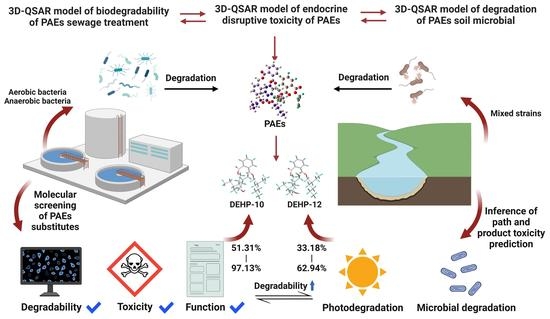
Molecular Design and Mechanism Analysis of Phthalic Acid Ester Substitutes: Improved Biodegradability in Processes of Sewage Treatment and Soil Remediation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Screening and Data Source of Molecular Biodegradable Enzymes for PAE Degradation during Sewage and Soil Treatment Processes
2.2. Characterization of the Binding Ability of the PAE Molecules to Microbial Degrading Enzymes Used in the Sewage Treatment Process and Present in the Soil Environment—The Molecular Docking Method
2.3. Calculation of Comprehensive Effect Value of Microbial Degradability—The CRITIC Method
2.4. Construction of the 3D-QSAR Model for Comprehensive Effects of Biodegradation in PAEs Molecular Sewage Treatment Process—The SYBYL-X Software Method
3. Results and Discussion
3.1. Construction and Evaluation of a 3D-QSAR Model to Study the Comprehensive Effects of Microbial Degradation Associated with PAE Removal during Water Treatment
3.1.1. Calculation and Model Construction to Study the Comprehensive Effect Value Associated with Microbial Degradability for the PAE Systems Associated with the Sewage Treatment Process
3.1.2. Evaluation of the 3D-QSAR Model to Study the Comprehensive Effect of Microbial Degradability of PAEs Associated with the Sewage Treatment Process
3.2. Molecular Design and Characterization of Alternatives of Readily Biodegradable PAEs Associated with the Sewage Treatment Process
3.2.1. Molecular Design Obtained Based on the 3D-QSAR Models for the Readily Biodegradable Substitutes of PAEs
3.2.2. Aerobic/Anaerobic Biodegradability, Endocrine Disturbance, and Functional Evaluation of PAE Substitutes
3.2.3. Analysis of the Mechanism of Differential Microbial Degradation before and after Molecular Modification of PAEs
3.3. Evaluation and Mechanism Analysis of the Process of Photodegradation and Microbial Degradation Potential of DEHP Substitutes in the Soil Environment
3.3.1. Soil Environmental Photodegradation and Microbial Degradation Pathways (before and after DEHP Molecular Modification)
3.3.2. Evaluation of the Extent of Photodegradation and Microbial Degradation Responses Produced and Product Toxicity in Soils before and after Molecular Modification by DEHP
3.3.3. Analysis of the DEHP Substitutes Suitable for Sewage Treatment and the Soil Microbial Degradation Mechanism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- González-Sálamo, J.; González-Curbelo, M.Á.; Socas-Rodríguez, B.; Hernández-Borges, j.; Rodríguez-Delgado, M.Á. Determination of phthalic acid esters in water samples by hollow fiber liquid-phase microextraction prior to gas chromatography tandem mass spectrometry. Chemosphere 2018, 201, 254–261. [Google Scholar] [CrossRef]
- Kaewlaoyoong, A.; Vu, C.T.; Lin, C.; Liao, C.S.; Chen, J.R. Occurrence of phthalate esters around the major plastic industrial area in southern Taiwan. Environ. Earth Sci. 2018, 77, 457. [Google Scholar] [CrossRef]
