wien: Vienna, Austria, 2012; pp. 1–21. [Google Scholar]
- Nilsson, L. Separation and characterization of food macromolecules using field-flow fractionation: A review. Food Hydrocoll. 2013, 30, 1–11. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Balance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised staticin vitrodigestion method suitable for food–an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Hadváry, P.; Sidler, W.; Meister, W.; Vetter, W.; Wolfer, H. The lipase inhibitor tetrahydrolipstatin binds covalently to the putative active site serine of pancreatic lipase. J. Biol. Chem. 1991, 266, 2021–2027. [Google Scholar]
- Tsuzuki, W.; Ue, A.; Kitamura, Y. Effect of dimethylsulfoxide on hydrolysis of lipase. Biosci. Biotechnol. Biochem. 2001, 65, 2078–2082. [Google Scholar] [CrossRef] [PubMed]
- Zimm, B.H. The Scattering of Light and the Radial Distribution Function of High Polymer Solutions. J. Chem. Phys. 1948, 16, 1093. [Google Scholar] [CrossRef]
- Theisen, A. Refractive Increment Data Book for Polymer and Biomolecular Scientists; Nottingham Univ. Press: Nottingham, UK, 2000. [Google Scholar]
- Håkansson, A.; Magnusson, E.; Bergenstahl, B.; Nilsson, L. Hydrodynamic radius determination with asymmetrical flow field-flow fractionation using decaying cross-flows. Part I. A theoretical approach. J. Chromatogr. A 2012, 1253, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; A Case, D. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Aronsson, A.; Güler, F.; Petoukhov, M.V.; Crennell, S.J.; Svergun, D.I.; Linares-Pastén, J.A.; Karlsson, E.N. Structural insights of Rm Xyn10A–A prebiotic-producing GH10 xylanase with a non-conserved aglycone binding region. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2018, 1866, 292–306. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef] [Green Version]
- Nosé, S.; Klein, M. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Bustos, A.-S.; Håkansson, A.; Linares-Pastén, J.A.; Penarrieta, J.M.; Nilsson, L. Interaction Between Phenolic Compounds and Lipase: The Influence of Solubility and Presence of Particles in the IC50 Value. J. Food Sci. 2018, 83, 2071–2076. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, Y.; Gao, F.; Zhao, Y.; Cai, S.; Pang, M. The free, esterified, and insoluble-bound phenolic profiles of Rhus chinensis Mill. fruits and their pancreatic lipase inhibitory activities with molecular docking analysis. J. Funct. Foods 2018, 40, 729–735. [Google Scholar] [CrossRef]
- Pohjala, L.; Tammela, P. Aggregating Behavior of Phenolic Compounds—A Source of False Bioassay Results? Molecules 2012, 17, 10774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, A.; Gray, N.M.; Lowe, P.N.; Chung, C.-W. Effect of Detergent on “Promiscuous” Inhibitors. J. Med. Chem. 2003, 46, 3448–3451. [Google Scholar] [CrossRef] [PubMed]




| Sample Name | Water (µL) | SIF Stock (µL) | CaCl2 Stock (µL) | Saturated Lipase Stock (µL) | Myricetin-Stock (µL) |
|---|---|---|---|---|---|
| Myricetin-intestinal solution | 95 | 0 | 20 | 80 | 5 |
| Myricetin-water | 195 | - | - | - | 5 |
| Myricetin-control | 95 | 80 | 20 | - | 5 |
| Lipase-control | 95 | 0 | 20 | 80 | 5 * |
| Analyte | rh (nm) | Retention Time (min) | Retention Level |
|---|---|---|---|
| Lipase | 3.1 | 4.8 | 15 |
| Small myricetin aggregates | 1.3 | 2.1 | 7 |
| Myricetin–lipase aggregates | 2.2–4.2 | 3.4–6.5 | > 11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bustos, A.-S.; Håkansson, A.; Linares-Pastén, J.A.; Nilsson, L. Interaction between Myricetin Aggregates and Lipase under Simplified Intestinal Conditions. Foods 2020, 9, 777. https://doi.org/10.3390/foods9060777
Bustos A-S, Håkansson A, Linares-Pastén JA, Nilsson L. Interaction between Myricetin Aggregates and Lipase under Simplified Intestinal Conditions. Foods. 2020; 9(6):777. https://doi.org/10.3390/foods9060777
Chicago/Turabian StyleBustos, Atma-Sol, Andreas Håkansson, Javier A. Linares-Pastén, and Lars Nilsson. 2020. "Interaction between Myricetin Aggregates and Lipase under Simplified Intestinal Conditions" Foods 9, no. 6: 777. https://doi.org/10.3390/foods9060777
APA StyleBustos, A.-S., Håkansson, A., Linares-Pastén, J. A., & Nilsson, L. (2020). Interaction between Myricetin Aggregates and Lipase under Simplified Intestinal Conditions. Foods, 9(6), 777. https://doi.org/10.3390/foods9060777