Application of Recombinase Polymerase Amplification with Lateral Flow for a Naked-Eye Detection of Listeria monocytogenes on Food Processing Surfaces
Abstract
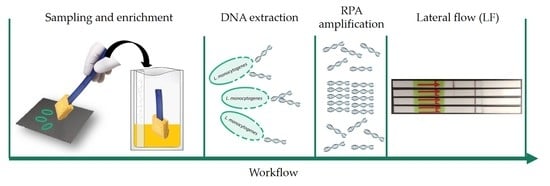
:1. Introduction
2. Materials and Methods
2.1. Pure Culture Preparation
2.2. RPA Primers Design
2.3. Surface Contamination and Sampling
2.4. Optimization of Enrichment Step
2.5. DNA Extraction
2.5.1. Pure Culture Cell Lysis
2.5.2. Surface Samples
2.6. Food Contact Surface Sample Analysis
2.6.1. RPA-LF
2.6.2. qPCR
2.6.3. Culture-Based Methodology
2.7. Evaluation
2.7.1. RPA-LF Evaluation
2.7.2. Methodology Evaluation
3. Results
3.1. Evaluation of RPA-LF Reaction and Comparison of Primers
3.1.1. Dynamic Range of RPA-LF
3.1.2. Inclusivity/Exclusivity
3.2. Sample Pre-Treatments Optimization
3.2.1. Sampling Procedure
3.2.2. Enrichment Broth
3.2.3. Enrichment Time
3.3. Evaluation of the Methodology
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef] [Green Version]
- European Commission. Commission Regulation (EC) COMMISSION REGULATION (EC) No 1441/2007 of 5 December 2007 amending Regulation (EC) No 2073/2005 on microbiological criteria for foodstuffs. Off. J. Eur. Union 2007, 322, 12–29. [Google Scholar]
- Castro-Ibáñez, I.; Gil, M.I.; Allende, A. Ready-to-eat vegetables: Current problems and potential solutions to reduce microbial risk in the production chain. LWT Food Sci. Technol. 2017, 85, 284–292. [Google Scholar] [CrossRef]
- Lin, C.-M.; Takeuchi, K.; Zhang, L.; Dohm, C.B.; Meyer, J.D.; Hall, P.A.; Doyle, M.P. Cross-contamination between processing equipment and deli meats by Listeria monocytogenes. J. Food Prot. 2006, 69, 71–79. [Google Scholar] [CrossRef]
- Walker, S.J.; Archer, P.; Banks, J.G. Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol. 1990, 68, 157–162. [Google Scholar] [CrossRef]
- Da Silva, E.P.; De Martinis, E.C.P. Current knowledge and perspectives on biofilm formation: The case of Listeria monocytogenes. Appl. Microbiol. Biotechnol. 2013, 97, 957–968. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef]
- Melero, B.; Manso, B.; Stessl, B.; Hernández, M.; Wagner, M.; Rovira, J.; Rodríguez-Lázaro, D. Distribution and Persistence of Listeria monocytogenes in a Heavily Contaminated Poultry Processing Facility. J. Food Prot. 2019, 82, 1524–1531. [Google Scholar] [CrossRef]
- Hitchens, A.D.; Jinneman, K. Laboratory Methods-BAM: Detection and Enumeration of Listeria monocytogenes. FDA Bacteriol. Anal. Man. 2013. [Google Scholar]
- ISO. Microbiology of Food and Animal Feeding Stuffs—Polymerase Chain Reaction (PCR) for the Detection of Food-Borne Pathogens—Requirements for Sample Preparation for Qualitative Detection; ISO 20837:2006; International Organization for Standardization: Geneva, Switzerland, 2006. [Google Scholar]
- Mangal, M.; Bansal, S.; Sharma, S.K.; Gupta, R.K. Molecular Detection of Foodborne Pathogens: A Rapid and Accurate Answer to Food Safety. Crit. Rev. Food Sci. Nutr. 2016, 56, 1568–1584. [Google Scholar] [CrossRef] [PubMed]
- Alía, A.; Andrade, M.J.; Córdoba, J.J.; Martín, I.; Rodríguez, A. Development of a multiplex real-time PCR to differentiate the four major Listeria monocytogenes serotypes in isolates from meat processing plants. Food Microbiol. 2020, 87, 103367. [Google Scholar] [CrossRef]
- Jana, M.; Adriana, V.; Eva, K. Evaluation of DNA Extraction Methods for Culture-Independent Real-Time PCR-Based Detection of Listeria monocytogenes in Cheese. Food Anal. Methods 2020, 13, 667–677. [Google Scholar] [CrossRef]