- Tan, S.; Wang, D.; Chi, Z.; Li, W.; Shan, Y. Study on the interaction between typical phthalic acid esters (PAEs) and human haemoglobin (hHb) by molecular docking. Environ. Toxicol. Pharmacol. 2017, 53, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Waller, C.L.; Griffiths, H.J.; Waluda, C.M.; Thorpe, S.E.; Loaiza, L.; Moreno, B.; Pacherres, C.O.; Hughes, K.A. Microplastics in the Antarctic marine system: An emerging area of research. Sci. Total Environ. 2017, 598, 220–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Liu, C.; He, D.; Xu, J.; Sun, J.; Li, J.; Pan, X. Environmental behaviors of microplastics in aquatic systems: A systematic review on degradation, adsorption, toxicity and biofilm under aging conditions. J. Hazard. Mater. 2022, 423, 126915. [Google Scholar] [CrossRef]
- Isobe, A.; Iwasaki, S.; Uchida, K.; Tokai, T. Abundance of non-conservative microplastics in the upper ocean from 1957 to 2066. Nat. Commun. 2019, 10, 417. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Chen, Y.; Chen, C.; Feng, L.; Dong, Y.; Chen, J.; Lan, J.; Hou, H. High-efficiency degradation of phthalic acid esters (PAEs) by Pseudarthrobacter defluvii E5: Performance, degradative pathway, and key genes. Sci. Total Environ. 2021, 794, 148719. [Google Scholar] [CrossRef]
- Eleonora, G.; Antoni, S.; Silvia, T.; Caterina, F. Microplastic in marine organism: Environmental and toxicological effects. Environ. Toxicol. Pharmacol. 2018, 64, 164–171. [Google Scholar] [CrossRef]
- Das, M.T.; Kumar, S.S.; Ghosh, P.; Shah, G.; Malyan, S.K.; Bajar, S.; Thakur, I.S.; Singh, L. Remediation strategies for mitigation of phthalate pollution: Challenges and future perspectives. J. Hazard. Mater. 2021, 409, 124496. [Google Scholar] [CrossRef]
- Becky, M.I.; Anbalagan, K.; Magesh, K.M. Phthalates removal from wastewater by different methods–a review. Water Sci. Technol. 2022, 85, 2581–2600. [Google Scholar] [CrossRef]
- Tran, H.T.; Lin, C.; Bui, X.T.; Nguyen, M.K.; Cao, N.D.T.; Mukhtar, H.; Hoang, H.G.; Varjani, S.; Ngo, H.H.; Nghiem, L.D. Phthalates in the environment: Characteristics, fate and transport, and advanced wastewater treatment technologies. Bioresour. Technol. 2022, 344, 126249. [Google Scholar] [CrossRef]
- Perpetuo, E.A.; Silva, E.C.N.D.; Karolski, B.; Nascimento, C.A.O.D. Biodegradation of diethyl-phthalate (DEP) by halotolerant bacteria isolated from an estuarine environment. Biodegradation 2020, 31, 331–340. [Google Scholar] [CrossRef]
- Wei, L.; Li, Z.; Sun, J.; Zhu, L. Pollution characteristics and health risk assessment of phthalate esters in agricultural soil and vegetables in the Yangtze River Delta of China. Sci. Total Environ. 2020, 726, 137978. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, J.; Liu, H.; Guo, X.; Zhang, X.; Yao, X.; Cao, Z.; Zhang, T. A review of the removal of microplastics in global wastewater treatment plants: Characteristics and mechanisms. Environ. Int. 2021, 146, 106277. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, H.; Kannan, K. A review of biomonitoring of phthalate exposures. Toxics 2019, 7, 21. [Google Scholar] [CrossRef] [Green Version]
- Lü, H.; Mo, C.H.; Zhao, H.M.; Xiang, L.; Katsoyiannis, A.; Li, Y.W.; Cai, Q.Y.; Wong, M.H. Soil contamination and sources of phthalates and its health risk in China: A review. Environ. Res. 2018, 164, 417–429. [Google Scholar] [CrossRef]
- He, M.; Yang, T.; Yang, Z.; Zhou, H.; Wei, S. Current state, distribution, and sources of phthalate esters and organophosphate esters in soils of the three gorges reservoir region, China. Arch. Environ. Contam. Toxicol. 2018, 74, 502–513. [Google Scholar] [CrossRef]