- Wei, S.; Park, B.-J.; Kim, S.-H.; Seo, K.-H.; Jin, Y.-G.; Oh, D.-H. Detection of Listeria monocytogenes using Dynabeads® anti-Listeria combined with real-time PCR in soybean sprouts. LWT 2019, 99, 533–539. [Google Scholar] [CrossRef]
- D’Agostino, M.; Wagner, M.; Vazquez-Boland, J.A.; Kuchta, T.; Karpiskova, R.; Hoorfar, J.; Novella, S.; Scortti, M.; Ellison, J.; Murray, A.; et al. A Validated PCR-Based Method to Detect Listeria monocytogenes Using Raw Milk as a Food Model—Towards an International Standard. J. Food Prot. 2004, 67, 1646–1655. [Google Scholar] [CrossRef] [PubMed]
- Hein, I.; Flekna, G.; Krassnig, M.; Wagner, M. Real-time PCR for the detection of Salmonella spp. in food: An alternative approach to a conventional PCR system suggested by the FOOD-PCR project. J. Microbiol. Methods 2017, 66, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Gianfranceschi, M.V.; Rodríguez-Lázaro, D.; Hernández, M.; González-García, P.; Comin, D.; Gattuso, A.; Delibato, E.; Sonnessa, M.; Pasquali, F.; Prencipe, V.; et al. European validation of a real-time PCR-based method for detection of Listeria monocytogenes in soft cheese. Int. J. Food Microbiol. 2014, 184, 128–133. [Google Scholar] [CrossRef]
- Malorny, B.; Hoorfar, J.; Hugas, M.; Heuvelink, A.; Fach, P.; Ellerbroek, L.; Bunge, C.; Dorn, C.; Helmuth, R. Interlaboratory diagnostic accuracy of a Salmonella specific PCR-based method. Int. J. Food Microbiol. 2003, 89, 241–249. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Isothermal Amplification Technologies for the Detection of Foodborne Pathogens. Food Anal. Methods 2018, 11, 1543–1560. [Google Scholar] [CrossRef]
- D’Agostino, M.; Robles, S.; Hansen, F.; Ntafis, V.; Ikonomopoulos, J.; Kokkinos, P.; Álvarez-Ordóñez, A.; Jordan, K.; Delibato, E.; Kukier, E.; et al. Validation of a Loop-Mediated Amplification/ISO 6579-Based Method for Analysing Soya Meal for the Presence of Salmonella enterica. Food Anal. Methods 2016, 9, 2979–2985. [Google Scholar] [CrossRef]
- Bird, P.; Flannery, J.; Crowley, E.; Agin, J.R.; Goins, D.; Monteroso, L.; Barnes, C.; Bastin, B.; Blumfield, J.; Bonilla, T.; et al. Evaluation of the 3M™ Molecular Detection Assay (MDA) 2—Salmonella for the Detection of Salmonella spp. in Select Foods and Environmental Surfaces: Collaborative Study, First Action 2016.01. J. AOAC Int. 2016, 99, 980–997. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; A Armes, N. DNA Detection Using Recombination Proteins. PLoS Boil. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Fuciños, P.; Prado, M. Development and evaluation of loop-mediated isothermal amplification, and Recombinase Polymerase Amplification methodologies, for the detection of Listeria monocytogenes in ready-to-eat food samples. Food Control 2017, 86, 27–34. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, J.-L. Development of a multiplex real-time recombinase polymerase amplification (RPA) assay for rapid quantitative detection of Campylobacter coli and jejuni from eggs and chicken products. Food Control 2017, 73, 1247–1255. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.P.; Agarwal, R.K.; Thomas, P.; Sailo, B.; Prasannavadhana, A.; Kumar, A.; Kataria, J.L.; Singh, D. Rapid Detection of Salmonella enterica Subspecies enterica serovar Typhimurium by Loop Mediated Isothermal Amplification (LAMP) Test From Field Chicken Meat Samples. Food Biotechnol. 2014, 28, 50–62. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Fuciños, P.; Azinheiro, S.; Carvalho, J.; Prado, M. Systematic loop-mediated isothermal amplification assays for rapid detection and characterization of Salmonella spp., Enteritidis and Typhimurium in food samples. Food Control 2017, 80, 297–306. [Google Scholar] [CrossRef]