- Wang, D.; Xi, Y.; Shi, X.Y.; Zhong, Y.J.; Guo, C.L.; Han, Y.N.; Li, F.M. Effect of plastic film mulching and film residues on phthalate esters concentrations in soil and plants, and its risk assessment. Environ. Pollut. 2021, 286, 117546. [Google Scholar] [CrossRef]
- Lee, Y.M.; Lee, J.E.; Choe, W.; Kim, T.; Lee, J.Y.; Kho, Y.; Choi, K.; Zoh, K.D. Distribution of phthalate esters in air, water, sediments, and fish in the Asan Lake of Korea. Environ. Int. 2019, 126, 635–643. [Google Scholar] [CrossRef]
- Sun, J.; Zhu, H.; Yang, X.; Zheng, Y.; Sun, T.; Xu, H.; Meng, J.; Zhang, A. Carboxylesterase and lipase-catalyzed degradation of phthalate esters in soil and water: Congener structure selectivity and specificity. Environ. Technol. Innov. 2022, 28, 102571. [Google Scholar] [CrossRef]
- Zeng, X.; Qu, R.; Feng, M.; Chen, J.; Wang, L.; Wang, Z. Photodegradation of polyfluorinated dibenzo-p-dioxins in organic solvents: Experimental and theoretical studies. Environ. Sci. Technol. 2016, 50, 8128–8134. [Google Scholar] [CrossRef] [PubMed]
- Courtene-Jones, W.; Quinn, B.; Gary, S.F.; Mogg, A.O.M.; Narayanaswamy, B.E. Microplastic pollution identified in deep-sea water and ingested by benthic invertebrates in the Rockall Trough, North Atlantic Ocean. Environ. Pollut. 2017, 231, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Meng, L.; Luo, J.; Zhang, M.; Wang, Y.; Dai, Y.; Sun, C.; Wang, Z.; Yang, S.; He, H.; et al. Self-assembled ultrathin CoO/Bi quantum dots/defective Bi2MoO6 hollow Z-scheme heterojunction for visible light-driven degradation of diazinon in water matrix: Intermediate toxicity and photocatalytic mechanism. Appl. Catal. B-Environ. 2021, 293, 120231. [Google Scholar] [CrossRef]
- Zhou, B.; Zhao, L.; Wang, Y.; Sun, Y.; Li, X.; Xu, H.; Weng, L.; Pan, Z.; Yang, S.; Chang, X.; et al. Spatial distribution of phthalate esters and the associated response of enzyme activities and microbial community composition in typical plastic-shed vegetable soils in China. Ecotoxicol. Environ. Saf. 2020, 195, 110495. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, T.; Wang, F.; Dai, Y.; Liang, W. Removal efficiency and enzymatic mechanism of dibutyl phthalate (DBP) by constructed wetlands. Environ. Sci. Pollut. Res. 2018, 25, 23009–23017. [Google Scholar] [CrossRef]
- Sarkar, J.; Dutta, A.; Chowdhury, P.P.; Chakraborty, J.; Dutta, T.K. Characterization of a novel family VIII esterase EstM2 from soil metagenome capable of hydrolyzing estrogenic phthalates. Microb. Cell Factories 2020, 19, 77. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, M.; Basu, S.; Dhar, R.; Dutta, T.K. Phthalate hydrolase: Distribution, diversity and molecular evolution. Environ. Microbiol. Rep. 2022, 14, 333–346. [Google Scholar] [CrossRef]
- Wang, X.; Han, B.; Wu, P.; Li, S.; Lv, Y.; Lu, J.; Yang, Q.; Li, J.; Zhu, Y.; Zhang, Z. Dibutyl phthalate induces allergic airway inflammation in rats via inhibition of the Nrf2/TSLP/JAK1 pathway. Environ. Pollut. 2020, 267, 115564. [Google Scholar] [CrossRef]
- Hu, R.; Zhao, H.; Xu, X.; Wang, Z.; Yu, K.; Shu, L.; Yan, Q.; Wu, B.; Mo, C.; He, Z.; et al. Bacteria-driven phthalic acid ester biodegradation: Current status and emerging opportunities. Environ. Int. 2021, 154, 106560. [Google Scholar] [CrossRef]
- Mesdaghinia, A.; Azari, A.; Nodehi, R.N.; Yaghmaeian, K.; Bharti, A.K.; Agarwal, S.; Gupta, V.K.; Sharafi, K. Removal of phthalate esters (PAEs) by zeolite/Fe3O4: Investigation on the magnetic adsorption separation, catalytic degradation and toxicity bioassay. J. Mol. Liq. 2017, 233, 378–390. [Google Scholar] [CrossRef]
- Fan, S.; Wang, J.; Yan, Y.; Wang, J.; Jia, Y. Excellent degradation performance of a versatile phthalic acid esters-degrading bacterium and catalytic mechanism of monoalkyl phthalate hydrolase. Int. J. Mol. Sci. 2018, 19, 2803. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Hou, Y.; Li, Y. Multi-directional selective toxicity effects on farmland ecosystems: A novel design of green substitutes for neonicotinoid insecticides. J. Clean. Prod. 2020, 272, 122715. [Google Scholar] [CrossRef]
- Wang, X.; Xiang, W.; Wang, S.; Ge, J.; Qu, R.; Wang, Z. Oxidative oligomerization of phenolic endocrine disrupting chemicals mediated by Mn (III)-L complexes and the role of phenoxyl radicals in the enhanced removal: Experimental and theoretical studies. Environ. Sci. Technol. 2019, 54, 1573–1582. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, C.; Na, H. PAEs Derivatives’ Design for Insulation: Integrated In-Silico Methods, Functional Assessment and Environmentally Friendly Molecular Modification. Int. J. Environ. Res. Public Health 2022, 19, 3232. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, B. A hybrid MADM algorithm based on attribute weight and utility value for heterogeneous network selection. J. Netw. Syst. Manag. 2019, 27, 756–783. [Google Scholar] [CrossRef]
- Li, Q.; Qiu, Y.; Li, Y. Molecular design of environment-friendly PAE derivatives based on 3D-QSAR assisted with a comprehensive evaluation method combining toxicity and estrogen activities. Water Air Soil Pollut. 2020, 231, 194. [Google Scholar] [CrossRef]
- Gu, W.; Zhao, Y.; Li, Q.; Li, Y. Environmentally friendly polychlorinated naphthalenes (PCNs) derivatives designed using 3D-QSAR and screened using molecular docking, density functional theory and health-based risk assessment. J. Hazard. Mater. 2019, 363, 316–327. [Google Scholar] [CrossRef]
- Hou, Y.; Zhao, Y.; Li, Q.; Li, Y. Highly biodegradable fluoroquinolone derivatives designed using the 3D-QSAR model and biodegradation pathways analysis. Ecotoxicol. Environ. Saf. 2020, 191, 110186. [Google Scholar] [CrossRef]
- Li, X.; He, W.; Zhao, Y.; Chen, B.; Zhu, Z.; Kang, Q.; Zhang, B. Dermal exposure to synthetic musks: Human health risk assessment, mechanism, and control strategy. Ecotoxicol. Environ. Saf. 2022, 236, 113463. [Google Scholar] [CrossRef]
- Li, X.; Zhao, Y.; Chen, B.; Zhu, Z.; Kang, Q.; Husain, T.; Zhang, B. Inhalation and ingestion of synthetic musks in pregnant women: In silico spontaneous abortion risk evaluation and control. Environ. Int. 2022, 158, 106911. [Google Scholar] [CrossRef]