- Lv, X.; Wang, L.; Zhang, J.; Zeng, H.; Chen, X.; Shi, L.; Cui, H.; He, X.; Zhao, L. Rapid and sensitive detection of VBNC Escherichia coli O157: H7 in beef by PMAxx and real-time LAMP. Food Control 2020, 115, 107292. [Google Scholar] [CrossRef]
- Dao, T.N.T.; Yoon, J.; Jin, C.E.; Koo, B.; Han, K.; Shin, Y.; Lee, T.Y. Rapid and sensitive detection of Salmonella based on microfluidic enrichment with a label-free nanobiosensing platform. Sens. Actuators B Chem. 2018, 262, 588–594. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Prado, M. Combination of Immunomagnetic Separation and Real-Time Recombinase Polymerase Amplification (IMS-qRPA) for Specific Detection of Listeria monocytogenes in Smoked Salmon Samples. J. Food Sci. 2019, 84, 1881–1887. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Abalde-Cela, S.; Carbó-Argibay, E.; Diéguez, L.; Piotrowski, M.; Kolen’Ko, Y.V.; Prado, M. Combination of Microfluidic Loop-Mediated Isothermal Amplification with Gold Nanoparticles for Rapid Detection of Salmonella spp. in Food Samples. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K.-I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 2009, 46, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of Loop-Mediated Isothermal Amplification Reaction by Turbidity Derived from Magnesium Pyrophosphate Formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Du, X.-J.; Zang, Y.-X.; Liu, H.-B.; Li, P.; Wang, S. Recombinase Polymerase Amplification Combined with Lateral Flow Strip for Listeria monocytogenes Detection in Food. J. Food Sci. 2018, 83, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Prado, M. Rapid and sensitive detection of viable Listeria monocytogenes in food products by a filtration-based protocol and qPCR. Food Microbiol. 2018, 73, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Fuciños, P.; Prado, M. Optimized sample treatment, combined with real-time PCR, for same-day detection of E. coli O157 in ground beef and leafy greens. Food Control 2020, 108. [Google Scholar] [CrossRef]
- Wilrich, C.; Wilrich, P.-T. Estimation of the POD Function and the LOD of a Qualitative Microbiological Measurement Method. J. AOAC Int. 2009, 92, 1763–1772. [Google Scholar] [CrossRef] [Green Version]
- NordVal. NordVal International Protocol for the Validation of Microbiological Alternative (Proprietary) Methods against a Reference Method; NMKL: Lyngby, Denmark, 2017. [Google Scholar]
- Mikš-Krajnik, M.; Lim, H.S.Y.; Zheng, Q.; Turner, M.; Yuk, H.-G. Loop-mediated isothermal amplification (LAMP) coupled with bioluminescence for the detection of Listeria monocytogenes at low levels on food contact surfaces. Food Control 2016, 60, 237–240. [Google Scholar] [CrossRef]
- Firstenberg-Eden, R.; A Shelef, L. A new rapid automated method for the detection of Listeria from environmental swabs and sponges. Int. J. Food Microbiol. 2000, 56, 231–237. [Google Scholar] [CrossRef]
- Liu, H.-B.; Du, X.-J.; Zang, Y.-X.; Li, P.; Wang, S. SERS-Based Lateral Flow Strip Biosensor for Simultaneous Detection of Listeria monocytogenes and Salmonella enterica Serotype Enteritidis. J. Agric. Food Chem. 2017, 65, 10290–10299. [Google Scholar] [CrossRef]
- Escadafal, C.; Faye, O.; Sall, A.A.; Faye, O.; Weidmann, M.; Strohmeier, O.; Von Stetten, F.; Drexler, J.; Eberhard, M.; Niedrig, M.; et al. Rapid Molecular Assays for the Detection of Yellow Fever Virus in Low-Resource Settings. PLoS Negl. Trop. Dis. 2014, 8, e2730. [Google Scholar] [CrossRef]