- Le, T.M.; Nguyen, H.M.N.; Nguyen, V.K.; Nguyenet, A.V.; Vu, N.D.; Yen, N.T.H.; Hoang, A.Q.; Minh, T.B.; Kannan, K.; Tran, T.M. Profiles of phthalic acid esters (PAEs) in bottled water, tap water, lake water, and wastewater samples collected from Hanoi, Vietnam. Sci. Total Environ. 2021, 788, 147831. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gu, W.; Chen, B.; Zhu, Z.; Zhang, B. Functional modification of HHCB: Strategy for obtaining environmentally friendly derivatives. J. Hazard. Mater. 2021, 416, 126116. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, H.; Xiao, X.; Zhao, Z.; Liu, C. Effects of phthalate esters (PAEs) on cell viability and Nrf2 of HepG2 and 3D-QSAR studies. Toxics 2021, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Yang, S.; Zhang, S. Occurrence, health risks and soil-air exchange of phthalate acid esters: A case study in plastic film greenhouses of Chongqing, China. Chemosphere 2021, 268, 128821. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Yang, L.; Du, M.; Li, Y. A novel pharmacophore model on PAEs’ estrogen and thyroid hormone activities using the TOPSIS and its application in molecule modification. Environ. Sci. Pollut. Res. 2020, 27, 38805–38818. [Google Scholar] [CrossRef]
- Liao, X.; Lu, R.; Xia, L.; Liu, Q.; Wang, H.; Zhao, K.; Wang, Z.; Zhao, Y. Density functional theory for electrocatalysis. Energy Environ. Mater. 2022, 5, 157–185. [Google Scholar] [CrossRef]
- Abdel-Kader, N.S.; Abdel-Latif, S.A.; El-Ansary, A.L.; Sayed, A.G. Spectroscopic studies, density functional theory calculations, non-linear optical properties, biological activity of 1-hydroxy-4-((4-(N-(pyrimidin-2-yl) sulfamoyl) phenyl) diazenyl)-2-naphthoic acid and its chelates with Nickel (II), Copper (II), Zinc (II) and Palladium (II) metal ions. J. Mol. Struct. 2021, 1223, 129203. [Google Scholar] [CrossRef]
- Geng, H.; Zhang, P.; Peng, Q.; Cui, J.; Hao, J.; Zeng, H. Principles of Cation−π Interactions for Engineering Mussel-Inspired Functional Materials. Acc. Chem. Res. 2022, 55, 1171–1182. [Google Scholar] [CrossRef]
- Liu, J.; Hua, D.; Zhang, Y.; Japip, S.; Chung, T.S. Precise molecular sieving architectures with Janus pathways for both polar and nonpolar molecules. Adv. Mater. 2018, 30, 1705933. [Google Scholar] [CrossRef]
- Wang, W.; Liang, Y.; Jin, Y.; Zhang, J.; Su, J.; Li, Q. E484K mutation in SARS-CoV-2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: Binding free energy calculation studies. J. Mol. Graph. Model. 2021, 109, 108035. [Google Scholar] [CrossRef]
- Gontrani, L. Choline-amino acid ionic liquids: Past and recent achievements about the structure and properties of these really “green” chemicals. Biophys. Rev. 2018, 10, 873–880. [Google Scholar] [CrossRef]
- Shen, W.; He, P.; Xiao, C.; Chen, X. From Antimicrobial Peptides to Antimicrobial Poly (α-amino acid) s. Adv. Healthc. Mater. 2018, 7, 1800354. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, H.; Zhang, S.; Li, J.; Zhu, X.; Jin, H.; Xu, J. Improvement of protein emulsion stability through glycosylated black bean protein covalent interaction with (−)-epigallocatechin-3-gallate. RSC Adv. 2021, 11, 2546–2555. [Google Scholar] [CrossRef]
- Kolesov, B.A. Hydrogen bonds: Raman spectroscopic study. Int. J. Mol. Sci. 2021, 22, 5380. [Google Scholar] [CrossRef]
- Feng, J.; Deng, Q.; Ni, H. Photodegradation of phthalic acid esters under simulated sunlight: Mechanism, kinetics, and toxicity change. Chemosphere 2022, 299, 134475. [Google Scholar] [CrossRef]
- Ye, Q.; Liu, C.; Wu, P.; Wu, J.; Lin, L.; Li, Y.; Ahmed, Z.; Rehman, S.; Zhu, N. Insights into photocatalytic degradation of phthalate esters over MSnO3 perovskites (M=Mg, Ca): Experiments and density functional theory. J. Environ. Manag. 2022, 307, 114511. [Google Scholar] [CrossRef]
- Mphahlele, I.J.; Malinga, S.P.; Dlamini, L.N. Combined biological and photocatalytic degradation of dibutyl phthalate in a simulated wastewater treatment plant. Catalysts 2022, 12, 504. [Google Scholar] [CrossRef]
- Du, M.; Li, X.; Cai, D.; Zhao, W.; Wang, J.; Li, Y. In-silico study of reducing human health risk of POP residues’ direct (from tea) or indirect exposure (from tea garden soil): Improved rhizosphere microbial degradation, toxicity control, and mechanism analysis. Ecotoxicol. Environ. Saf. 2022, 242, 113910. [Google Scholar] [CrossRef]
- Kaur, R.; Kumari, A.; Sharma, G.; Singh, D.; Kaur, R. Biodegradation of endocrine disrupting chemicals benzyl butyl phthalate and dimethyl phthalate by Bacillus marisflavi RR014. J. Appl. Microbiol. 2021, 131, 1274–1288. [Google Scholar] [CrossRef]
- Gu, W.; Zhao, Y.; Li, Q.; Li, Y. Plant-microorganism combined remediation of polychlorinated naphthalenes contaminated soils based on molecular directed transformation and Taguchi experimental design-assisted dynamics simulation. J. Hazard. Mater. 2020, 396, 122753. [Google Scholar] [CrossRef]
- Han, J.; Zhang, X. Evaluating the comparative toxicity of DBP mixtures from different disinfection scenarios: A new approach by combining freeze-drying or rotoevaporation with a marine polychaete bioassay. Environ. Sci. Technol. 2018, 52, 10552–10561. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, J.; Nakamura, M. DNA-protein crosslink formation by endogenous aldehydes and AP sites. DNA Repair 2020, 88, 102806. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, J.; Xu, M.; Yang, Z.; Yue, S.; Zhou, W.; Gui, C.; Zhang, H.; Li, S.; Wang, P.G.; et al. Advances in glycopeptide enrichment methods for the analysis of protein glycosylation over the past decade. J. Sep. Sci. 2022, 45, 3169–3186. [Google Scholar] [CrossRef] [PubMed]
- Gonos, E.S.; Kapetanou, M.; Sereikaite, J.; Bartosz, G.; Naparlo, K.; Grzesik, M.; Sadowska-Bartosz, I. Origin and pathophysiology of protein carbonylation, nitration and chlorination in age-related brain diseases and aging. Aging (Albany NY) 2018, 10, 868. [Google Scholar] [CrossRef]
- Fu, L.; Chen, Y.; Xu, C.; Wu, T.; Guo, H.; Lin, Z.; Wang, R.; Shu, M. 3D-QSAR, HQSAR, molecular docking, and new compound design study of 1, 3, 6-trisubstituted 1, 4-diazepan-7-ones as human KLK7 inhibitors. Med. Chem. Res. 2020, 29, 1012–1029. [Google Scholar] [CrossRef]
- Shah, B.M.; Modi, P.; Trivedi, P. Pharmacophore-based virtual screening, 3D-QSAR, molecular docking approach for identification of potential dipeptidyl peptidase IV inhibitors. J. Biomol. Struct. Dyn. 2021, 39, 2021–2043. [Google Scholar] [CrossRef]







| No. | Compounds | Docking Scores | Score | Docking Scores | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1HZV | 1P80 | 7CUZ | 1NIC | 2IOF | 3FMX | TVSP | |||
| 1 | Butyl benzyl phthalate (BBP) | 60.75 | 92.62 | 57.81 | 101.16 | 102.90 | 67.45 | 0.69 | 78.48 |