- Rosser, A.; Rollinson, D.; Forrest, M.S.; Webster, B.L. Isothermal Recombinase Polymerase amplification (RPA) of Schistosoma haematobium DNA and oligochromatographic lateral flow detection. Parasites Vectors 2015, 8, 446. [Google Scholar] [CrossRef] [Green Version]
- Powell, M.L.; Bowler, F.R.; Martinez, A.J.; Greenwood, C.J.; Armes, N.; Piepenburg, O. New Fpg probe chemistry for direct detection of recombinase polymerase amplification on lateral flow strips. Anal. Biochem. 2018, 543, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Gómez, D.; Ariño, A.; Carramiñana, J.J.; Rota, C.; Yangüela, J. Comparison of Sampling Procedures for Recovery of Listeria monocytogenes from Stainless Steel Food Contact Surfaces. J. Food Prot. 2012, 75, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Lahou, E.; Uyttendaele, M. Evaluation of Three Swabbing Devices for Detection of Listeria monocytogenes on Different Types of Food Contact Surfaces. Int. J. Environ. Res. Public Health 2014, 11, 804–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ISO. Microbiology of the Food Chain—Horizontal Methods for Surface Sampling; ISO 18593:2018; International Organization for Standardization: Geneva, Switzerland, 2018. [Google Scholar]
- Mafu, A.A.; Roy, D.; Goulet, J.; Magny, P. Attachment of Listeria monocytogenes to Stainless Steel, Glass, Polypropylene, and Rubber Surfaces After Short Contact Times. J. Food Prot. 1990, 53, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Martinon, A.; Cronin, U.P.; Quealy, J.; Stapleton, A.; Wilkinson, M.G. Swab sample preparation and viable real-time PCR methodologies for the recovery of Escherichia coli, Staphylococcus aureus or Listeria monocytogenes from artificially contaminated food processing surfaces. Food Control 2012, 24, 86–94. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, Y.; Zhang, Z.; Chen, M.; Su, Y.; Yuan, Y.; Alam, M.J.; Yan, H.; Shi, L. Rapid detection of food-borne Listeria monocytogenes by real-time quantitative loop-mediated isothermal amplification. Food Sci. Biotechnol. 2012, 21, 101–106. [Google Scholar] [CrossRef]





| Bacteria Species | Source | N | qPCR Primers | RPA Primers |
|---|---|---|---|---|
| Salmonella spp. | Mollusk (AMC 90, 238, 253, 255) | 4 | - | -* 238 |
| Salmonella spp. | Unknown (AMC 260, 261) | 2 | -* 261 | - |
| Salmonella Oranienburg | Mollusk (AMC 28) | 1 | - | - |
| Salmonella Veneziana | Mollusk (AMC 200) | 1 | - | - |
| Salmonella Anatum | Proficiency test (AMC 60) | 1 | - | - |
| Salmonella Enteritidis | Proficiency test (AMC 82) | 1 | - | - |
| Salmonella Enteritidis | University of Bristol | 1 | - | - |
| Salmonella Wentworth | Proficiency test (AMC 84) | 1 | -* | - |
| Salmonella Wentworth | Mollusk (AMC 200) | 1 | - | - |
| Salmonella Liverpool | Proficiency test (AMC 198) | 1 | - | -* |
| Salmonella Typhimurium | Proficiency test (AMC 96) | 1 | - | - |
| Salmonella Typhimurium | Unknown (WDCM 00031) | 1 | - | - |
| Listeria monocytogenes | Spinal fluid, serotype 4b (WDCM 00021) | 1 | + | + |
| Listeria monocytogenes | Mollusk | 4 | + | + |
| Listeria monocytogenes | Chestnut | 3 | + | + |
| Listeria monocytogenes | Chicken | 3 | + | + |
| Listeria ivanovii | WDCM 00018 | 1 | - | - |
| Listeria innocua | WDCM 00017, CECT 5376, 4030, 1325, 1141, 2110 | 6 | -* 1141 | - |
| Staphylococcus aureus | WDCM 00034, 00033 | 2 | - | -* 00033 |