| 2 | Diallyl phthalate (DAP) | 56.31 | 71.36 | 52.66 | 78.92 | 76.78 | 59.60 | 0.40 | 63.02 |
| 3 | Dibutyl phthalate (DBP) | 66.41 | 82.54 | 64.56 | 101.04 | 85.22 | 75.90 | 0.66 | 84.09 |
| 4 | Diethyl phthalate (DEP) | 43.52 | 60.46 | 50.31 | 83.82 | 68.76 | 49.11 | 0.32 | 59.74 |
| 5 | Dihexyl phthalate (DHP) | 76.02 | 106.04 | 78.84 | 62.19 | 101.86 | 78.31 | 0.60 | 104.63 |
| 6 | Diisobutyl phthalate (DIBP) | 68.90 | 83.48 | 54.34 | 89.15 | 81.14 | 73.42 | 0.56 | 70.46 |
| 7 | Diisoheptyl phthalate (DIHP) | 82.08 | 52.24 | 90.46 | 55.95 | 105.16 | 81.75 | 0.53 | 106.61 |
| 8 | Bis (4-methylpentyl) phthalate (DIHXP) | 72.91 | 99.31 | 85.17 | 40.05 | 99.77 | 70.39 | 0.44 | 81.29 |
| 9 | Diisopentyl phthalate (DIPP) | 73.03 | 90.29 | 62.19 | 99.48 | 93.98 | 64.95 | 0.67 | 80.99 |
| 10 | Diisopropylo-phthalate (DIPRP) | 48.66 | 65.12 | 48.70 | 81.83 | 69.97 | 49.44 | 0.33 | 65.17 |
| 11 | Bis (2-methoxyethyl) phthalate (DMEP) | 61.98 | 78.64 | 63.56 | 84.66 | 86.95 | 68.18 | 0.53 | 86.18 |
| 12 | Dimethyl phthalate (DMP) | 52.85 | 52.25 | 47.59 | 73.86 | 61.86 | 38.89 | 0.21 | 52.74 |
| 13 | Di-n-octylo-phthalate (DNOP) | 90.08 | 123.21 | 99.19 | 83.44 | 100.47 | 79.24 | 0.82 | 83.55 |
| 14 | Di-N-pentyl phthalate (DPP) | 74.17 | 97.39 | 66.60 | 50.16 | 97.95 | 68.34 | 0.44 | 90.28 |
| 15 | Dipropyl phtalate (DPRP) | 57.54 | 72.99 | 56.65 | 71.04 | 76.31 | 65.00 | 0.38 | 69.68 |
| 16 | Diisotridecyl phthalate (DTDP) | 100.64 | 101.47 | 109.35 | 90.05 | 91.38 | 71.89 | 0.81 | 98.39 |
| 17 | Diundecyl phthalate (DUP) | 99.87 | 131.46 | 105.18 | 72.27 | 92.33 | 75.26 | 0.76 | 102.92 |
| 18 | Bis (2-ethylhexyl) phthalate (DEHP) | 86.29 | 103.40 | 88.43 | 57.21 | 100.61 | 83.03 | 0.61 | 80.77 |
| 19 | Dibenzyl phthalate (DBZP) | 74.74 | 100.09 | 78.57 | 60.45 | 99.27 | 61.87 | 0.69 | 76.01 |
| 20 | Dicyclohexyl phthalate (DCHP) | 68.10 | 87.82 | 59.03 | 64.61 | 85.92 | 42.15 | 0.40 | 73.77 |
| 21 | Diisodecyl phthalate (DIDP) | 103.39 | 128.09 | 82.34 | 76.60 | 105.08 | 101.72 | 0.66 | 122.40 |
| 22 | Diisononyl phthalate (DINP) | 87.46 | 125.33 | 79.90 | 50.32 | 100.11 | 93.76 | 0.32 | 86.11 |
| 23 | Diisooctyl phthalate (DIOP) | 82.03 | 126.98 | 73.58 | 76.60 | 100.42 | 86.34 | 0.59 | 84.44 |
| 24 | Didecyl phthalate (DNDP) | 112.76 | 128.69 | 81.37 | 102.99 | 101.64 | 100.05 | 0.56 | 111.87 |
| 25 | Dinonyl phthalate (DNP) | 97.61 | 126.02 | 88.44 | 80.63 | 100.48 | 110.94 | 0.53 | 117.49 |
| 26 | Dioctyl Phthalate (DOP) | 86.30 | 123.21 | 74.87 | 83.44 | 103.36 | 87.53 | 0.44 | 102.07 |
| 27 | Diphenyl phthalate (DPHP) | 46.80 | 81.72 | 87.88 | 90.44 | 71.16 | 31.56 | 0.67 | 55.16 |
| 28 | Octyldecyl phthalate (ODP) | 89.10 | 127.87 | 96.76 | 92.52 | 100.45 | 96.62 | 0.33 | 101.73 |
| 3D-QSAR Model | n | q2 | r2 | F | SEE | r2pred | SEP | S | E |
|---|---|---|---|---|---|---|---|---|---|
| Sewage treatment process Comprehensive effect of microbial degradability | 8 | 0.76 | 0.999 | 1159.179 | 0.009 | 0.646 | 0.162 | 76.50% | 23.50% |
| Soil microbial degradability | 9 | 0.88 | 1.000 | 24128.603 | 0.188 | 0.957 | 15.719 | 65.48% | 32.52% |
| Molecular | Substitution Sites and Substituents | Predictive Value | Change Rate | Molecular | Substitution Sites and Substituents | Predictive Value | Change Rate |