| Staphylococcus coagulase positive | Proficiency test | 1 | - | - |
| Campylobacter coli | University of Minho | 1 | -* | -* |
| Escherichia coli | WDCM 00013, 00012 | 2 | - | - |
| Escherichia coli | WDCM 00014 | 1 | - | - |
| Enterococcus faecalis | WDCM 00009 | 1 | - | - |
| Methodology | Sequence 5′-3′ | Modifications | Reference | |
|---|---|---|---|---|
| RPA | hly-RPA F (forward primer) | TTACACTTATATTAGTTAGTCTACCAATTGCG | - | [35] |
| hly-RPA R (reverse primer) | TCCAATCCTTGTATATACTTATCGATTTCATC | 5′-Biotin/ | ||
| hly-P3F (forward primer) | GCAACAAACTGAAGCAAAGGAT | - | [34] | |
| hly-P3R (reverse primer) | CGATTGGCGTCTTAGGACTTGC | 5′-Biotin/ | ||
| hly-LF- P (lateral flow probe) | TCTGCATTCAATAAAGAAAATTCAATTTCATCZATGGCACCACCAGCATC | 5′-FAM/THF/SpC3 *-3′ | ||
| qPCR | hly-P3P (hly probe) | CATGGCACC//ACCAGCATCTCCG | 5′-FAM/ZEN/IABkFQ-3′ | [34] |
| IAC-P3P (IAC probe) | AGTGGCGGT//GACACTGTTGACCT | 5′-YY/ZEN/IABkFQ-3′ | ||
| IAC-DNA | GGATTACCCTAGAGTGGCGGTGACACTGTTGACCTTCTATTACCTC |
| Contamination Level (CFU/cm2) | N | RPA-LF | qPCR | COMPASS | Full Fraser 24 h | Full Fraser 48 h |
|---|---|---|---|---|---|---|
| 8 × 105 | 2 | + | + | + | + | + |
| 8 × 104 | 2 | + | + | + | + | + |
| 8 × 103 | 2 | + | + | + | + | + |
| 8 × 102 | 2 | + | + | + | + | + |
| 8 × 101 | 1 | + | + | + | + | + |
| ** 7 × 101 | 1 | + | + | + | + | + |
| ** 5 × 101 | 3 | + | + | + | + | + |
| ** 2 × 101 | 2 | + | + | + | + | + |
| 1 | + | - | - | - | - | |
| ** 9 | 2 | + | + | + | + | + |
| 2 | - | - | - | - | - | |
| ** 8 | 2 | + | - | +/- * | + | + |
| 2 | + | + | + | + | + | |
| 7 | 1 | - | - | - | - | - |
| ** 4 | 2 | + | + | + | + | + |
| 3 | - | - | - | - | - | |
| 1 | 1 | - | - | - | - | - |
| 1 | 1 | - | - | - | - | - |
| Methodology | LoD50 * | LoD95 * | ||||
|---|---|---|---|---|---|---|
| LoD | Lower Conf. Limit | Upper Conf. Limit | LoD | Lower Conf. Limit | Upper Conf. Limit | |
| RPA-LF without enrichment | 117.9 | 26.9 | 516.0 | 509.3 | 116.3 | 2230.0 |
| RPA-LF with enrichment | 4.2 | 2.1 | 8.5 | 18.2 | 9.1 | 36.5 |
| qPCR | 8.3 | 4.1 | 16.8 | 36.1 | 17.9 | 72.5 |
| Approach | N | PA | NA | FN | TP | FP | AC (%) | SE (%) | SP (%) | p0 | pe | k |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qPCR | 32 | 21 | 10 | 1 | 0 | 0 | 97 | 95 | 100.0 | 0.97 | 0.44 | 0.94 |
| RPA-LF | 32 | 23 | 8 | 0 | 0 | 1 | 97 | 100 | 89 | 0.97 | 0.40 | 0.95 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azinheiro, S.; Carvalho, J.; Prado, M.; Garrido-Maestu, A. Application of Recombinase Polymerase Amplification with Lateral Flow for a Naked-Eye Detection of Listeria monocytogenes on Food Processing Surfaces. Foods 2020, 9, 1249. https://doi.org/10.3390/foods9091249
Azinheiro S, Carvalho J, Prado M, Garrido-Maestu A. Application of Recombinase Polymerase Amplification with Lateral Flow for a Naked-Eye Detection of Listeria monocytogenes on Food Processing Surfaces. Foods. 2020; 9(9):1249. https://doi.org/10.3390/foods9091249
Chicago/Turabian StyleAzinheiro, Sarah, Joana Carvalho, Marta Prado, and Alejandro Garrido-Maestu. 2020. "Application of Recombinase Polymerase Amplification with Lateral Flow for a Naked-Eye Detection of Listeria monocytogenes on Food Processing Surfaces" Foods 9, no. 9: 1249. https://doi.org/10.3390/foods9091249
APA StyleAzinheiro, S., Carvalho, J., Prado, M., & Garrido-Maestu, A. (2020). Application of Recombinase Polymerase Amplification with Lateral Flow for a Naked-Eye Detection of Listeria monocytogenes on Food Processing Surfaces. Foods, 9(9), 1249. https://doi.org/10.3390/foods9091249