|---|---|---|---|---|---|---|---|
| DBP | - | 0.586 | - | DEHP | - | 0.567 | - |
| DBP-1 | 5-F-6-C | 0.614 | 4.78% | DEHP-1 | 5-B-8-B | 0.617 | 8.82% |
| DBP-2 | 5-G-6-C-8-C | 0.643 | 9.73% | DEHP-2 | 5-C-8-D | 0.59 | 4.06% |
| DBP-3 | 5-C-6-C | 0.596 | 1.71% | DEHP-3 | 5-B-8-K | 0.568 | 0.18% |
| DBP-4 | 5-F | 0.591 | 0.85% | DEHP-4 | 5-E-8-L | 0.569 | 0.35% |
| DBP-5 | 6-C | 0.588 | 0.34% | DEHP-5 | 5-E-8M | 0.618 | 8.99% |
| DBP-6 | 5-G-6-F-8-C | 0.679 | 15.87% | DEHP-6 | 5-E-8-E | 0.57 | 0.53% |
| DBP-7 | 5-C | 0.589 | 0.51% | DEHP-7 | 5-B-8-F | 0.604 | 6.53% |
| DBP-8 | 5-I-8-C | 0.636 | 8.53% | DEHP-8 | 5-C-8-G | 0.574 | 1.23% |
| DBP-9 | 8-G | 0.591 | 0.85% | DEHP-9 | 5-H-8-C | 0.572 | 0.88% |
| DBP-10 | 5-I-8-I | 0.832 | 41.98% | DEHP-10 | 5-E-8-E | 0.633 | 11.64% |
| DBP-11 | 5-F-6-E | 0.595 | 1.54% | DEHP-11 | 5-B-8-G | 0.613 | 8.11% |
| DBP-12 | 5-C-6-F | 0.605 | 3.24% | DEHP-12 | 5-H-8-E | 0.629 | 10.93% |
| DBP-13 | 5-G-6-F | 0.617 | 5.29% | DEHP-13 | 5-E-8-H | 0.63 | 11.11% |
| DBP-14 | 5-F-6-C-8-F | 0.666 | 13.65% | DEHP-14 | 5-E-8-J | 0.647 | 14.11% |
| DBP-15 | 6-C-8-C | 0.587 | 0.17% | DEHP-15 | 5-B-8-N | 0.666 | 17.46% |
| DBP-16 | 5-G-6-F-8-C | 0.714 | 21.84% | DEHP-16 | 5-E-8-K | 0.648 | 14.29% |
| DBP-17 | 5-B-8-C | 0.589 | 0.51% | DEHP-17 | 5-B-8-K | 0.621 | 9.52% |
| DBP-18 | 5-C-6-C-8-C | 0.599 | 2.22% | DEHP-18 | 5-C-8-K | 0.573 | 1.06% |
| DBP-19 | 5-G-8-A | 0.632 | 7.85% | DEHP-19 | 5-C-8-L | 0.576 | 1.59% |
| Molecular | The Scoring Function Value of Docking with AEDECP | Change Rate | The Scoring Function Value of Docking with ANDECP | Change Rate | Toxicity | Change Rate | Frequency | Energy Gap |
|---|---|---|---|---|---|---|---|---|
| DEHP | 84.91 | - | 49.53 | - | 0.66 | - | 8.49 | 0.20 |
| DEHP-10 | 119.70 | 40.97% | 79.59 | 60.69% | 0.64 | −3.03% | 6.16 | 0.20 |
| DEHP-12 | 121.00 | 42.50% | 95.95 | 93.72% | 0.67 | 1.52% | 4.47 | 0.20 |
| DBP | 99.90 | - | 64.20 | - | 0.53 | - | 10.70 | 0.20 |
| DBP-5 | 108.91 | 9.02% | 68.35 | 6.46% | 0.58 | 9.43% | 14.32 | 0.21 |
| DBP-7 | 107.11 | 7.22% | 64.78 | 0.90% | 0.56 | 5.67% | 9.78 | 0.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Zuo, Q.; Du, M.; Li, Y. Molecular Design and Mechanism Analysis of Phthalic Acid Ester Substitutes: Improved Biodegradability in Processes of Sewage Treatment and Soil Remediation. Toxics 2022, 10, 783. https://doi.org/10.3390/toxics10120783
Sun S, Zuo Q, Du M, Li Y. Molecular Design and Mechanism Analysis of Phthalic Acid Ester Substitutes: Improved Biodegradability in Processes of Sewage Treatment and Soil Remediation. Toxics. 2022; 10(12):783. https://doi.org/10.3390/toxics10120783
Chicago/Turabian StyleSun, Shuhai, Qilin Zuo, Meijin Du, and Yu Li. 2022. "Molecular Design and Mechanism Analysis of Phthalic Acid Ester Substitutes: Improved Biodegradability in Processes of Sewage Treatment and Soil Remediation" Toxics 10, no. 12: 783. https://doi.org/10.3390/toxics10120783
APA StyleSun, S., Zuo, Q., Du, M., & Li, Y. (2022). Molecular Design and Mechanism Analysis of Phthalic Acid Ester Substitutes: Improved Biodegradability in Processes of Sewage Treatment and Soil Remediation. Toxics, 10(12), 783. https://doi.org/10.3390/toxics10120783